Abstract
Shal K+ (Kv4) channels across species carry the major A-type K+ current present in neurons. Shal currents are activated by small EPSPs and modulate post-synaptic potentials, backpropagation of action potentials, and induction of LTP. Fast inactivation of Shal channels regulates the impact of this post-synaptic modulation. Here, we introduce SKIP3, as the first protein interactor of Drosophila Shal K+ channels. The SKIP gene encodes three isoforms with multiple protein-protein interaction domains. SKIP3 is nervous system specific and co-localizes with Shal channels in neuronal cell bodies, and in puncta along processes. Using a genetic deficiency of SKIP, we show that the proportion of neurons displaying a very fast inactivation, consistent with Shal channels exclusively in a “fast” gating mode, is increased in the absence of SKIP3. As a scaffold-like protein, SKIP3 is likely to lead to the identification of a novel regulatory complex that modulates Shal channel inactivation.
Introduction
Shal (Kv4) is the most highly conserved of the voltage-gated K+ channel gene subfamilies, sharing 82% amino acid identity between Drosophila and mouse (Pak et al. 1991) and generating a transient A-type K+ current from C elegans to humans (Fawcett et al. 2006; Isbrandt et al. 2000; Jerng et al. 2004; Salkoff et al. 1992). Molecular, genetic, and pharmacological studies have identified Shal channels as underlying the somato-dendritic A-type K+ current in most neurons (Jerng et al. 2004). With a hyperpolarized voltage operating range typical of A-type currents, Kv4.2 currents in hippocampal neurons have been shown to act at subthreshold potentials, regulating the integration of high-frequency trains of synaptic input (Ramakers and Storm 2002), the shape of mEPSCs (Kim et al. 2007), backpropagating action potentials in dendrites (Cai et al. 2004; Chen et al. 2006; Kim et al. 2005), and induction of long-term potentiation (LTP) (Chen et al. 2006; Kim et al. 2007). Rapid inactivation of Shal currents directly affects the duration of its effect on membrane potential, and therefore plays a critical role in Shal channel modulation of post-synaptic potentials.
Regulation of inactivation rates then is likely to be an important mechanism for modulating neuronal firing frequency and integration of post-synaptic potentials. Inactivation of mammalian Kv4 channels has been shown to be regulated by auxillary subunits, including K+ channel interacting proteins (KChIPs) (An et al. 2000) and dipeptidyl aminopeptidase-like proteins (DPPX) (Nadal et al. 2003). In Drosophila, there is only one Shal K+ channel gene, and the encoded protein underlies the predominant transient A-type current present in virtually all neurons (Tsunoda and Salkoff 1995a). Interestingly, the inactivation rate of these currents varies over several orders of magnitude (Tsunoda and Salkoff 1995a). Single Shal channels in Drosophila have been shown to adopt either a “fast” or “slow” gating mode, giving rise to whole-cell currents with different rates of inactivation (Tsunoda and Salkoff 1995a). These Shal channels have been proposed to switch between the two gating modes (Tsunoda and Salkoff 1995a). The molecular mechanisms regulating gating mode changes, or proportions of channels in each gating mode, however, are still unknown. Thus, Drosophila Shal K+ channels present an interesting model system for studying additional mechanisms underlying the regulation of K+ current inactivation.
In this study, we identify a novel protein, SKIP3 (Shal K+ Channel Interacting Protein-3), as the first interactor of Drosophila Shal K+ channels. We show that SKIP isoforms are expressed specifically in the nervous system, and that SKIP3 is likely the only isoform that interacts with Shal channels. To examine the function of SKIP3 in neurons, we identify a genetic deficiency of SKIP (Df(3R)Exel6190). We show that a higher percentage of homozygous Df(3R)Exel6190 neurons display Shal currents with inactivation rates that correspond to Shal channels exclusively in the “fast” gating mode. Altogether, our study introduces SKIP3 as a novel Shal K+ channel interactor that regulates the inactivation of Shal K+ channels.
Results
Identification of a Novel Shal K+ Channel Interactor, SKIP3
Little is known about the mechanism(s) underlying the variable inactivation rate of Drosophila Shal K+ channels. We hypothesized that protein interactor(s) might function in the regulation of Shal channel inactivation. We first examined whether homologs of known proteins that bind mammalian Shal (Kv4) channels also interact with Drosophila Shal K+ channels. We tested Drosophila frequenin and neurocalcin, which have sequence similarity to the recoverin/NCS (neuronal calcium sensor) subfamily of Ca2+ binding proteins (Burgoyne and Weiss 2001), which include mammalian KChIPs that have been shown to regulate mammalian Kv4 channels (An et al. 2000), as well as Drosophila Hyperkinetic (Kaplan and Trout 1969), the homolog of the mammalian K+ channel β-subunit, which regulate Shaker (Kv1) channels (Rettig et al. 1994; Chouinard et al. 1995; Wang and Wu 1996) and has been shown to bind Shal channels (Nakahira et al. 1996). We used direct yeast-two-hybrid (Y2H) assays to test for interaction of these candidate proteins with Shal. We found that none of these proteins interacted with the large cytoplasmic N- or C- termini of Shal (data not shown). Although these and other protein interactors have been shown to play important roles in regulating mammalian Kv4 channels (Jerng et al. 2004; Vacher et al. 2008), they have not been able to account for the interesting regulation of Drosophila Shal channel inactivation (Tsunoda and Salkoff 1995a).
We set out to identify new protein interactors that might function in the regulation of Shal K+ channels. Using the Y2H approach, we sought to identify new channel regulators that interact with the cytoplasmic C-terminus of Shal channel subunits. Drosophila Shal encodes two alternatively spliced isoforms, Shal1 and Shal2, differing only by an extended C-terminal tail present in Shal1. Using the entire cytoplasmic C-terminus of Shal1 (Shal1C, residues 411 to 571) as “bait”, we screened for interacting “prey” expressed from an embryonic cDNA library (see Experimental Methods). An embryonic cDNA library was chosen since Shal channels with variable inactivation rates have been genetically identified in embryonic neurons (Tsunoda and Salkoff 1995a). Positive interactions were detected by expression of three reporter genes. From 4.8 × 106 clones screened, ~800 positives were selected using a low stringency test for expression of one reporter gene. ~150 positives were re-selected, using more stringent media, for the expression of all three reporter genes; interaction was indicated by growth of co-transformed yeast on stringent media, and a blue color when incubated with the substrate X-α-gal. These positive clones were sequenced and analyzed. Partial sequences were used to obtain full-length sequences, which were then subjected to homology searches on Flybase (http://flybase.bio.indiana.edu/) and NCBI (http://www.ncbi.nlm.nih.gov/BLAST/) sequence databases. Sequences encoding transcription factors, nuclear proteins, ribosomal proteins, or mitochondrial proteins were discarded as false-positives. The 24 remaining candidate sequences were assayed again in a direct Y2H assay with the N- and C-termini of Shal to test for specificity of binding the C-terminus. Clones that tested positive for both “baits” were discarded. Nine sequences showed strong binding to Shal1C and no interaction with the N-terminus (ShalN). One of these protein interactors is the protein we now designate as SKIP3 for Shal K+ Channel Interacting Protein-3 (Fig. 1).
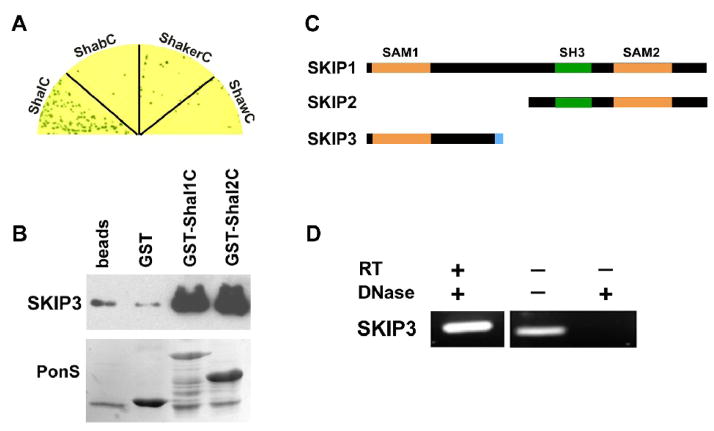
Fig. 1. The Novel Protein, SKIP3, Interacts with the C-terminus of Shal K+ Channels.
