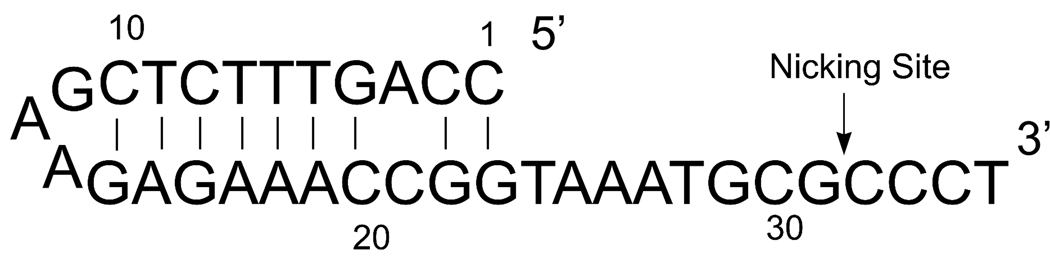Abstract
The MobA protein encoded by plasmid R1162 plays an important role in conjugative mobilization between bacterial cells. It has two functional domains, the N-terminal relaxase domain and C-terminal primase domain. The N-terminal 186 residues (minMobA) is the minimal domain required for relaxase activity. We investigated the effects of different divalent metallic cations on minMobA activity measuring DNA binding, DNA nicking, and protein denaturation experiments. The results show that divalent cations are not required for DNA binding but are required for DNA nicking. The range of metals that function in minMobA suggests the cation role is largely structural. The most tightly binding cation is Mn2+, but the expressed protein shows roughly equal amounts of Mg2+ and Ca2+, both of which facilitate substrate binding and catalysis. Surprisingly, Zn2+ does not facilitate DNA binding nor allow nicking activity.
Keywords: Relaxase, Bacterial conjugation, Plasmid mobilization, Metallic cations, Transesterification
INTRODUCTION
Bacterial conjugation is the direct transfer of genetic material between bacterial cells through temporary cell-to-cell contact. It is not only an important mechanism in the evolution of bacteria but is also one of major causes of antibiotics resistance. Bacteria acquire resistance to antibiotics by the conjugative transfer of plasmids or transposable elements.
There are two classes of plasmids involved in conjugative transfer. One is called the self-transmissible element; the other is the mobilizable plasmid [1–3]. The self-transmissible element encodes a self-transmissible conjugative transfer system, while the mobilizable plasmid is transmissible only in the presence of a type IV secretion system (T4SS), which can be encoded by a self-transmissible plasmid. Interestingly, the mobilization plasmids are exceptionally promiscuous, in that they can be transferred into a very diverse range of organisms including yeast, plant, and animal cells [4–6]. This promiscuity of mobilizable plasmids can constitute a great danger to human health, by spreading antibiotic resistance among potential pathogens [7, 8].
Among the most studied mobilizable plasmids is R1162, which is isolated from Pseudomonas aeruginosa [9]. The R1162 plasmid is the archetype for the so-called incompatibility group Q (IncQ), whose members are characterized by their broad host range and relatively small size [2]. The plasmid encodes resistance to the antibiotics streptomycin (strA and strB) and sulfonamide (sulII). It also contains an origin of transfer (oriT), and encodes three proteins: MobA, B and C, which are all required for plasmid mobilization.
The oriT DNA sequence of R1162 is a 35mer oligonucleotide, which was mapped by Becker and Meyer (see Figure 1) [10, 11]. Sequence alignments of oriT with other IncQ family members reveals that a 12-base core region is highly conserved. The 23-base oligonucleotide 5’ to the core region is an imperfectly inverted repeat, which is commonly found in this family of plasmids. The Mob proteins assemble at oriT to form a complex called the relaxosome [9].
Figure 1.
OriT sequence of R1162. The nicking site is marked by a black arrow.
The most important protein of the relaxosome is the 78 kDa (709 residues) MobA protein, which has two functional domains. The C-terminal domain encodes a 43 kDa protein, called primase, which can also be expressed separately. Both forms of primase have been found in host cells. The function of the primase is to lay down primers within the origin of replication, oriV, which can be further extended by host cell DNA polymerases. The N-terminal domain of MobA is a dsDNA-nicking enzyme, called a nickase or relaxase. This 250 residue enzyme cleaves one of the DNA strands within oriT and forms a covalent adduct with the 5’ end. Tyr25 of the N-terminal domain has been identified as the critical residue [10]; it carries out a nucleophilic attack on the target phosphodiester bond at the nick site and forms a covalent phosphodiester bond with the 5’ end of the nicked product. Once the covalent adduct is formed, the DNA-protein complex is recognized by a T4SS system. The cleaved strand is unwound from its complement strand and then transferred in the 5’ to 3’ direction to the host cell. When the transfer is completed, the MobA protein carries out a second transesterification reaction that rejoins the two DNA ends and releases the protein. Once the circular plasmid DNA is released in the host cell, a complementary strand is synthesized by the host polymerase [10].
The nickase enzymes are part of the HUH superfamily, in which the amino acid motif His-hydrophobe-His is conserved and participates in cation binding [12, 13]. Nickases may also exhibit a second amino acid motif near their N termini, called a YYXXXYY sequence. The first Tyr is the nucleophile in the nicking reaction, and the first Y in the second cluster may function in the release and recirularization of the nicked DNA [14]. MobA contains the HUH motif, but lacks a second Tyr and is the only member of this subgroup for which an X-ray structure currently exists.
Like other HUH proteins, MobA is known to have a requirement for divalent cations, initially thought to be Mg2+, although Mn2+, Ca2+, or Ba2+ could substitute at lesser efficiencies [15]. The related relaxase TraI is also said to have Mg2+ in the active site of its crystal structure [16]. This contrasts with the situation reported for the relaxase TrwC. An X-ray structure and binding data suggested that the most likely active site ion was Zn2+, although Ni2+ and Cu2+ could also sustain nickase activity. These authors saw no evidence that Mg2+ or Mn2+ could bind [14].
It has been shown that the N-terminal 186 residues of R1162 MobA, referred to as minMobA, is competent to cleave DNA [10]. The X-ray structure of minMobA has been solved [17]. It consists of five antiparallel beta-strands connected by four helices lying on both sides of the sheet. The critical active site residue, Tyr25, lies on the first helix on the front side of the molecule, close to an electron-dense metal ion. This metal ion is bound on the front side of the beta sheet, chelated by His120 and His122 (the HUH motif), His112, and a water molecule in a pseudo-tetrahedral geometry. The His cluster of residues is conserved in other relaxase structures [14, 18]. Since the minMobA crystallization condition included 10mM MnCl2, the metal ion in the crystal structure was thought to be Mn2+. Anomalous X-ray scattering confirmed that this was the case. However, the physiologically relevant metal for relaxase is still unknown. It is also uncertain if the metal is structural or catalytic in nature. To further explore the role of a metal ion in mobA’s cleavage reaction, we have assessed the ability of minMobA to cleave single-strand oligo-DNA substrate and to bind ssDNA. We have also examined its circular dichroism spectrum in the presence of Ni2+, Mg2+, Zn2+, Mn2+ and Ca2+. The results suggest that the protein flexibility and the conformational change of active site residues produced by divalent ion binding are essential for the catalytic cycle to occur.