(A) SKIP3 was tested in direct Y2H tests with the C-termini of representative isoforms of voltage-gated K+ channels from the four major subfamilies, including Shal1 (ShalC), ShA1 (ShakerC), Shab-RB (ShabC), and Shaw2 (ShawC). Shown are co-transfected yeast grown on stringent media and X-α-Gal to select for interaction. Note that growth and a blue color, indicating interaction, is specific for ShalC. (B) Shown are results from GST-pull down assays using GST-fused to the C-termini of Shal1 (residues 411 to 571) and Shal2 (residues 411 to 490), then incubated with purified SKIP3 protein; glutathione-agarose beads alone (beads) and GST were used as negative controls. A representative immunoblot of the proteins pulled-down by GST-fusion proteins (top) shows that large amounts of SKIP3 are pulled-down by GST-Shal1C and GST-Shal2C, with much lower background levels pulled-down by beads alone or GST. Immunoblot was incubated with Ponceau S (PonsS) to confirm that GST protein(s) were indeed pulled-down by glutathione-beads. (C) SKIP encodes three alternatively spliced isoforms, SKIP1 (CG31163-PB/PC), SKIP2 (CG31163-PA), and SKIP3. Each isoform contains a different combination of protein binding domains, including two different SAM domains (orange) and an SH3 domain (green) domain. SKIP3 contains a unique eight residue C-terminal sequence (blue), which is required for binding Shal channels (see Fig. 2). (D) RT-PCR of SKIP3 from wild-type embryos. When RNA treated with DNase was used as template, SKIP3 was successfully reverse-transcribed and amplified by PCR (RT-PCR, left lane), suggesting that SKIP3 is a true isoform expressed in the embryo. When RNA was not treated with DNase, as indicated (−), SKIP3 could be amplified even without reverse transcription (middle lane), suggesting that DNase treatment is essential to degrade contaminating genomic DNA. A mock RT-PCR performed from DNase-treated RNA in the absence of reverse transcriptase (RT) confirmed that SKIP3 was not amplified without RT.
To examine the specificity of the Shal-SKIP3 interaction, the C-termini of three other voltage-gated K+ channels, Shaker (ShA1), Shaw (Shaw2) and Shab (Shab-PB), were used as “bait” in direct Y2H assays with SKIP3. In contrast to the strong interaction of SKIP3 with Shal1C, represented by growth on high stringency media and blue color, Y2H results for SKIP3 with the C-termini of Shaker, Shaw, or Shab, showed little to no growth or blue color (Fig. 1A). These results suggest that SKIP3 interacts specifically with the C-terminus of Shal K+ channels, and not other voltage-gated K+ channels.
To confirm the SKIP3-Shal interaction by another independent method, we tested whether SKIP3 and Shal interact in GST-pull down assays. We generated GST-Shal1C and GST-Shal2C fusion proteins and incubated them with purified SKIP3 protein. We found that both GST-Shal1C and GST-Shal2C pulled-down large quantities of SKIP3, while GST alone pulled-down only background levels of SKIP3, also seen with agarose beads (Fig. 1B). These results validate the SKIP3-Shal interaction by a second independent approach.
SKIP Encodes Three Isoforms with Multiple Protein-Protein Interaction Domains
To determine if there are other alternatively spliced isoforms related to SKIP3, we searched available databases using the SKIP3 sequence obtained in our Y2H screen. We found that the gene we now call SKIP (CG31163) is predicted to produce two polypeptides, CG31163-PA (778 aa) and CG31163-PB/PC (952 aa). CG31163-PB and CG31163-PC contain the same amino acid sequences, which we now term SKIP1 (Fig. 1C). We refer to CG31163-PA as SKIP2 (Fig. 1C). The cDNA identified in our Y2H screen appears to represent a third isoform of SKIP, that we named SKIP3 (Fig. 1C). Since SKIP3 was the only isoform not previously found as an expressed sequence tagged (EST) clone, we examined whether SKIP3 is a true isoform expressed in Drosophila by performing RT-PCR using primers specific for SKIP3. We isolated RNA from embryos (0–24 hr, 25°C), and performed RT-PCR from DNase-treated RNA. We found that SKIP3 could indeed be amplified by RT-PCR, and only in the presence of reverse transcriptase (RT; Fig. 1D). These results confirmed the presence of SKIP3 RNA, and therefore suggest that SKIP3 is a true isoform expressed in the Drosophila embryo.
Sequence analysis of the three SKIP isoforms revealed that they all contain one or two sterile-alpha-motif (SAM) domains (Fig. 1C). SKIP1 contains both SAM1 and SAM2 domains, while SKIP2 contains only the SAM2 domain and SKIP3 contains only the SAM1 domain. Additionally, SKIP1 and SKIP2 contain the same SH3 domain. Given that SAM and SH3 domains have been shown to mediate protein-protein interactions, SKIP proteins are likely to function as a family of adaptor or scaffolding proteins. Since SAM domains have been shown to form homomeric and heteromeric interactions with other SAM domains (Bhattacharjya et al. 2005; Kim and Bowie 2003), one possibility was that SKIP isoforms interact through their SAM domains. We tested whether SAM1 or SAM2 domains of SKIP proteins were able to homo- or hetero-dimerize. Using direct Y2H assays, we tested for any interaction between SAM1 and SAM1, SAM1 and SAM2, or SAM2 and SAM2. None of these combinations of SAM domains tested positive for an interaction (data not shown). Thus, the SAM domains of SKIP proteins are likely to interact with yet unidentified proteins.
Mapping the SKIP3-Shal Interaction Sites Predicts that SKIP3 is the Only Isoform that Interacts with Shal Channels
We next mapped the SKIP3 binding site on Shal channel subunits. We first examined whether SKIP3 interacts with both Shal1 and Shal2 isoforms. A similarly strong interaction was observed when SKIP3 was tested with the C-terminus of either isoform of Shal (Fig. 2A). We then tested various deletion constructs of Shal2C to further narrow the SKIP3 binding site. N-terminal truncations up to 40 amino acids in length, as well as deletions removing the conserved di-leucine motif on the C-terminal end, did not affect the SKIP3-Shal interaction (Fig. 2A). These results suggest that the SKIP3 binding site lies within the 23 amino acids between residues 451 and 474 of Shal. Since no consensus sequence for any protein-protein interaction domain appeared to overlap with these 23 amino acids, binding is likely to occur via a novel binding site specific for SKIP3.
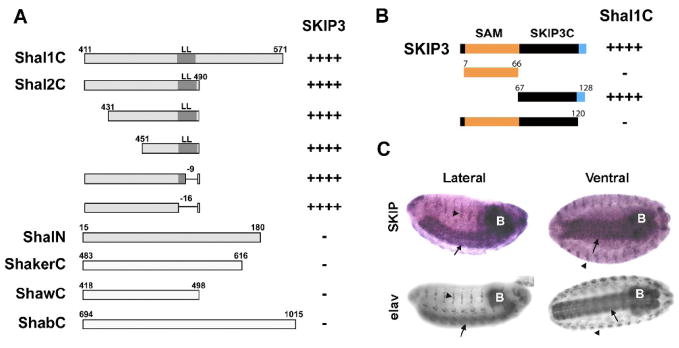
Fig. 2. Mapping the SKIP3-Shal Interaction, and Nervous System Specific Expression of SKIP3.
(A) Direct Y2H results are shown for “bait” fusion proteins, including C-terminal sequences from Shal1 (Shal1C), Shal2 (Shal2C), ShA1 (ShakerC), Shab-RB (ShabC), and Shaw2 (ShawC), and the N-terminus of Shal (ShalN), co-expressed with the SKIP3 “prey” fusion protein identified in our Y2H screen. Positive interactions, assayed by growth on stringent media and a blue color when grown on X-α-gal, are indicated by the number of “+” symbols; “−” indicates no/minimal growth or blue color. SKIP3 interacted specifically with the C-terminus of Shal. Binding is mediated by the sequence between residues 451 and 474 since deletions outside of this region, including the conserved di-leucine motif (LL), did not affect the interaction. (B) Direct Y2H results are shown for the Shal1C “bait” co-expressed with various truncations of SKIP3 as “prey”. Full-length SKIP3, containing a 60 amino acid N-terminal SAM domain (orange), followed by a 54 amino acid domain (SKIP3C; black), and a C-terminal eight amino acids (blue) unique to SKIP3, showed strong interaction with Shal1C, as expected. Protein-protein interaction, indicated by the number of “+” symbols, was assayed by growth on stringent media and a blue color when grown on X-α-Gal. Note that truncation of the C-terminal eight amino acids (blue) of SKIP3 completely disrupted interaction with Shal1C, while deletion of the SAM domain did not. Similar results were seen for Shal2C (data not shown). (C) Representative RNA in situ hybridization for SKIP (top) and immunostaining for the neuron-specific elav protein (bottom) in embryos 12 hours AEL. The single-stranded digioxygenin-labeled probe for in situs, amplified from SKIP3 template, likely hybridizes with both SKIP1 and SKIP3. After hybridization, the embryos were incubated with anti-Dig antibody and developed by NBT/BCIP. SKIP1/3 displays expression primarily in the central nervous system (brain indicated by “B”, ventral nerve cord indicated by arrows), and peripheral nervous system (indicated by arrowheads), similar to elav.
To map the Shal binding site in SKIP3, we first designed “prey” vectors containing either the N-terminal SAM domain or the remaining 61 amino acids of SKIP3 (SKIP3C; Fig. 2B). In direct Y2H assays, we found that SKIP3C, but not the SAM domain, showed strong interaction with Shal1C. Since SKIP1 shares all of the SKIP3 sequence except for the C-terminal eight amino acids, which are unique to SKIP3 (Fig. 1C, 2B), we generated and tested a deletion construct of SKIP3 that was missing these eight amino acids. We found that truncation of these eight residues completely disrupted the interaction between SKIP3 and Shal1C (Fig. 2B), suggesting that Shal binding requires these unique C-terminal residues. Thus, SKIP3 is likely to be the only SKIP isoform that interacts with Shal channels.