MATERIALS AND METHODS
Protein purification
Wild-type and mutant minMobA were produced and purified as described [17].
Construction of minMobA variants
Site directed mutations were introduced according to the Stratagene protocol (Stratagene, La Jolla, CA). Around 50 ng of plasmid and 150 ng of each primer were combined with reaction buffer (20 mM Tris-HCI (pH 7.5), 8 mM MgCl2, 7.5 mM DTT, 50 µg/ml of bovine serum albumin (BSA)), 150 µM dNTP mix, 1 unit of KOD Hot Start DNA Polymerase (Novagen), and deionized water to final volume of 50 uI. Reactions were cycled with a denaturing temperature of 95 °C for 50 s, a re-annealing temperature of 65 °C for 50 s, and an extension temperature of 72 °C for 9 min for 18 cycles on a GeneAmp 2400 thermocycler (Perkin-Elmer, Norwalk, CT). The reaction mixture was further treated with 10 units of DpnI (New England Biolabs, Ipswich, MA) at 37 °C for 2hrs. Then 1 µl of treated reaction mixture was transformed into E. coli DH5α competent cells. The presence of the expected minMobA mutations was confirmed by DNA sequencing.
Fluorescence polarization assay
All fluorescence measurements were made using an Envision spectrofluorometer (PerkinElmer). A 35mer oligonucleotide (Integrated DNA Technologies) with sequence CCAGTTTCTGAAGAGAAACCGGTAAATGCGCCCT was labeled at the 3’ end with fluorescein. Samples were excited at 490 nm, and emitted light was collected through orange glass filters (OG 515, Schott). The binding affinities for the minMobA (Y25F) and oligo-DNA complexes were determined in the presence and absence of 1 mM metallic cations by measurement of the steady-state anisotropy of fluorescence as a function of added protein. The binding buffer contained 10 mM HEPES (pH 7.5), 20 mM KCl. The concentration of protein was plotted against anisotropy of fluorescence and fit to a hyperbolic equation in order to compute the dissociation constants. The nonlinear regression analysis was performed with the program GraFit 5.0 (Erithacus Software).
DNA cleavage assay
DNA cleavage assays were performed as described [17]. Briefly, a 35mer oligonucleotide (Integrated DNA Technologies) with sequence CCAGTTTCTGAAGAGAAACCGGTAAATGCGCCCT was (3′-33P)-labeled with terminal transferase, according to the manufacturer's instructions (New England Biolabs). The assay reaction mixtures contained 0.03 µM labeled 35mer oligonucleotide, 0.1 µM minMobA, 40 mM Tris–HCl (pH 8.0), 20 mM NaCl, and in the presence and absence of 1 mM metallic cations. For minMobA mutants, the reaction mixtures also contained 50 mM MgCl2. The reactions were terminated by addition of EDTA to 40 mM after 0, 1, 2, 4, 6, 9, 11, 15, 20, 25, 28, 36, 45, 55, and 60 min. Reaction products were separated by 12 % SDS–PAGE and imaged on a Molecular Imager FX system (BioRad Laboratories, Inc., Hercules, CA) to quantify the bound radiolabeled adduct. The concentration of protein-DNA covalent adduct was plotted against time and fit to a single exponential equation: [protein-DNA] =A*(1-exp(–kobs* t))+C. The nonlinear regression analysis was performed with the program GraFit 5.0 (Erithacus Software).
To explore the nicking activity over a range of metallic cations concentrations , DNA cleavage assay was also carried out by using a 49mer oligonucleotide (Integrated DNA Technologies) with sequence CCAGTTTCTCGAAGAGAAACCGGTAAGTGCGCCCTCCCCTTCAAAGTAG. Cleavage experiments were performed by incubation of 5 µM 49mer oligonucleotide with 10 µM minMobA at 37 °C in 10 mM HEPES (pH 7.5), 20 mM KCl plus a one of the divalent metal salts (CaCl2, MgCl2, MnCl2, NiCl2 and ZnCl2). Metal ion concentrations ranged from 1 µM to 1 mM (1 µM, 10 µM, 1000 µM and 1000 µM). The reactions were terminated by addition of EDTA to 40 mM after 60 min. Reaction products were separated on 12 % SDS–PAGE, followed by silver staining with SilverXpress staining kit (Invitrogen), and imaged on a Molecular Imager FX system (BioRad Laboratories, Inc., Hercules, CA) to quantify the protein and protein-18mer oligonucleotide adduct.
Isothermal titration calorimetry
A MicroCal VP-ITC calorimeter (MicroCal, Northampton, MA) was used to measure the binding affinity of minMobA to metallic cation. Protein was dialyzed against 10 mM Na-HEPES (pH 7.5), 25 mM NaCl, with 10 g/l of Chelex beads in the dialysis buffer. Protein, metal, and buffer solutions were degassed before loading into the calorimeter. For the measurement of Mn2+ binding, 4mM MnCl2 was injected into 158 µM minMobA, with a stirring speed at 200 rpm. Data analysis was carried out by fitting to both single site and multiple binding sites models as defined in the Origin software package (OriginLab, Northampton, MA).
Inductively coupled plasma mass spectrometry (ICP-MS)
ICP-MS studies were carried out on an Agilent 7500ce ICP mass spectrometer (Agilent Technologies, Santa Clara, CA) with an Octopole Reaction System for interference reduction, and an electron multiplier detector that operates simultaneously in pulse counting and analog modes. All the samples were prepared in a class 100 clean laboratory with all reagents being ultra pure. The supernatant of cell lysate was dissolved in 50% HNO3 and diluted to final 1% HNO3 before analysis. Protein was dialyzed against 10 mM Tris-HCl (pH 8.0), 20 mM NaCl, with 10 g/l of Chelex beads in the dialysis buffer. Concentrations of all elements were calculated from the calibration curve after proper correction for control blank, matrix and drift effects.
Circular dichroism measurements
Circular dichroism spectra were recorded using a JASCO J-715 spectropolarimeter (JASCO Inc., Easton, MD), equipped with a thermostated cell holder and a NesLab-111 circulating water bath. CD spectra were recorded in cells with an optical path length of 0.1 cm. Experiments were performed in 10 mM HEPES (pH 7.5), 20 mM KCl, in the presence or in the absence of different concentrations of metal ions. Three scans were repeated for each experimental condition. Data were analyzed using the software program k2d [19] from an online server (http://www.embl-heidelberg.de/~andrade/k2d/).
Thermal denaturation experiments
Thermal denaturation profiles were obtained by recording the temperature dependence of the ellipticity at 220 nm in the range 6–80 °C. The temperature was continuously changed at a rate of 0.5 °C/min. Experiments were performed in 10 mM HEPES (pH 7.5), 20 mM KCI, in the presence or absence of 1 mM metallic cations. Tm was determined by locating the maxima/minima of the first derivative of the curve describing the melting profile (CD versus T).
RESULTS
minMobA nicking activity
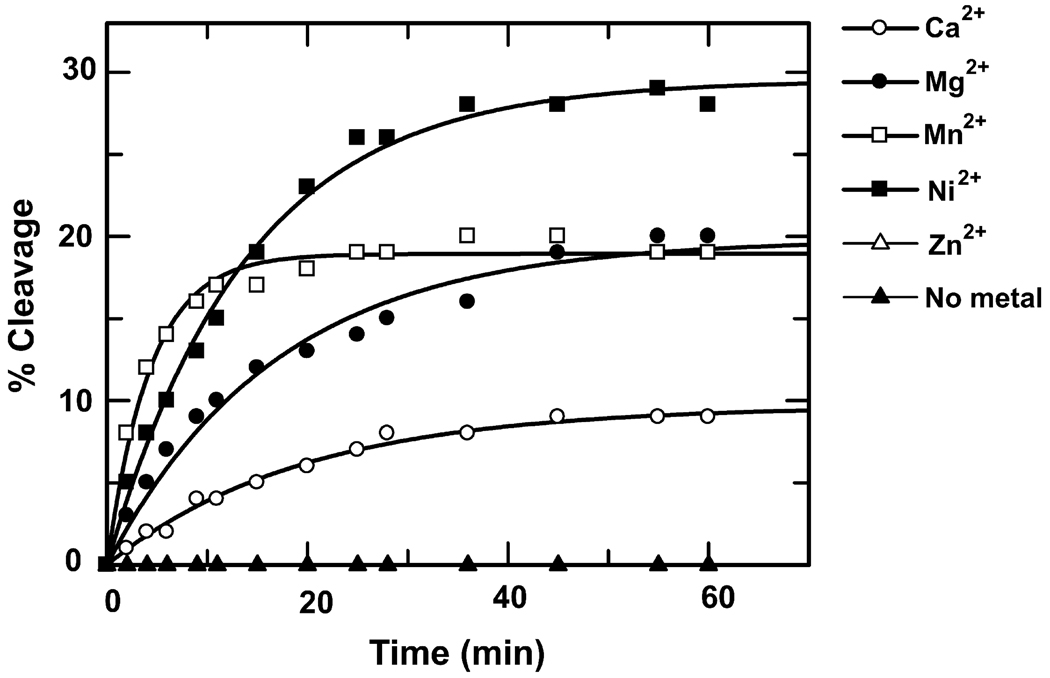
The metal ions were replaced in minMobA by an initial exhaustive dialysis against EDTA followed by dialysis against the divalent cation of choice. The nicking reaction transfers the radiolabeled 3’ tetranucleotide to the enzyme time courses for the reactions with different cations are shown in Figure 2a. As expected, no covalent DNA adduct formed in the absence of any divalent cations; that is, nicking activity has an absolute requirement for cations. As shown in Figure 2a, Mg2+, Mn2+, Ni2+ and Ca2+ are all able to restore the enzyme activity, generating labeled protein as a function of time. At 1 mM concentration, Mn2+ gives the highest reaction rate, while Ni2+ gives the most complete reaction; fitted parameters are shown in Table 1. Surprisingly, 1 mM Zn2+ can not restore enzyme activity, as no detectable covalent adduct is formed. The crystal structure of minMobA shows that the metal binds with three histidine residues (H112, H120 and H122) and a water molecule in a pseudo-tetrahedral conformation. The mutation of any of these three histidine residues to alanine completely abolishes enzyme activity, in spite of the presence of high concentration of Mg2+ (data not shown). This suggests that the loss of any of the histidines precludes metal binding at this site.
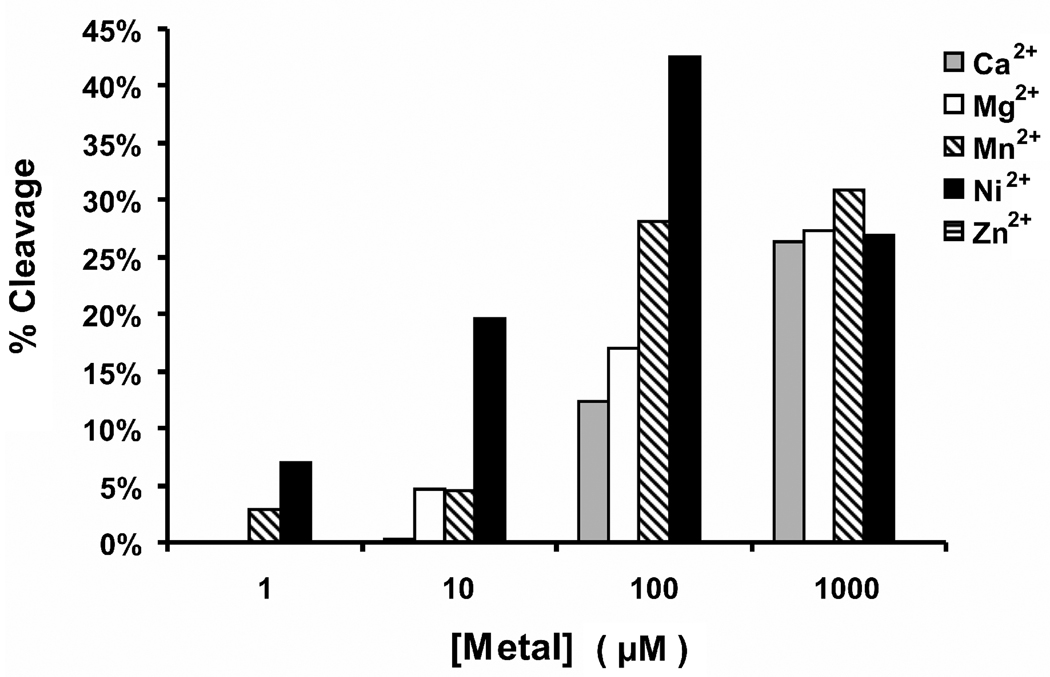
Figure 2.
a. Time course for in vitro cleavage of a 35mer oligonucleotide by minMobA in the presence of different metallic cations at 1 mM. The curve for Zn2+ superimposes with the no metal plot; that is there was no detectable cleavage by this method. b. Cleavage of a 49mer by varying concentration of divalent cations. The bar for Zn2+ is at the right hand of each cluster, but there was no detectable cleavage at any concentration.
Table 1.