Nervous System Specific Expression, and Subcellular Co-Localization of myc-SKIP3 and GFP-Shal Channels in Embryonic Neurons
To examine the endogenous expression pattern of SKIP3 in the embryo, we used an anti-sense DNA probe for SKIP3 in RNA in situ hybridization studies. Since SKIP1 contains nearly all of the SKIP3 sequence, our results likely represent the combined expression of SKIP1 and SKIP3. We found strong SKIP1/3 expression primarily in the central nervous system, including the brain (Fig. 2C, see “B”) and ventral nerve cord (Fig. 2C, see arrows), as well as in the peripheral nervous system (Fig. 2C, see arrowheads). This localization pattern is similar to the immunostaining pattern of the neuron-specific elav protein shown as a reference (Fig. 2C).
To examine the subcellular localization of SKIP3 and SKIP1 in neurons, we used the Drosophila UAS/GAL4 system to attain tissue-specific expression of transgenes (Brand and Perrimon 1993). We generated transgenic fly lines expressing either a 6Xmyc-tagged SKIP3 or SKIP1 fusion construct (myc-SKIP3/1) under the control of the upstream-activating-sequence, UAS. Expression of UAS-myc-SKIP3/1 transgenes were driven by crossing these transgenic lines to the neuron-specific elav-GAL4 fly line. We then examined the subcellular localization of myc-SKIP3 and myc-SKIP1 proteins in primary neuronal cultures from these animals. Single neurons, as well as clusters of neurons derived from successive divisions of dissociated neuroblasts, are present in these cultures. We found that myc-SKIP3 and myc-SKIP1 are localized to neuronal cell bodies and in puncta along neuronal processes (Fig. 3); anti-myc staining in wild-type cultures showed little to no background staining (Fig. 3A, bottom-left). To investigate whether this subcellular localization pattern was perhaps due to the expression of either protein in excess of the endogenous levels of SKIP3 and SKIP1, we also expressed myc-SKIP3 and myc-SKIP1 in a genetic background that lacks the SKIP gene (ΔSKIP, also see Fig. 5). myc-SKIP3 and myc-SKIP1 in the ΔSKIP background were again localized to cell bodies and puncta along neuronal processes (Fig. 3A and data not shown), suggesting that this is indeed the normal subcellular localization pattern of SKIP3 and SKIP1.
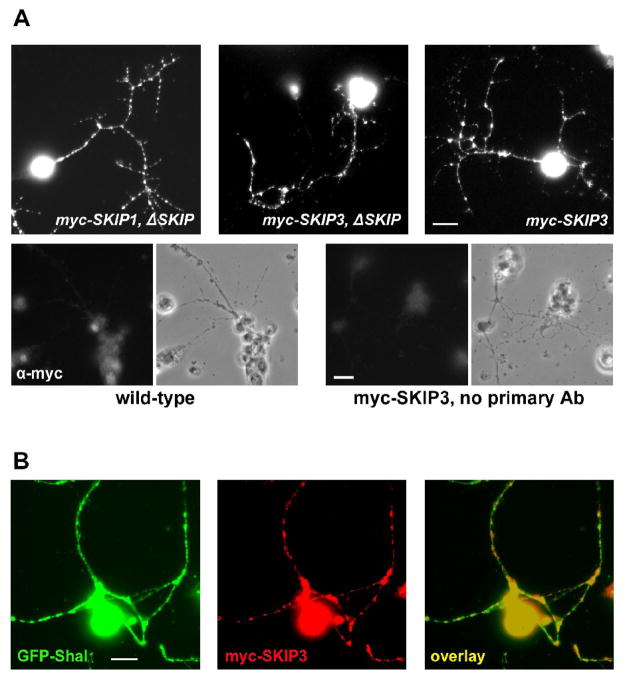
Fig. 3. Subcellular Localization of myc-SKIP1, myc-SKIP3, and GFP-Shal in Embryonic Neurons.
(A) Transgenic lines were generated expressing the neuron-specific elav-GAL4 driver and either the transgene UAS-myc-SKIP1 or UAS-myc-SKIP3 (noted as elav:myc-SKIP1 or elav:myc-SKIP3). Additionally, elav:myc-SKIP1 was present either in a wild-type background or a homozygous Df(3R)Exel6190 (ΔSKIP) background (see text and Fig. 5). Primary cultures were made from embryos of these genotypes and immunostained for myc. Shown are representative single neurons expressing myc-SKIP1/3 in a ΔSKIP background (myc-SKIP1/3, ΔSKIP) and myc-SKIP3 in a wild-type background (myc-SKIP3). myc-SKIP1 and myc-SKIP3 are localized in cell bodies and in puncta along neuronal processes. As negative controls, clusters of wild-type neurons were immunostained with the anti-myc antibody neurons (bottom, left) and myc-SKIP3 expressing clusters of neurons were stained with only secondary antibody (bottom, right); phase-contrast images are shown to the right of each stained panel. (B) Co-localization of myc-SKIP3 and GFP-Shal was examined in cultures from transgenic lines in which elav-GAL4 is driving expression of UAS-myc-SKIP3 and UAS-GFP-Shal2. Shown is a representative image of neurons immunostained for myc-SKIP3 (red) and GFP-Shal (green). Co-localization (yellow) of myc-SKIP3 and GFP-Shal is observed in cell bodies, and in many, although not all, puncta in neuronal processes (right). All scale bars represent 5 μm.
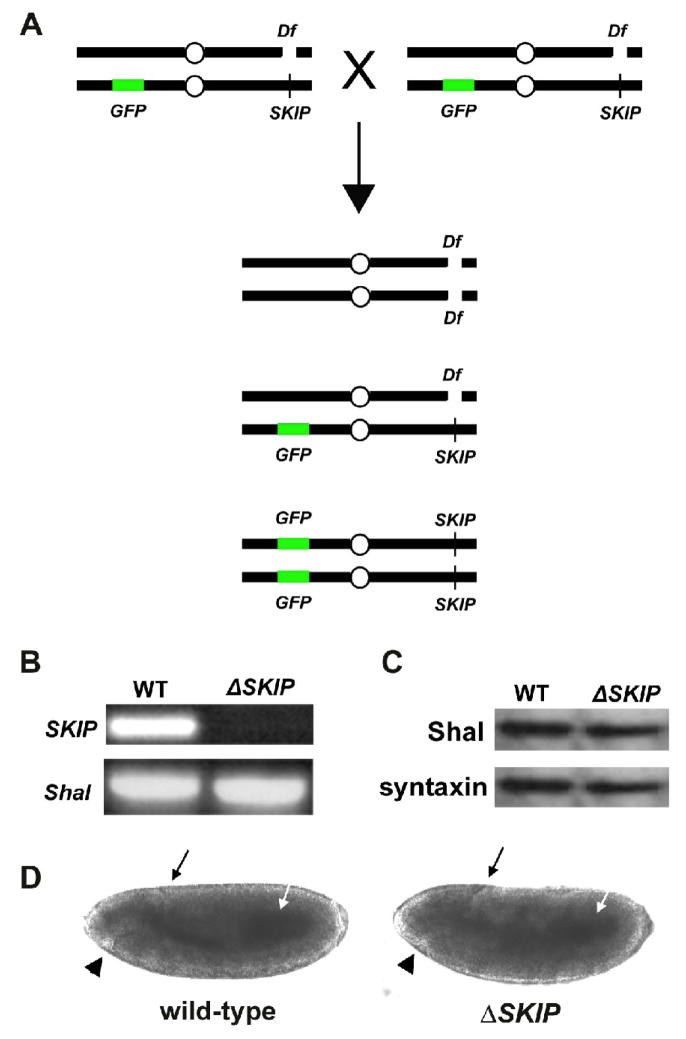
Fig. 5. Genetic Deficiency Df(3R)Exel6190 of SKIP as a Tool to Study the Role of SKIP.
(A) Shown is the genetic strategy used to identify embryos and embryonic cultures that are homozygous for the deficiency Df(3R)Exel6190 (Df). Since the Df(3R)Exel6190 deficiency is homozygous lethal in adult flies, heterozygotes are maintained over a balancer chromosome containing a transgene for GFP expression. Heterozygous adult flies crossed together produce viable embryos of the three genotypes shown. Homozygous Df(3R)Exel6190 embryos (and primary embryonic cultures) can be identified by the lack of GFP expression. (B) PCR amplifications of SKIP and Shal from genomic DNA isolated from wild-type and homozygous Df(3R)Exel6190 (ΔSKIP) embryos. Df(3R)Exel6190 was maintained as a heterozygote over a GFP balancer chromosome. Heterozygotes were crossed to each other, embryos were collected 13–14 hours AEL, and homozygous Df(3R)Exel6190 embryos were selected based on the absence of GFP expression (ΔSKIP). GFP-positive embryos, containing at least one copy of the genes uncovered by the deficiency, were used as a wild-type control (WT). PCR using primers for a common SKIP sequence and Shal show that SKIP could not be amplified from homozygous Df(3R)Exel6190 embryos, verifying that Df(3R)Exel6190 does indeed remove the SKIP gene. (C) Representative immunoblot analysis for Shal protein in wild-type (WT) and homozygous SKIP deficiency (ΔSKIP) embryos. Embryos 13–14 hours AEL were identified as in (B), and analyzed by immunoblot analysis (20 embryos/lane), using antibodies against Shal and syntaxin. Levels of Shal protein are similar in wild-type and ΔSKIP embryos. (D) Representative wild-type and homozygous Df(3R)Exel6190 embryos at stage 10 (5–6 hours AEL), at which time cells are dissociated to make primary cultures, are shown. No gross differences can be observed in the developing embryos at this stage. Black arrows indicate the dorsal invagination of the posterior midgut primordium, white arrows indicate the midgut primordium, and black arrowheads indicate the stomodium.