Nicking rate constants and DNA binding in the presence of different metallic cations. The kobs values and % DNA cleavage are derived from a single exponential fitting of product formation. Kds are derived from a hyperbolic fitting of fluorescence polarization curves as a function of minMobA concentration.
| Metallic Cation | kobs (1/min) | % Cleavage | DNA Kd (nM) |
|---|---|---|---|
| Mn2+ | 0.2272 | 19 | 13 |
| Ni2+ | 0.0723 | 29 | 24 |
| Mg2+ | 0.0628 | 20 | 14 |
| Ca2+ | 0.0519 | 10 | 15 |
| Zn2+ | 0 | 0 | 27 |
| No metal | 0 | 0 | 57 |
We also explored the nicking reaction at a single time point using a 49mer substrate and varying divalent cation concentrations. The results are shown in Figure 2b. In general, the reaction is facilitated by increasing cation concentrations. At 1 mM, all the cations support nicking of the 49mer, except Zn2+, which shows no activity at any concentration between 1 µM and 1 mM.
minMobA ssDNA binding activity
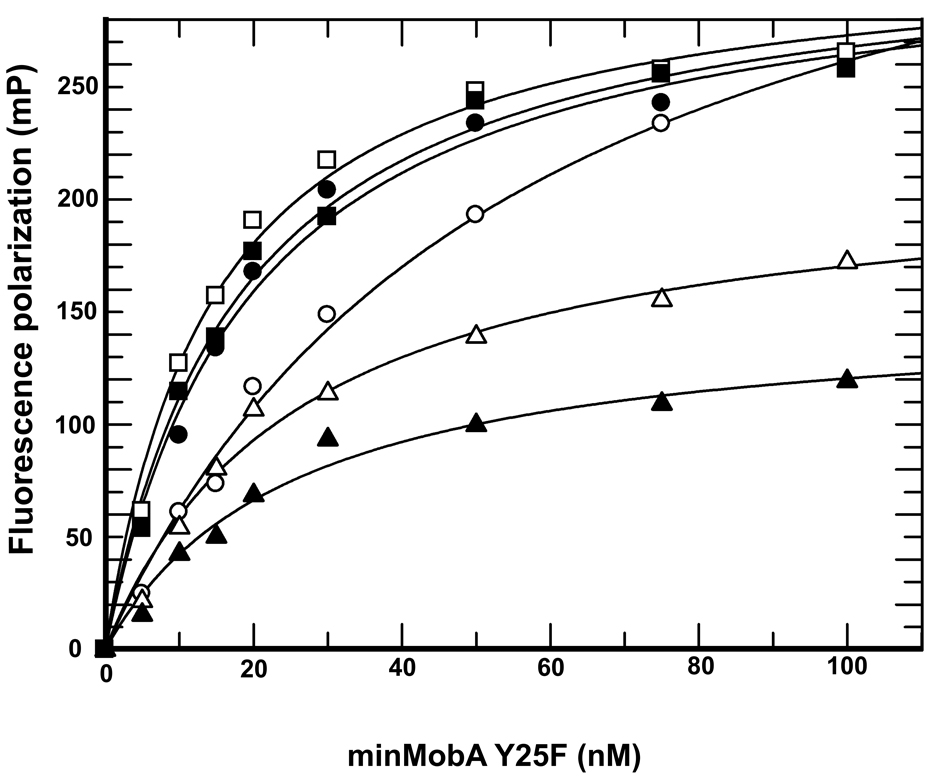
To measure the equilibrium binding strength of ssDNA to MobA independent of covalent adduct formation, we mutated Tyr25 to Phe (Y25F). This removes the attacking nucleophile from the active site, but should allow the DNA substrate to bind in a manner similar to that for nicking activity. A fluorescence polarization assay was used to analyze the binding affinity of the 35mer oligo-DNA to mutant minMobA (Y25F), in the presence of 1 mM Mg2+, Zn2+, Mn2+, Ca2+ and 0.2 mM Ni2+. As shown in Figure 3, the binding of oligonucleotide to minMobA does not require added divalent citrons, but the presence of any of the divalent cations improved binding affinity by 2 to 3 fold. The oligonucleotide D35 binds to minMobA (Y25F) with Kd ~ 20 nM in the presence of Mg2+, Mn2+, Ca2+, Ca2+or Zn2+(Table 1). We used 0.2 mM Ni2+ in our experiments because Ni2+ at concentration greater than 0.2 mM causes significant fluorescence quenching in this system.
Figure 3.
Fluorescence anisotropy assay of binding of minMobA (Y25F) to 3’-fluorescein-labeled oligonucleotide in the presence of different metallic cations. All cation concentrations are 1 mM, except Ni2+ at 0.2 mM. The symbols are as follows: open circle, no metal; filled circle, Mg2+; open square, Mn2+; filled square Ca2+; open triangle, Zn2+; filled triangle, Ni2+.
minMobA Mn2+ binding affinity by ITC
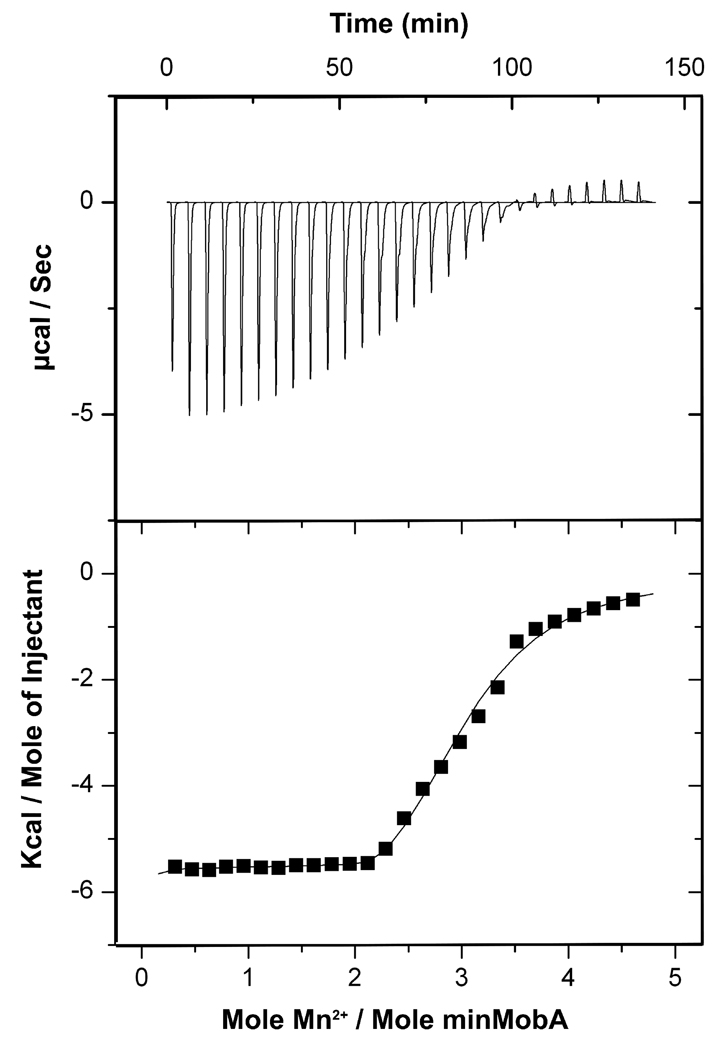
Affinities of wild-type and variant minMobA proteins for Mn2+ were measure by isothermal titration calorimetry (ITC). As shown in Figure 4, the stoichiometry of Mn2+ to minMobA appears to be 3:1. This is consistent with what we found in the X-ray structure, in which three metal ions were bound to minMobA; one is chelated by three histidine residues in the active site, and the other two are chelated by His46 and His98, respectively. The integrated curve was fit to an independent multiple sites binding model, and gave a Kd of 0.2 µM for the active site Mn2+, 40 µM and 45 µM for the Mn2+ outside the active site. This assignment is based on X-ray crystal results of the Mn2+ soaked crystal, which shows the HUH site Mn2+ to be the most highly occupied and to have the lowest temperature factor [17].
Figure 4.
Calorimetry data for the titration of 4 mM MnCl2 into 180 µM wild-type minMobA. The upper panel shows the heat change observed with titration. The lower panel plots the integration of the peaks using the Origin software package.
Inductively coupled plasma mass spectrometry (ICP-MS) analysis
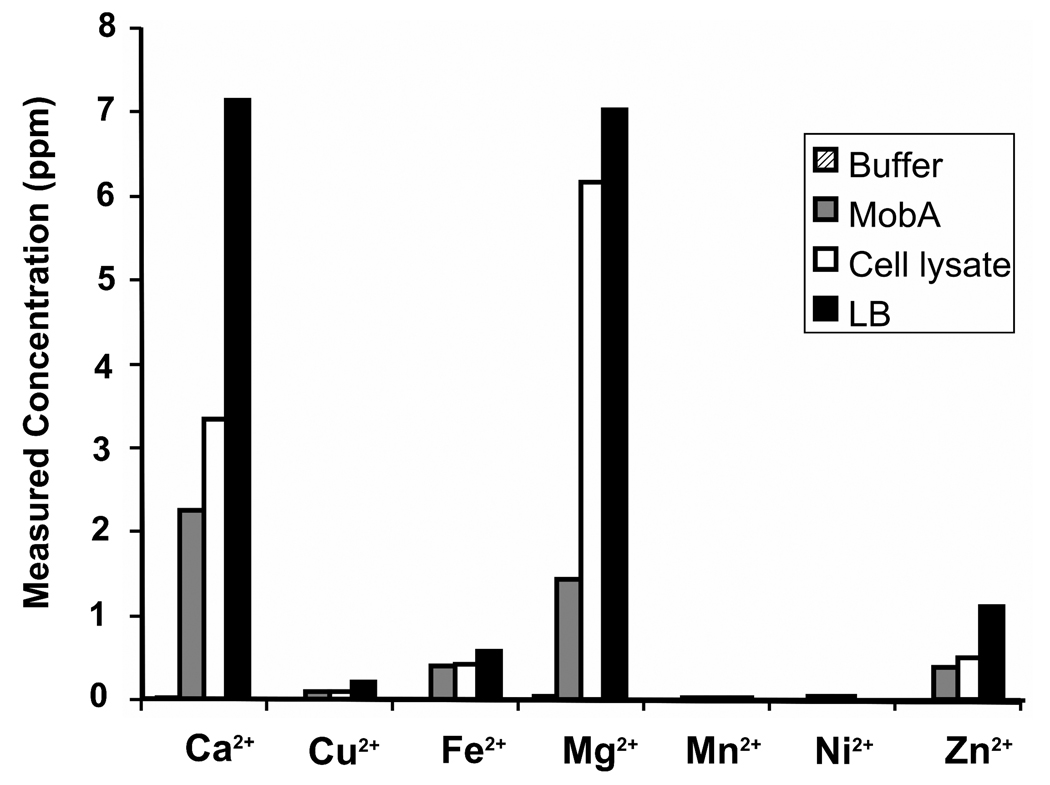
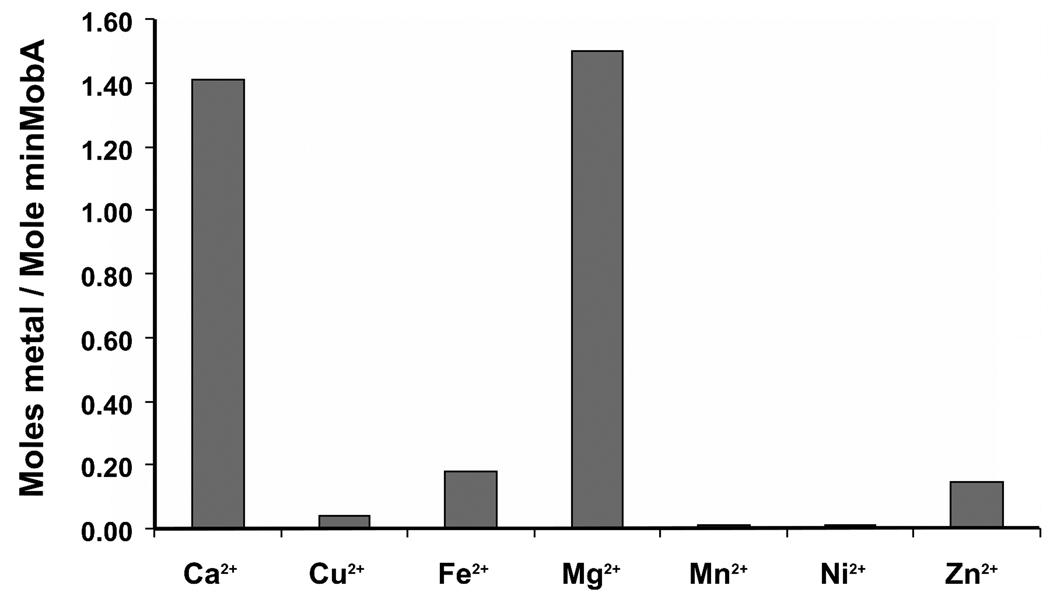
ICP-MS studies were used to identify the most abundant metallic cations in LB growth media, in cell lysate, and in purified minMobA protein solution. As shown in Figure 5a, Mg2+ and Ca2+ are the two most abundant metallic cations in LB media, in the supernatant of cell lysates, and in the protein. The molar ratio of metal to protein for purified minMobA is roughly the same for Mg2+ and Ca2+, around 1.4:1 (Figure 5b). In other words, Mg2+ and Ca2+ are equally likely to be bound to MobA protein in vivo and greatly exceed the concentration of other divalent cations.
Figure 5.