Since SKIP3 is the isoform that interacts with Shal channels, we next examined whether myc-SKIP3 co-localizes with GFP-tagged Shal channels. We generated a transgenic line expressing a GFP-tagged Shal2 construct under the control of UAS (UAS-GFP-Shal), then used the elav-GAL4 transgene to drive expression of UAS-GFP-Shal and UAS-myc-SKIP3 in all neurons. Primary cultures dissociated from these embryos were examined for the subcellular localization of GFP-Shal and myc-SKIP3. We found that both GFP-Shal and myc-SKIP3 were expressed in neuronal cell bodies and in puncta along processes (Fig. 3B). Double-labeling showed that many puncta displayed signal for both GFP-Shal and myc-SKIP3 (Fig. 3B), suggesting that these proteins co-localize in neuronal process as well as in the soma.
Expression of myc-SKIP3 Does Not Alter Shal Channel Properties in Sf9 Cells
We next examined whether expression of myc-SKIP3 altered the biophysical properties of Shal K+ channels when co-expressed in the insect Sf9 cell line. We chose this system because Drosophila proteins are expected to exhibit functional properties in this insect cell line which are more similar to their native function than if expressed in a vertebrate heterologous system (Brock et al. 2001). Baculovirus infection of Sf9 cells has indeed proven to be an effective system for analyzing ion channel function and modulation (Brock et al. 2001; Gaymard et al. 1996; Gubitosi-Klug and Gross 1996; Klaiber et al. 1990; Mikhailov et al. 1998; Nehrke et al. 2000). Recombinant baculovirus particles expressing Shal or SKIP3 were generated, amplified, and used to infect Sf9 cells (see Experimental Methods). Expression of Shal and SKIP3 in Sf9 cells was confirmed by both immunoblot analysis (data not shown) and confocal imaging (Fig. 4A,B). Infected cells were fixed, immunostained, and imaged by confocal microscopy. Immunostaining was performed using our newly generated Shal and SKIP antibodies (see Experimental Methods). We double-labeled cells with FITC-conjugated phalloidin which binds F-actin. Uninfected cells displayed no expression of Shal or SKIP3 (Fig. 4A,B–top panels), showing that Sf9 cells do not express endogenous Shal or SKIP3 proteins. In contrast, infected cells showed Shal and SKIP3 signal localized to the periphery of cells (Fig. 4A,B).
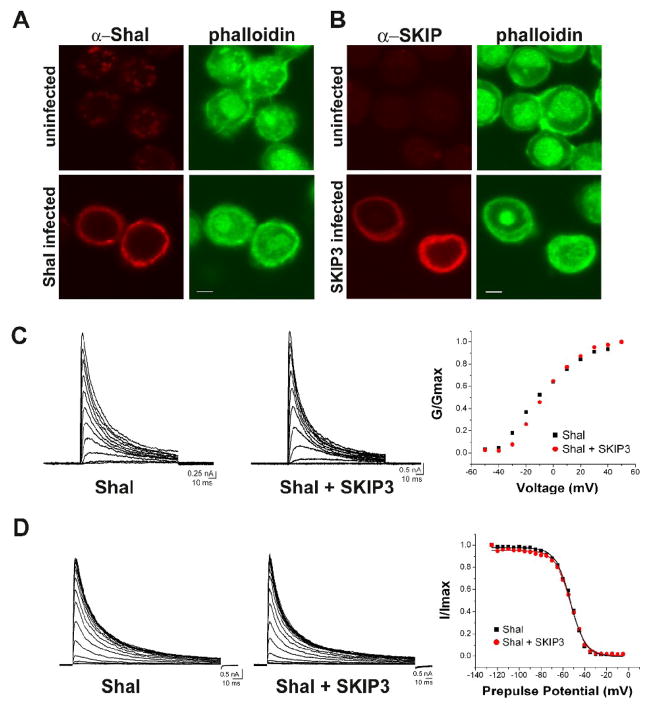
Fig. 4. Co-Expression of Shal and SKIP3 in Sf9 Cells.
(A–B) Sf9 cells were infected with baculovirus expressing either Shal2 (A) or SKIP3 (B). Representative confocal images of Sf9 cells immunostained for Shal/SKIP3, and FITC-conjugated phalloidin, show expression of Shal and SKIP3. Uninfected cells are shown for comparison. Scale bars represent 5 μm. (C) Whole-cell voltage-clamp recordings from representative Sf9 cells infected with the Shal-expressing baculovirus alone (Shal), or both Shal-expressing and SKIP3-expressing baculoviruses (Shal + SKIP3); capacitative transients have been clipped for illustration purposes. Left and middle, Current traces in response to 120 ms voltage-jumps were taken from a holding potential of −100 mV to voltages between −50 mV and +50 mV, in 10 mV increments. Right, G/Gmax-Voltage plots are shown for each cell; no significant difference was observed in any cells. (D) Left and middle, Representative current traces in response to a 140 ms test potential to +50 mV, following a 500 ms pre-pulse at potentials from −125 to −40 mV, in 5 mV increments. Right, Resulting steady-state inactivation plots and fits are shown from these two cells. No significant differences were observed in any cells. Also see Table 1.
We then examined the electrophysiological properties of Shal channels expressed with and without SKIP3. While uninfected Sf9 cells displayed no voltage-dependent currents (data not shown), cells infected with Shal2 baculovirus consistently expressed (in over 30 cells recorded) a transient A-type K+ current activated by depolarization (Fig. 4C). In another set of Sf9 cell cultures, Shal2 was co-expressed with SKIP3 (Shal2+SKIP3 infected) for comparison. Figure 4C shows representative current traces in response to voltage jumps to depolarized potentials from Shal2-infected and Shal2+SKIP3-infected cells. Conductance-voltage (G–V) plots from these two representative cells are indistinguishable from one another (Fig. 4C, right). Times to peak Shal current were also similar (Shal alone at 3.68 +/− 0.83 ms, Shal+SKIP3 at 3.45 +/− 0.72; see Table 1). Inactivation of Shal currents was analyzed by either 1) fitting the entire inactivation time-course with a double exponential function to obtain τ1 and τ2, or 2) by fitting the initial 30 ms of the inactivation time-course with a single exponential decay function to obtain τfast. τ1 and τ2 values were not significantly different for Shal currents expressed with or without SKIP3 (τ1 was 8.65 +/− 1.65 ms for Shal alone and 8.30 +/− 1.46 ms for Shal+SKIP3; τ2 was 38.02 +/− 7.93 ms for Shal alone and 35.74 +/− 7.93 ms for Shal+SKIP3; see Table 1). τfast values were also similar (13.55 +/− 2.75 ms for Shal alone, 12.21 +/− 1.91 ms for Shal+SKIP3; see Table 1). Rates of recovery from inactivation were also not significantly different (see Table 1).
Table 1.
Analysis of Shal currents expressed in Sf9 cells alone (Shal) or with SKIP3 (Shal + SKIP3).
| Shal | Shal + SKIP3 | |
|---|---|---|
| Time to peak at 50 mV | 3.68 +/− 0.83 ms (N=8) | 3.45 +/− 0.72 (N=8) |
| T1 inactivation at 50 mV | 8.65 +/− 1.65 ms (N=7) | 8.30 +/− 1.46 (N=6) |
| T2 inactivation at 50 mV | 38.02 +/− 7.93 ms (N=7) | 35.74 +/− 7.93 ms (N=6) |
| Tfast inactivation at 50 mV | 13.55 +/− 2.75 ms (N=7) | 12.21 +/− 1.91 ms (N=8) |
| Trecovery from inactivation | 125.54 +/− 20.34 ms (N=3) | 135.60 +/− 31.67 (N=3) |
|
Steady-State Inactivation: V1/2 |
−55.95 +/− 5.95 mV (N=5) | −55.11 +/− 4.79 mV (N=5) |
| k (mV/e-fold shift) | 7.09 +/− 1.12 (N=5) | 7.25 +/− 0.54 (N=5) |
Baculovirus particles expressing Shal and SKIP3 were used to infect Sf9 cells. Whole-cell voltage-clamp recordings were performed 2 days post-infection. Voltage protocols applied for analyzing times to peak, inactivation, and steady-state inactivation are described in the text. Averages, standard deviations, and number of cells (N) are shown.