(a) Measured concentrations of different metallic cations in lysis buffer, minMobA, supernatant of cell lysate, and LB media. The concentrations of Ca2+, Cu2+, Fe2+, Mg2+, Mn2+, Ni2+ and Zn2+ found by inductively coupled plasma mass spectrometry are shown; ppm stands for parts per million by weight. (b) The molar ratios of cations to protein in purified minMobA.
Conformational change by circular dichroism
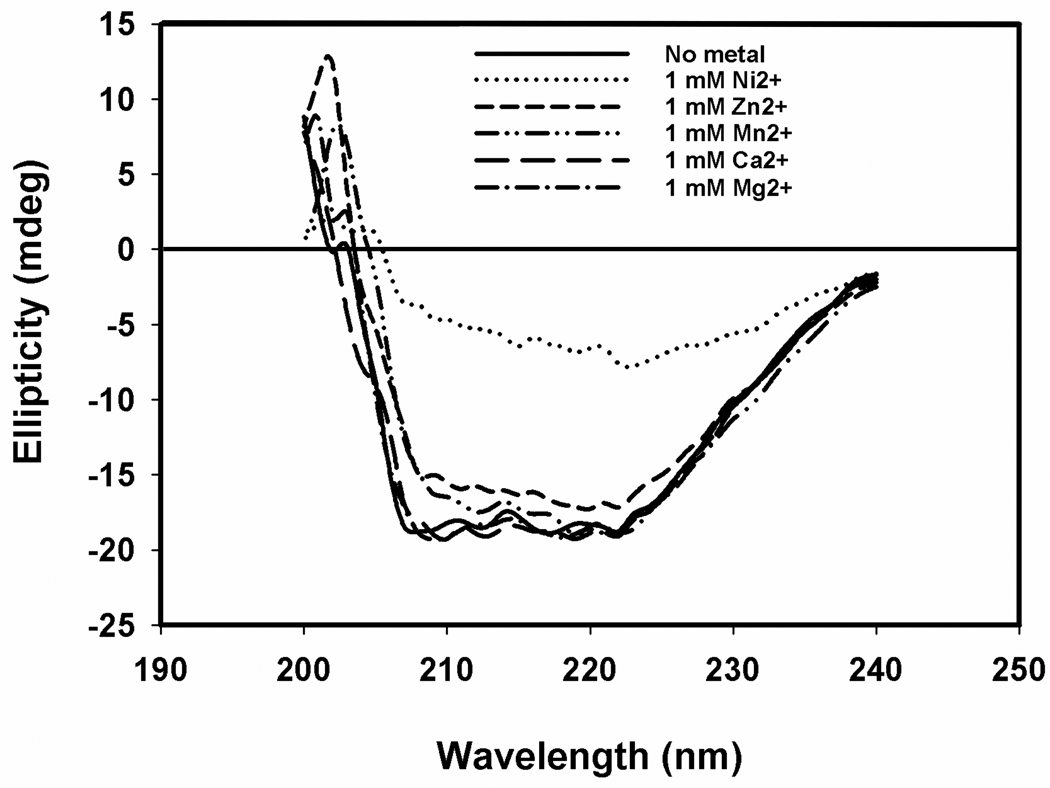
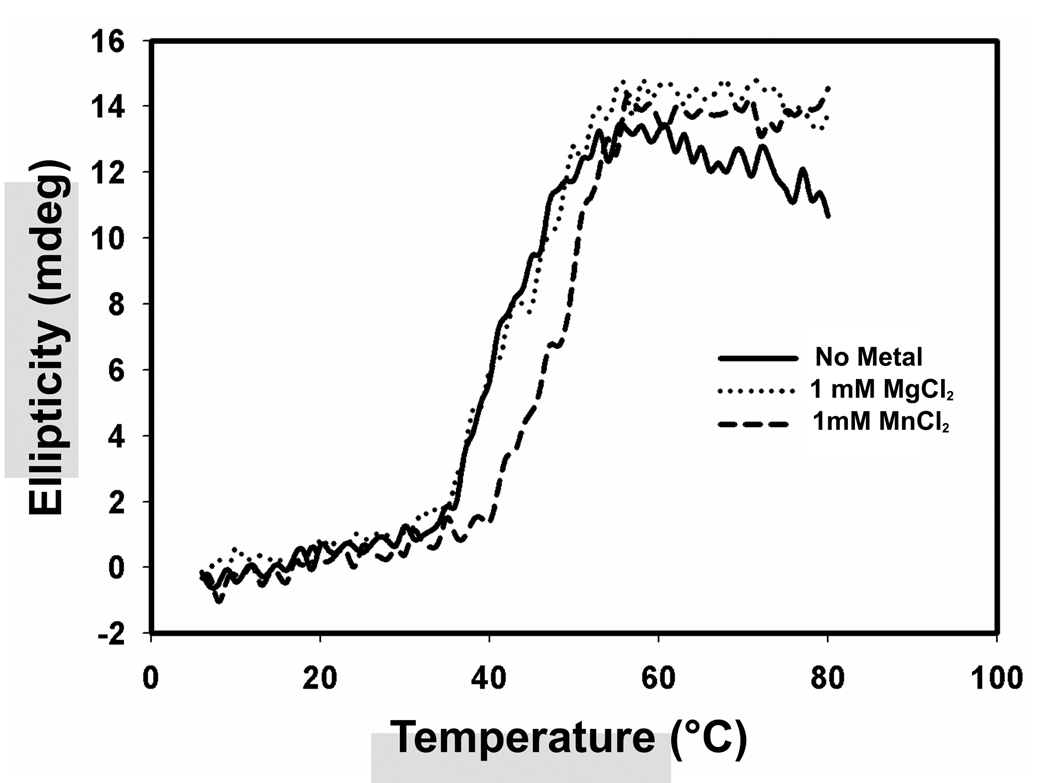
The structure of minMobA was characterized chiroptically by circular dichroism. A quantitative analysis of the experimental CD profile suggests the structure is 36% α-helix and 21% (β-sheet. This is in reasonable agreement with the observed X-ray structure of minMobA, which is 31% α-helix and 17% (β-sheet [17]. We then assessed the effect of divalent cations on the structure, by CD. Ions like Mg2+ and Ca2+ at a concentration of 1 mM did not induce any changes in the CD spectra at 25 °C. However, 1 mM Mn2+ and Zn2+ led to slight perturbations of the CD spectra, as shown in Figure 6. By contrast, 1 mM Ni2+ produced a significant decrease in the apparent a-helical content (~8%) at room temperature. CD spectra were measured as a function of concentration for Ni2+ and Zn2+; they both showed essentially native conformations at 0.5 mM (data not shown). To explore metal induced structural changes more fully, we carried out thermal denaturation experiments in the presence of different cations. The partial unfolding was assessed by monitoring ellipticity at 220 nm. The spectra for Mn2+ and Mg2+ are shown in Figure 7. We observed that Mn2+ appears to stabilize the structure, increasing the Tm by 6 °C at a concentration of 1 mM. Other cations like Mg2+, Ca2+ and Zn2+ at 1 mM or Ni2+ at 0.5 mM, do not alter the Tm of minMobA significantly; the ΔTm values are shown in Table 2.
Figure 6.
Effects of different metallic cations on the minMobA CD spectra at room temperature.
Figure 7.
The effect of Mn2+ and Mg2+ on the thermal denaturation profile of minMobA. The cation concentrations were 1 mM; partial protein unfolding was monitored by following CD spectrum at 220 nm.
Table 2.
Shift of Tm of the thermal melting profile of minMobA in the presence of different metallic cations.
| Metallic Cation | Tm (˚C) | ΔTm (˚C) |
|---|---|---|
| Mn2+ | 48.57 | + 5.86 |
| Ni2+ | 42.56 | − 0.15 |
| Mg2+ | 44.28 | + 1.57 |
| Ca2+ | 43.69 | + 0.98 |
| Zn2+ | 42.34 | − 0.37 |
| No metal | 42.71 |
DISCUSSION
MobA carries out metal ion-dependent DNA cleavage and rejoining reactions as part of its mobilization function. Like TrwC and TraI, it is an HUH relaxase; but unlike those proteins, it has only a single Tyr residue in its mechanism. Our X-ray analysis shows MinMobA is structurally related to TrwC and TraI, but sequence analysis suggests that it is a fairly distant relationship; minMobA has about 11% identity with each of those enzymes, although they share 42% identify between themselves [17]. It has been shown that two tyrosine relaxases, like TraI, can be specifically inhibited by bisphosphonates [20]. We have tested some of these on minMobA (data not shown) and found they have no effect on our single Tyr relaxase. Despite this particular difference, however, the HUH sites of the relaxases are very similar; and it is likely that results from our analysis of metal binding will help elucidate the properties of the entire nickase family.
Previously, Scherzinger reported that the in vitro nicking activity of MobA depended on the presence of a divalent cation, such as Mg2+, Mn2+, Ca2+, or Ba2+ [15]. The X-ray structure of minMobA showed that the active site metal ion is chelated by three histidine residues and a water molecule in a pseudo-tetrahedral geometry. This arrangement is commonly observed for Zn2+ or Ni2+ binding proteins [21–23]. However, the biologically relevant MobA cation, and the role of that cation in the nicking reaction, is uncertain. The literature for related HUH-type nickases illustrates this uncertainty. The structure of TrwC has been solved bound to DNA and in the presence of several different trial cations [14]. They showed that Zn2+ and Cu2+ bind in a tetrahedral geometry but failed to bind Mg2+ to the crystalline protein, even though kinetic measurements showed it would enable the nicking reaction. TraI is another nickase, related to TrwC. The X-ray structure of the apo protein showed the Mg2+ was bound in an octahedral geometry, using the three conserved His residues as ligands, along with three waters [18]. Subsequently, a complex with DNA showed an active site Mg2+ bound in a pentagonal arrangement with two phosphate oxygens joining the histidines as ligands. This geometry was also consistent with Ca2+ binding [24].
In this paper we observed that a bound cation is not necessary for minMobA to physically bind DNA, but we see that such cation-independent binding does not allow the nicking reaction to proceed. Addition of any of a variety of cations increased the magnitude of DNA binding around 3-fold, and generally allows nicking to occur (Zn2+ being the exception). We cannot say if DNA binds in a different orientation in the presence of cations. However, it seems likely that cations organize the MobA active site in a way that improves DNA binding and favors an orientation that is potentially productive. X-ray structures of DNA complexes to TrwC [14] and Tral [24] both show that the substrates are oriented in a way that the scissile phosphate is near the attacking Tyr nucleophile, and that a non-bridging oxygen of the phosphate acts as a ligand to the cation (Zn2+ and Mg2+ in the respective models).
Based on our CD measurements the magnitude of this organization may be small, since it has little effect on the backbone structure. (Ni2+ is the exception, causing significant structural changes). In the X-ray structure of minMobA, the critical Tyr25 lies at the edge of an a-helix, surrounded by four acidic residues (Asp35, Asp37, Glu38 and Glu74) and four basic residues (Arg28, Arg34, Lys22 and Lys31). The presence of a divalent metallic cation may affect the conformation of those residues, altering the local protein conformation, stability, and reactivity.
It is worth considering that the bound cation may participate in the chemistry of the nicking reaction in addition to increasing the affinity for DNA. The cation may function to sterically align the scissile bond for nucleophilic attack by Tyr25, or it may participate in the chemistry of the bond cleavage, perhaps by stabilizing negative charge in catalytic intermediates. Indeed, it is generally considered that the cation polarizes the scissile phosphate, increasing the electrophilicity of the phosphorous atom and facilitating attack by the active site Tyr [20, 25]. However, it is important to recall that MobA is a very poor enzyme; and indeed, its biological function requires it to carry out a single turnover, forming a covalent adduct that persists during transmission out of the cell. As a consequence, there may be little or no selective pressure to optimize the rate of this reaction.
Our data show that Mn2+ binds minMobA more avidly than other tested cations, with a Kd–0.2 M. Mn2+ also binds tightly to TraI, with a Kd~0.5 µM, about ten thousand times more tightly than Mg2+ or Ca2+ [24]. For minMobA, Mn2+ also gives the fastest reaction, the strongest DNA complex, and the greatest thermal stabilization. Given this, it might seem that Mn2+ would be the major cation for the enzyme, but this does not seem to be the case. Its catalytic advantage over Mg2+ and Ca2+ is small, and it may be that any of these cations will function in the mobilization scheme once they occupy the HUH site. Since so many cations can function in this scheme, at roughly the same efficiency, it seems unlikely that the particular cation plays a very specific catalytic role in the reaction, say based on its Lewis acid or other physico-chemical properties. The lack of cation selectivity, and the variable concentrations of free cations may account for the diversity of metals, seen by X-ray crystallography, for this family of enzymes [14, 17, 18]. Not all divalent cations can function in Mob A, however. We showed that Ni2+ seems to create structural aberrations that diminish substrate binding. Most surprisingly we found that Zn2+, a cation that should bind strongly to the tetrahedral geometry of the MobA active site, is unable to support the catalytic reaction.
The HUH metal site itself does not seem particularly well suited to Mg2+ or Ca2+ binding. Metal ligation is affected by the hardness of the metals and the ligands. The known order of the hardness of metallic cations used here is: Mg2+>Ca2+ > Mn2+ > Ni2+ > Zn2+ [3]. The nitrogens of the histidine ligands are soft and should favor the softer metals, like Zn2+, while oxygen (from water or phosphate) is a hard ligand. A survey of protein-metal sites shows that other Mg2+ sites in proteins have at least one carboxylate ligand, favor hard ligands in general, and tend to favor octahedral geometries; in contrast Zn2+ favors soft ligands, like His nitrogens, in a tetrahedral geometry [14, 21].