Finally, steady-state inactivation properties of Shal currents were compared between Shal currents expressed with and without SKIP3. We applied 500 ms pre-pulses to voltages between −125 mV and −40 mV, in 5 mV increments, and recorded Shal currents elicited with a voltage jump to a subsequent test potential of +50 mV. Normalized peak currents were plotted against pre-pulse potentials, and data points were fit with a Boltzman function. Current traces and steady-state inactivation plots/fits from two representative cells are shown in Figure 4D. We found that Shal currents were half-inactivated (V1/2) with a pre-pulse potential of −55.95 +/− 5.95 mV for Shal channels expressed alone, and similarly −55.11 +/−4.79 mV for Shal channels co-expressed with SKIP3 (see Table 1). Altogether, these results suggest that SKIP3 does not directly modulate the biophysical properties of Shal channels in Sf9 cells.
Genetic Deficiency of SKIP as a Tool to Study the Role of SKIP3 in Neurons
Since SKIP3 appears to be a scaffolding/adaptor protein, SKIP3 may recruit protein regulator(s) to Shal channels. One possibility is that Sf9 cells do not express the SKIP3 interactor(s) required to regulate Shal channels. The role of SKIP3 for Shal channels might then be revealed more clearly in native cells. Ideally, we would like to examine a single gene mutant of SKIP, however no such mutant yet exists. Thus, we searched the fly databases for available genetic deletions (deficiencies) that would remove the SKIP gene. We identified one deficiency, Df(3R)Exel6190, which is predicted to uncover 18 genes, including the SKIP gene. Although homozygous Df(3R)Exel6190 adult animals are lethal, homozygous Df(3R)Exel6190 embryos are viable and develop normally until very late embryogenesis (16 hours after egg laying (AEL); stages 16–17). Thus, embryos collected 5–6 hours AEL (stage 10) for dissociation into cell cultures included healthy embryos homozygous for Df(3R)Exel6190. Whole embryos and single-embryo cultures, which are homozygous for Df(3R)Exel6190, can be identified based on the absence of a GFP-marked balancer chromosome (Fig. 5A). To determine whether Df(3R)Exel6190 deletes the SKIP gene, we performed PCR amplification of SKIP from GFP-positive (wild-type and heterozygous Df(3R)Exel6190) and GFP-negative (homozygous Df(3R)Exel6190) embryos. We found that SKIP could indeed be amplified from GFP-positive (WT) embryos, but not from GFP-negative embryos (ΔSKIP; Fig. 5B). These results confirm that the deficiency Df(3R)Exel6190 does indeed remove the SKIP gene, and that homozygous Df(3R)Exel6190 embryos can be reliably identified by this method.
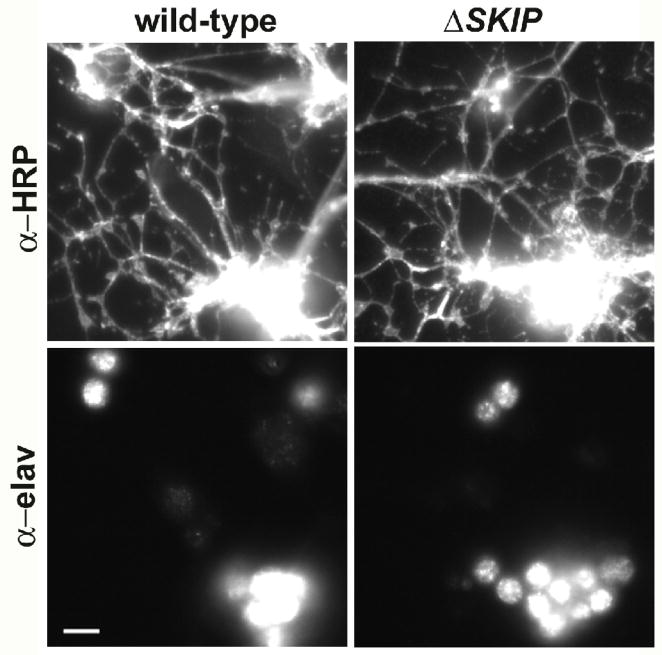
Developing embryos homozygous for Df(3R)Exel6190 are indistinguishable from wild-type embryos. Figure 5D shows wild-type and homozygous Df(3R)Exel6190 embryos in late gastrulation (stage 10), at which time neuroblasts have segregated and cells are dissociated into cultures. At this stage, the dorsal invagination of the posterior midgut primordium (Fig. 5D, black arrows) and the onset of the stomodium (Fig. 5D, arrowheads) are clearly evident in both wild-type and homozygous Df(3R)Exel6190 embryos. Cell cultures homozygous for Df(3R)Exel6190 were also indistinguishable from wild-type. Figure 6 shows wild-type and homozygous Df(3R)Exel6190 cell cultures, immunostained for two different neuron specific markers. Immunostaining for the neuron specific RNA binding protein, elav, labels all neuronal cell bodies, while immunostaining with anti-horseradish peroxidase labels all neuronal membranes (Jan and Jan 1982). No visible differences were observed in the number of labeled neuronal cell bodies (data not shown), or in the extent of labeled neuronal processes (Fig. 6). Homozygous Df(3R)Exel6190 cells appeared as healthy and robust as wild-type cells. Cell sizes, measured by whole-cell capacitance (see next section) were typical for Drosophila neurons and not significantly different (2.85 +/− 0.84 pF for wild-type (N=22), 2.60 +/− 0.90 pF for ΔSKIP neurons (N=20).
Fig. 6. Primary Cultures from Df(3R)Exel6190 Embryos are Similar to Wild-Type.
Cells were dissociated from wild-type and homozygous Df(3R)Exel6190 (ΔSKIP) embryos, and grown for 2–5 days in culture. Cells were fixed and immunostained using α-elav and α-horseradish peroxidase (α-HRP) antibodies, which recognize a neuron-specific RNA binding protein and all neuronal membranes, respectively. Representative images of small clusters of neurons from these cultures are shown. The number of neuronal cell bodies recognized by α-elav and extent of neuronal processes recognized by α-HRP were indistinguishable from one another across many cultures.
We next examined whether the loss of SKIP had any effect on steady-state levels of Shal protein. We collected and isolated homozygous Df(3R)Exel6190 embryos and analyzed levels of Shal protein by immunoblot analysis. We found that levels of Shal protein in Df(3R)Exel6190 embryos were similar to wild-type (Fig. 5C), suggesting that the loss of SKIP does not affect the expression or stability of Shal protein. Thus, we have a verified deficiency of SKIP in hand, the ability to identify homozygous Df(3R)Exel6190 embryos and single-embryo cultures, and evidence that late-gastrulating Df(3R)Exel6190 embryos at stage 10, and primary cultures made from them, are indistinguishable from wild-type. Since homozygous Df(3R)Exel6190 embryos show similar levels of Shal protein as in wild-type, we set out to examine endogenous Shal currents in a SKIP mutant background.
Loss of SKIP3 Results in More Shal K+ Currents with Very Fast Inactivation
To examine whether the absence of SKIP3 alters the functional properties of Shal K+ currents in native cells, we compared Shal currents from wild-type and homozygous Df(3R)Exel6190 (now referred to as ΔSKIP) neurons. Since SKIP3 may recruit cytosolic regulator(s) to Shal channels, all recordings were performed using the perforated patch-clamp technique to avoid “washing out” SKIP3 or any other relevant molecules. Whole-cell currents exhibited variable inactivation rates, as expected from previous studies. Interestingly, the transient components of ΔSKIP neurons appeared to display faster rates of inactivation compared with wild-type. Whole-cell recordings from three representative wild-type and ΔSKIP neurons are shown in Figure 7A. To investigate this further, we set out to isolate and compare the Shal current component from wild-type and ΔSKIP neurons.
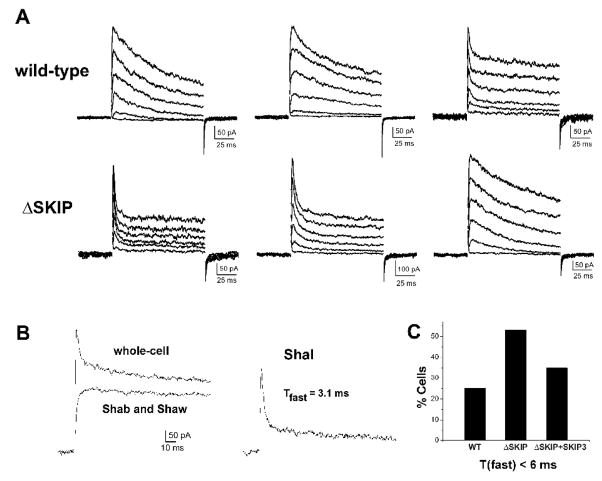
Fig. 7. The Inactivation Rate of Shal K+ Currents is Affected by the Loss of SKIP3.