The abundance of Mg2+ and Ca2+ in the media and the cell are consistent with our observation that they are the most common cations found in isolated minMobA. It appears that minMobA can, and does, bind both ions. Although Mn2+ binds much more tightly than Mg2+ or Ca2+, it has a much lower concentration in the cellular environment [26]. Furthermore, the effective free concentraton of Mn2+ or Zn2+ available for relaxase binding may be even less favorable than the total concentration in the bacteria. Under artifical conditions, the HUH site binds Mn2+ the most tightly, stabilizing the protein against thermal denaturation and allowing substrate binding and catalysis. This is consistent with our crystallization studies as well; our largest and most strongly diffracting crystals [17] grow in the presence of Mn2+.
In conclusion, divalent metallic ions affect the conformations of active site residues of minMobA; they may organize key side chains and the local backbone structure to favor DNA cleavage. The protein appears to function equally well with Mg2+ and Ca2+, which are the predominate ions in the expressed protein.
ACKNOWLEDGEMENTS
We thank Dustin Gross for technical assistance in the isothermal titration calorimetry experiments, and Dr. Nathan Miller for the technical assistance in ICP-MS experiments. This work was supported by NIH grant AI 75509, by the Robert A. Welch Foundation, and by the College of Natural Sciences support to the Center for Structural Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lanka E, Wilkins BM. DNA processing reactions in bacterial conjugation. Annu Rev Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 2.Rawlings DE, Tietze E. Comparative biology of IncQ and IncQ-like plasmids. Microbiol Mol Biol Rev. 2001;65:481–496. doi: 10.1128/MMBR.65.4.481-496.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zechner EL, Eisenbrandt FR, Grabn AM, Koraimann G, Lanka E, Muth G, pansegrau CM, Thomas CM, Wilkins BM, Zatyka M. Conjugative-DNA transfer processes. In: Thomas CM, editor. The horizontal gene pool bacterial plasmids and gene spread. Amsterdam: Harwood Academic Publishers; 2000. pp. 87–174. [Google Scholar]
- 4.Courvalin P. Transfer of antibiotic resistance genes between gram-positive and gram-negative bacteria. Antimicrob Agents Chemother. 1994;38:1447–1451. doi: 10.1128/aac.38.7.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gormley EP, Davies J. Transfer of plasmid RSF1010 by conjugation from Escherichia coli to Streptomyces lividans and Mycobacterium smegmatis. J Bacteril. 1991;173:6705–6708. doi: 10.1128/jb.173.21.6705-6708.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazodier P, Davies J. Gene transfer between distantly related bacteria. Annu Rev Genet. 1991;25:147–171. doi: 10.1146/annurev.ge.25.120191.001051. [DOI] [PubMed] [Google Scholar]
- 7.Lybarger SR, Sandkvist M. Microbiology. A hitchhiker's guide to type IV secretion. Science. 2004;304:1122–1123. doi: 10.1126/science.1098806. [DOI] [PubMed] [Google Scholar]
- 8.Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 9.Meyer R, Hinds M, Brasch M. Properties of R1162, a broad-host-range, high-copy-number plasmid. J Bacteriol. 1982;150:552–562. doi: 10.1128/jb.150.2.552-562.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker EC, Meyer RJ. MobA, the DNA strand transferase of plasmid R1162: the minimal domain required for DNA processing at the origin of transfer. J Biol Chem. 2002;277:14575–14580. doi: 10.1074/jbc.M110759200. [DOI] [PubMed] [Google Scholar]
- 11.Becker EC, Meyer RJ. Relaxed specificity of the R1162 nickase: a model for evolution of a system for conjugative mobilization of plasmids. J Bacteriol. 2003;185:3538–3546. doi: 10.1128/JB.185.12.3538-3546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hickman AB, Ronning DR, Kotin RM, Dyda F. Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep. Mol cell. 2002;10:327–337. doi: 10.1016/s1097-2765(02)00592-0. [DOI] [PubMed] [Google Scholar]
- 13.Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boer R, Russi S, Guasch A, Lucas M, Blanco AG, Perez-Luque R, Coll M, de la Cruz F. Unveiling the molecular mechanism of a conjugative relaxase: The structure of TrwC complexed with a 27-mer DNA comprising the recognition hairpin and the cleavage site. J Mol Biol. 2006;358:857–869. doi: 10.1016/j.jmb.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 15.Scherzinger E, Lurz R, Otto S, Dobrinski B. In vitro cleavage of double- and single-stranded DNA by plasmid RSF1010-encoded mobilization proteins. Nucleic Acids Res. 1992;20:41–48. doi: 10.1093/nar/20.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta S, Larkin C, Schildbach JF. Structural insights into single-stranded DNA binding and cleavage by F factor TraI. Structure (Camb) 2003;11:1369–1379. doi: 10.1016/j.str.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Monzingo AF, Ozburn A, Xia S, Meyer RJ, Robertus JD. The structure of the minimal relaxase domain of MobA at 2.1 A resolution. J Mol Biol. 2007;366:165–178. doi: 10.1016/j.jmb.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datta S, Larkin C, Schildbach JF. Structural insights into single-stranded DNA binding and cleavage by F factor TraI. Structure. 2003;11:1369–1379. doi: 10.1016/j.str.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Andrade MA, Chacon P, Merelo JJ, Moran F. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 1993;6:383–390. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- 20.Lujan SA, Guogas LM, Ragonese H, Matson SW, Redinbo MR. Disrupting antibiotic resistance propagation by inhibiting the conjugative DNA relaxase. Proc Natl Acad Sci U S A. 2007;104:12282–12287. doi: 10.1073/pnas.0702760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dudev T, Lim C. Principles governing Mg, Ca, and Zn binding and selectivity in proteins. Chem Rev. 2003;103:773–788. doi: 10.1021/cr020467n. [DOI] [PubMed] [Google Scholar]
- 22.Chivers PT, Tahirov TH. Structure of Pyrococcus horikoshii NikR: nickel sensing and implications for the regulation of DNA recognition. J Mol Biol. 2005;348:597–607. doi: 10.1016/j.jmb.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 23.Smith GD, Pangborn WA, Blessing RH. Phase changes in T(3)R(3)(f) human insulin: temperature or pressure induced? Acta Crystallogr D Biol Crystallogr. 2001;57:1091–1100. doi: 10.1107/s0907444901007685. [DOI] [PubMed] [Google Scholar]
- 24.Larkin C, Datta S, Harley MJ, Anderson BJ, Ebie A, Hargreaves V, Schildbach JF. Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the F factor relaxase. Structure. 2005;13:1533–1544. doi: 10.1016/j.str.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Larkin C, Haft RJ, Harley MJ, Traxler B, Schildbach JF. Roles of active site residues and the HUH motif of the F plasmid TraI relaxase. J Biol Chem. 2007;282:33707–33713. doi: 10.1074/jbc.M703210200. [DOI] [PubMed] [Google Scholar]
- 26.Outten CE, O'Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]