(A) Whole-cell voltage-clamp recordings of three representative wild-type (top row) and Df(3R)Exel6190 (ΔSKIP, bottom row) neurons are shown. K+ currents were activated by 120 ms voltage jumps to −50 to +50 mV, in 10 mV increments, from a holding potential of −100 mV; capacitative transients have been clipped for illustration purposes. Inactivation rates varied from cell to cell, however, almost twice as many Df(3R)Exel6190 neurons displayed the very fast inactivating A-type currents (bottom row, left and middle recordings). (B) Separation of Shal current from Shab/Shaw currents by pre-pulse potential. In one representative neuron, the whole-cell K+ current is shown in response to a voltage jump to +50 mV from a 500 ms pre-pulse of −125 mV (left, top trace). With a pre-pulse of −45 mV, the transient A-type current encoded by Shal is completely inactivated and only the delayed-rectifier currents encoded by Shab and Shaw are activated (left, bottom trace). Subtraction of the Shab/Shaw currents from the whole-cell current yields the Shal K+ current (right). In this neuron, the inactivation time course of the Shal current (first 30 ms after peak) is fit with a single exponential function (best fit with τfast =3.1 ms). (C) Quantification of the percentage of wild-type (WT) and Df(3R)Exel6190 (ΔSKIP) neurons containing a Shal current with an inactivation rate that corresponds to Shal channels exclusively in the “fast” gating mode (τfast< 6 ms). Shal currents were isolated as in (B), and the inactivation time course 30 ms following peak current was fit with the single exponential decay function, y(t)=Ae(−t/τfast) + c. In contrast to 25% of wild-type neurons (N=16), 55% of Df(3R)Exel6190 (ΔSKIP) neurons (N=20) contained a Shal current with τfast< 6 ms. Transgenic expression of myc-SKIP3 in Df(3R)Exel6190 neurons (ΔSKIP + SKIP3) showed partial rescue, with 35% (N=17) containing a Shal current with τfast< 6 ms.
Shal currents can usually be isolated based on their hyperpolarized operating range (Tsunoda and Salkoff 1995a). We first tested whether Shal currents showed any differences in steady-state inactivation properties between wild-type and ΔSKIP neurons. Voltage protocols to examine steady-state inactivation properties were as described above for Sf9 cell recordings. Shal currents were half-inactivated with pre-pulses to −68.15 +/−8.00 mV (N=14) in wild-type and, similarly, −68.14 +/− 7.81 mV (N=19) in ΔSKIP neurons (see Table 2). We were therefore able to isolate Shal currents based on their hyperpolarized operating range, as previously described (Tsunoda and Salkoff 1995a). We applied a 500 ms pre-pulse of −45 mV to completely inactivate the Shal current present in a given neuron, such that a subsequent voltage jump to +50 mV activated only the slowly-inactivating currents encoded by Shab and Shaw channels (Tsunoda and Salkoff 1995a; Tsunoda and Salkoff 1995b). The slowly-inactivating current was subtracted from the whole-cell current (activated from a pre-pulse of −125mV) to isolate the transient current component carried by Shal channels (Fig. 7B). Shal current densities in ΔSKIP neurons (117.17 +/− 49.96 pA/pF) were similar to those in wild-type neurons (120.23 +/−90.93 pA/pF) (see Table 2).
Table 2.
Analysis of wild-type and homozygous Df(3R)Exel6190 (ΔSKIP) Neurons.
| wild-type | ΔSKIP | |
|---|---|---|
| Shal current density (pA/pF) | 120.23 +/− 90.93 (N=17) | 117.17 +/− 49.96 (N=18) |
| Time to peak (ms) at 50 mV | 3.91 +/− 2.38 (N=16) | 2.69 +/− 1.14 (N=21) |
|
Steady-State Inactivation: V1/2 (mV) |
−68.15 +/− 8.00 N=14) | −68.14 +/− 7.81 (N=19) |
| k (mV/e-fold shift) | 9.02 +/− 2.25 (N=14) | 8.66 +/− 1.46 (N=19) |
| Tfast inactivation at 50 mV | 17.95 +/− 20.37 (N=16) | 9.86 +/− 9.75 (N=20) |
| % cells Tfast < 6 ms | 25% (N=16) | 55% (N=20) |
Whole-cell voltage-clamp recordings were performed on neurons in primary cultures grown for 2–4 days after embryonic dissociation. Voltage protocols applied for analyzing times to peak, inactivation, and steady-state inactivation are described in the text. Averages, standard deviations, and number of cells (N) are shown.
We next examined the rates of inactivation of Shal currents isolated from wild-type and ΔSKIP neurons. Whole-cell inactivation rates are thought to arise from variable proportions of single Shal channels in a “fast” or “slow” gating mode. Single Shal channels in the “fast” gating mode have been shown to give rise to ensemble averaged currents with a τfast of 3.9 ms, while Shal channels in the “slow” gating mode give rise to ensemble averaged currents with τfast of 38.2 to 92.0 ms (Tsunoda and Salkoff 1995a). Inactivation rates of whole-cell Shal currents isolated in wild-type neurons varied from cell to cell (Fig. 7A), with fast inactivation time constant (τfast) values that varied from 2.6 to 82.6 ms (Fig. 8). While inactivation rates were still variable among Shal currents in ΔSKIP neurons, there appeared to be more ΔSKIP neurons with a whole-cell τfast that would correspond to Shal channels exclusively in the “fast” gating mode, when compared with wild type (Fig. 7–8). When the average τfast from ΔSKIP neurons (τfast= 9.86 +/− 9.75 ms) was compared to wild-type (τfast= 17.95 +/− 20.37 ms), however, no significant difference (p =0.15) was calculated using a Student’s t-test. Since these cells represent a very heterogeneous population, having been dissociated from whole embryos, it was not surprising that the high variability in τfast values made it difficult to detect differences across the whole population. Because τfast values from this heterogeneous population would not be predicted to have a Gaussian distribution, we also compared τfast values using the non-parametric Mann-Whitney U-test. This test yielded a more significant p-value of 0.05. The distribution of τfast values from Shal currents in ΔSKIP neurons also showed a noticeable shift to faster τfast values when compared with wild-type (Fig. 8). We quantified the percentage of wild-type and ΔSKIP neurons that displayed whole-cell Shal currents with a τfast of inactivation equivalent to most Shal channels in the fast gating mode (τfast< 6 ms). In wild-type, we found that 25% of the neurons (N= 16) contained a Shal current with τfast< 6 ms, while 55% of ΔSKIP neurons (N= 20) displayed a Shal current with τfast< 6 ms (Fig. 7C). These results suggest that the loss of the SKIP gene results in a shift of Shal channels to the fast gating mode in some population of embryonic neurons.
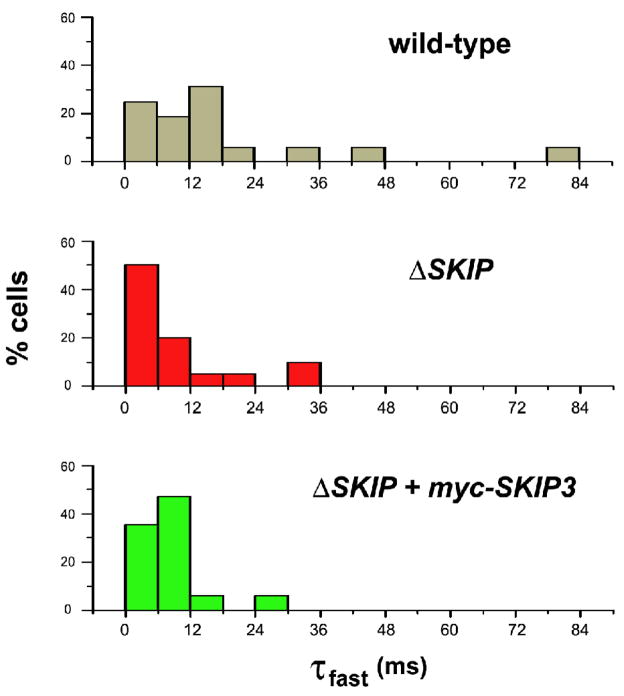
Fig. 8. Distribution of Fast Inactivation Rates of Shal K+ Currents in Wild-Type and ΔSKIP Neurons.
Histograms of fast inactivation time constants (τfast) from whole-cell Shal currents isolated from wild-type (top), Df(3R)Exel6190 (middle; ΔSKIP), and transgenic myc-SKIP3 in Df(3R)Exel6190 (bottom; ΔSKIP + myc-SKIP3) neurons. Shal K+ currents elicited with a voltage jump to +50 mV were isolated as described in Fig. 7B. Inactivation time courses 30 ms following peak current were fit with the single exponential decay function, y(t)=Ae(−t/τfast) + c. X-axis represents τfast values binned every 6 ms; y-axis represents the % cells (N=16 for wild-type, N=20 for ΔSKIP, N=17 for ΔSKIP + myc-SKIP3). Note the shift in τfast values to faster inactivation rates in SKIP neurons compared with wild-type.
In contrast, there was no significant difference in time to peak of Shal currents (TTP; TTP for wild-type was 3.91 +/− 2.38 ms, TTP for ΔSKIP was 2.69 +/− 1.14 ms), suggesting that only inactivation, and not activation, rates were altered. There was, however, no difference in the relative weights of fast and slow inactivation time constants when inactivation time courses were fit with a double exponential decay function (data not shown). Fast inactivation made up more than 90% of the total inactivation, suggesting that the loss of SKIP affects the major inactivation component/mechanism.
To test whether this phenotype was indeed due to the loss of SKIP3, we tested whether expression of myc-SKIP3 in neurons was able to rescue the number of ΔSKIP neurons with a very fast inactivation rate (τfast <6 ms). We generated a transgenic fly strain expressing myc-SKIP3 in ΔSKIP neurons. We found that myc-SKIP3 expression in ΔSKIP neurons indeed decreased the percentage of neurons with such a rapidly inactivating Shal current to 35% (N= 17, Fig. 7C, 8). Although full rescue of the ΔSKIP phenotype was not observed, this may be due to rather low transgenic expression levels of myc-SKIP3. Altogether, our results suggest that SKIP3 regulates the fast inactivation rate of whole-cell Shal currents in at least a subpopulation of embryonic Drosophila neurons.
Discussion
Mechanisms of fast inactivation of voltage-dependent K+ channels have been the focus of many studies. Fast inactivation of Kv4/Shal channels is conserved in all known Shal channels, and regulation of this inactivation rate is likely to play an important role in modulating the excitability of somato-dendritc compartments of neurons. Although Shal channel inactivation by “classical” N-type or C-type inactivation has remained unclear (Jerng et al. 2004), the gating properties of mammalian Kv4 channels have been shown to be regulated by two main interacting components, KChIPs and DPPX (An et al. 2000; Nadal et al. 2003). Although Drosophila Shal channels display variable rates of fast inactivation, the mechanism and participation of such auxiliary components has not been previously explored. We tested a few candidate proteins (frequenin, neurocalcin, and hyperkinetic) by Y2H, but found no obvious interaction. Although we could not find any Drosophila homolog of DPPX by sequence similarity, there is a putative Drosophila KChIP-like gene (cg5890) that was reported in a phylogenetic analysis of KChIP-like proteins across species (Salvador-Recatala et al. 2006). Interestingly, the lobster Shal channel has been shown to undergo modulation by a human KChIP when co-expressed in a heterologous system (Zhang et al. 2003). These results suggest that KChIP regulatory mechanisms are likely to be conserved in invertebrates. Since Drosophila Shal channels do contain the conserved N-terminal KChIP binding domain seen in mammalian Kv4 channels, it may be that this gene, cg5890, encodes a functional KChIP homolog; this possibility remains to be tested.
Since previous studies have shown that both the N- and C-termini of Kv4 channels are likely to function in fast inactivation of the channel (Jerng and Covarrubias 1997), we first focused on identifying new interactors of the C-terminus of Drosophila Shal channels. In this report, we introduce the nervous system specific SKIP3 protein as a new player in the regulation of Shal K+ channels. We show that SKIP3 is one of three SKIP isoforms, each of which contains multiple protein binding domains. The C-terminus of SKIP3 contains the Shal binding domain, with the last eight amino acids critical for interaction. Because these eight residues are unique to SKIP3, we propose that SKIP3 is the only isoform that binds Shal. SKIP3 binds the C-terminus of Shal channels via a unique binding site on the C-terminus of SKIP3. This sequence of Shal displays little identity with mammalian Kv4 channels, suggesting that SKIP3 may be a regulatory subunit unique to Drosophila. SKIP3 also contains a SAM domain (SAM1) on its N-terminal half, and a C-terminal Shal binding domain. Since SAM1 does not bind Shal or the SAM2 domain of SKIP1 and SKIP2, nor does it homomultimerize, SKIP3 is likely to recruit another, perhaps regulatory, protein via its SAM1 domain and bring it into close proximity with Shal channels. Thus, the role of SKIP3 for Shal channels likely depends on another key protein. Consistent with this hypothesis was our finding that a role for SKIP3 in Shal channel regulation could only be detected in neurons, and not when co-expressed in a heterologous system. We also tried introducing purified SKIP3 protein to neuronal Shal channels via the patch pipette in the conventional whole-cell mode. These attempts, however, were unsuccessful at altering Shal current inactivation and this may be because required cytosolic regulators were “washed out” in this recording mode.
What role, if any, do the other isoforms of SKIP play in Shal channel regulation? Because SKIP1 contains the same SAM1 domain as in SKIP3, any SAM1 interactor of SKIP3 would be subject to competitive binding by SKIP1. In this way, SKIP1 could potentially regulate the availability of a SAM1 interactor to SKIP3 and Shal channels. Alternatively, a SAM1 interactor that homomultimerizes could link SKIP1 and SKIP3 proteins, producing more extensive regulatory complexes for Shal channels. Similarly, SKIP1 and SKIP2 contain the identical SAM2 and SH3 domains and may regulate one another through competitive or simultaneous binding to a SAM2 or SH3 interactor.
An interesting feature of Drosophila Shal channels is that they adopt one of two gating modes, a “fast” or “slow” gating mode, each of which exhibits a very fast (τfast< 6 ms) or slower inactivation rate, respectively (Tsunoda and Salkoff 1995a). Such modal gating has also been described in other systems (Alzheimer et al. 1993; Delcour et al. 1993; Moorman et al. 1990), however, the underlying molecular mechanisms are largely unknown. Using a genetic deletion of the SKIP gene (ΔSKIP), we show that SKIP3, and therefore any regulatory complex it forms, results in a higher percentage of ΔSKIP neurons that display Shal currents with inactivation kinetics indicative of Shal channels exclusively in the fast gating mode (τfast< 6 ms). Interestingly, it didn’t seem to be the case that all neurons had an increased rate of inactivation. Instead, there appeared to be an increase in the subpopulation of neurons exhibiting this very fast inactivation rate. One possibility is that SKIP3 is expressed in a subset of neurons from this heterogeneous culture, and it is in those cells that we saw an effect on Shal K+ currents. It will be important for future studies to examine whether SKIP3-expressing neurons do indeed exhibit Shal currents with slower inactivation rates. This may require RNA in situ or single-cell RT-PCR correlation with patch-clamp recording. Unfortunately, such an ambitious undertaking, with the small size (3 to 5 μm diameter) of Drosophila neurons, was beyond the scope of the present study. Also noteworthy is the observation that the rates of Shal current inactivation in such a subpopulation of neurons predict that all, or most, Shal channels are in the “fast” gating mode. This might indicate that SKIP3 is required for Shal channels to be present in the “slow” gating mode in these neurons.
One possibility is that, in wild-type neurons, SKIP3 binds Shal channels to act as a “molecular switch”, setting channels in the slow gating mode. Interestingly, the SKIP3 binding site of Shal channels is just downstream of a highly conserved ~30 amino acid stretch that has been implicated in regulating some forms of inactivation of Shal channels (Hatano et al. 2004; Jerng and Covarrubias 1997). SKIP3 binding may perhaps alter the inactivation function of this sequence. Another possibility is that different isoforms or post-translationally modified forms of Shal channels exhibit only one gating mode, either fast or slow. Since the loss of SKIP3 results in Shal K+ channels in the fast gating mode, SKIP3 may up-regulate the proportion of Shal channels in the slow gating mode, or conversely, down-regulate the proportion of Shal channels in the fast gating mode. In short, another possibility is that SKIP3 might facilitate or inhibit the trafficking of fast or slow gating mode channels to the plasma membrane.
Since previous studies have shown that Shal channel gating may be regulated by protein kinase-A, protein kinase-C, and/or Ca2+/calmodulin-dependent protein kinase II (Hoffman and Johnston 1998; Sergeant et al. 2005), one hypothesis is that the SAM1 domain of SKIP3 may recruit a protein kinase/phosphatase to Shal channels to regulate its inactivation. Identifying protein interactors of the SAM1 domain should reveal whether SKIP3 indeed recruits a dynamic modulator of Shal channel gating, or a chaperone of Shal channels with an intrinsic gating mode. Since the N- and C-termini may function together in regulating fast inactivation (Jerng and Covarrubias 1997), future studies will also examine whether there is a functional KChIP homolog in Drosophila, and if it acts in concert with SKIP3. Studies examining single Shal channels in identified subpopulations of neurons will also aid our understanding of SKIP function. SKIP3 represents the first interactor of Shal K+ channels identified in Drosophila, and although there is no mammalian homolog that we can identify by sequence similarity, the SKIP3-Shal interaction is likely to be an important model for understanding an additional mechanism regulating fast inactivation of A-type K+ currents.
Experimental Methods
Yeast-Two-Hybrid (Y2H) Assays for cDNA Screening and Mapping Interaction Sites
The C-terminus of Shal1 (Shal1C; amino acids 411–571) was amplified from a cDNA clone of Shal1 and cloned into the “bait” vector, pGBKT7 (Matchmaker Gal4 Two-Hybrid System 3, Clontech Laboratories Inc., Palo Alto, CA). pGADT7 “prey” constructs contained cDNAs reverse transcribed from RNA isolated from Drosophila embryos (aged 0–24 hrs) fused to the yeast GAL4 activation domain (Clontech Laboraties Inc., Palo Alto, CA). Shal1C bait plasmid and cDNA prey plasmids were co-transformed into the yeast strain AH109, which contains three reporter-genes under the control of UAS: HIS3, Ade2, and MEL1.
For direct Y2H assays testing the specificity of the SKIP-Shal interaction, the C-termini of Shaker and Shaw were amplified from cDNA clones of ShA1 and Shaw2 by PCR, the C-terminus of Shab-RB was amplified by RT-PCR; amplified products were subcloned into the pGBKT7 bait vector. For mapping the SKIP-Shal interaction, deletion constructs of SKIP3 and Shal were amplified and cloned into pGADT7 and pGBKT7 vectors, respectively. All constructs were confirmed by DNA sequencing, co-transformed into AH109 yeast, and assayed for interaction by the expression of reporter genes.
GST-Pulldown Assays
Shal1C and Shal2C were amplified from Shal cDNA clones using appropriately designed primers. Full-length SKIP3 was amplified from genomic DNA; PCR amplification rounds were designed to fuse the epitope c-myc to the N-terminus of SKIP3. Shal and myc-SKIP3 products were then subcloned into pGEX-2TK and transformed into BL21 cells to express GST-fusion proteins. GST and GST-fusion proteins were purified using glutathione-agarose beads (Sigma Aldrich, St. Louis, MO). myc-SKIP3 protein was cut with biotinylated thrombin (Novagen Inc, San Diego, CA) from GST-myc-SKIP3, which contains an engineered thrombin site. Thrombin was removed using streptavidin-agarose beads. GST proteins were then incubated with purified myc-SKIP3 protein. Glutathione-agarose beads were washed, boiled in 4×SDS loading buffer, and run by standard SDS-PAGE techniques. Proteins were transferred to nitrocellulose and immunoblots were probed with anti-myc antibody.
Fly Stocks
cn bw and w1118 Drosophila were used as wild-type strains in this study. The Df(3R)Exel6190 deficiency, which we show uncovers the SKIP gene, was generated in the Exelixis deletion series (Parks et al. 2004; Thibault et al. 2004). The deficiency chromosome was maintained over one of two different GFP balancer chromosomes: TM3 P{GAL4-Kr.C}DC2 P{UAS-GFP.S65T}DC10, Sb1 (for identifying whole embryos 13–16 hours AEL), or TM6B, P{w[+mW.hs]=Ubi-GFP.S65T}PAD2, Tb1 (for identifying single-embryo cultures). pUAST-6×myc-SKIP1 and pUAST-6×myc-SKIP3 transgenic lines were constructed using the Gateway recombination technology-based system (Invitrogen Corp., Carlsbad, CA). SKIP1 and SKIP3 cDNAs were cloned into pENTR1A (Invitrogen, Carlsbad, CA) and recombined into the pTMW “destination” vector (Drosophila Gateway Vector collection, DGRC). Transgenic lines were generated by standard P-element mediated germline transformation.
In Situ Hybridization
Wild type (w1118) embryos aged 12 hours (at 25°C) were collected, washed with water, dechorionated with bleach, and fixed with 4% paraformaldehyde in PBS. After rehydration, embryos were treated with protease K and then hybridized with probe at 55°C. A single-stranded digioxygenin-labeled (DIG-11-UTP, DIG DNA Labeling and Detection Kit, Roche, Basel, Switzerland) probe was amplified by PCR. The linearized SKIP clone from the Y2H screen was used as template. A 3′ activation domain primer was used to amplify the SKIP probe. After hybridization and wash, embryos were incubated with anti-DIG antibody and developed by NBT/BCIP.
Antibodies
The anti-Shal antibody was generated using the peptide CGIELDDNYRD corresponding to sequence on the C-termini of Shal1 and Shal2. The anti-SKIP antibody was generated using the peptide CETPPNNELELVLRE. The N-terminal cysteines of both peptides were added for conjugation to the KLH carrier protein, which was then used to immunize rabbits (ProSci, Inc., Poway, CA). Anti-sera were then affinity-purified (Affi-gel 10 support matrix, Bio-Rad, Hercules, CA). The anti-Shal antibody was verified to recognize Shal protein on immunoblots, cell bodies of cultured neurons, and Sf9 cells expressing Shal2. The anti-SKIP antibody was verified to recognize SKIP1 and SKIP3 proteins expressed in Sf9 cells analyzed on immunoblots and in fixed cells. For Sf9 cell staining, the anti-Shal antibody was used at 1:50, and the anti-SKIP antibody was used at 1:50. Monoclonal anti-elav (9F8A9) and anti-myc (9E10) were obtained from the Developmental Studies Hybridoma Bank (DSHB, University of Iowa) and used at 1:50 and 1:5, respectively, for immunostaining. A second anti-c-myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was also used at 1:50. Polyclonal anti-GFP (Torrey Pines Biolabs, East Orange, NJ) was used at 1:100 for immunostaining. All primary antibodies used for immunostaining cells were incubated overnight at 4 °C.
Immunostaining Embryonic Cell Cultures
Embryos aged 5–6 hours (25°C; ~stage 10) were dissociated into culture media (Schneider’s Drosophila media supplemented with 18% fetal bovine serum, 100 units/mL penicillin, 100 ug/mL streptomysin) using a glass micropipet, as previously described (Tsunoda and Salkoff 1995a; Tsunoda and Salkoff 1995b). Cell cultures grown for 2 to 7 days were fixed (4% formaldehyde in PBS, 10 minutes), and incubated with primary antibody overnight at 4°C. Cell cultures were washed (0.1% sapponin in PBS), incubated with rhodamine or FITC-conjugated secondary antibody (Jackson Immunoresearch Laboratories, West Grove, PA; 2 hours, room temperature), washed again, and mounted in 90% glycerol with p-phenylenediamine (Sigma Aldrich, St. Louis, MO).
Baculovirus Infection of Sf9 Cells
The “Bac to Bac” Baculovirus Expression System (Ciccarone et al. 1997) was used to generate recombinant baculoviruses expressing Shal2 and SKIP3. pFastBac donor plasmids (Invitrogen, Carlsbad, CA) containing Shal2 and SKIP3 under the control of a baculovirus-specific promoter were constructed, transfected into an E. coli host strain, DH10Bac (Invitrogen, Carlsbad, CA), which contains a bacmid shuttle vector and a helper plasmid that promotes recombination, to generate recombinant baculovirus DNAs. Recombinant bacmid DNAs were transfected into Sf9 insect cells. Recombinant baculovirus particles produced by transfected Sf9 cells were harvested, amplified, titered, and used as a stock to infect Sf9 cells for functional studies.
Electrophysiological Studies
Whole-cell voltage-clamp recordings (Axopatch 200B, Molecular Devices Corp., Sunnyvale, CA) were performed on Sf9 cells two days post-infection with Shal2-expressing and SKIP3-expressing baculoviruses, and on embryonic Drosophila neurons grown in culture for 2 to 4 days. Internal solution was (in mM): 140 KCl, 2 MgCl2, 11 EGTA, 10 Hepes, pH 7.28. External solution was (in mM): 140 NaCl, 2 KCl, 6 MgCl2, 5 Hepes, pH 7.2. For neuronal recordings, NaCl in the external solution was replaced with Choline-Cl. Also for neuronal recordings, 200 ug/mL Amphoterocin B (Sigma-Aldrich, St. Louis, MO) was added fresh to the internal solution to obtain perforated-patch recordings; whole-cell mode was obtained within 1–5 minutes of seals. Electrode resistances were 5 to 7 MΩ. Average input resistances were 3.8 GΩ for embryonic neurons and 1.62 GΩ for Shal-expressing Sf9 cells. Series resistance was ~85% compensated for Sf9 cells; no compensation was applied for embryonic neurons. Since inactivation rates of Shal currents in these neurons have been shown to be relatively voltage-independent, we do not expect that the small voltage error from series resistance to affect our measurements. Recordings were digitized at 5 kHz and filtered using a lowpass Bessel filter, fc= 2 kHz.
Acknowledgments
We thank Dr. Lawrence Salkoff for Shal1, Shal2, ShA1, and Shaw2 cDNAs. S.T. is supported by The Clare Boothe Luce Program of the Henry Luce Foundation, and a grant from the National Institutes of Health (R01 GM083335).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alzheimer C, Schwindt PC, Crill WE. Modal gating of Na+ channels as a mechanism of persistent Na+ current in pyramidal neurons from rat and cat sensorimotor cortex. J Neurosci. 1993;13:660–673. doi: 10.1523/JNEUROSCI.13-02-00660.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An WF, Bowlby MR, Betty M, Cao J, Ling HP, Mendoza G, Hinson JW, Mattsson KI, Strassle BW, Trimmer JS, Rhodes KJ. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403:553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- Bhattacharjya S, Xu P, Chakrapani M, Johnston L, Ni F. Polymerization of the SAM domain of MAPKKK Ste11 from the budding yeast: implications for efficient signaling through the MAPK cascades. Protein Sci. 2005;14:828–835. doi: 10.1110/ps.041122105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brock MW, Lebaric ZN, Neumeister H, Detomaso A, Gilly WF. Temperature-dependent expression of a squid Kv1 channel in Sf9 cells and functional comparison with the native delayed rectifier. J Membr Biol. 2001;180:147–161. doi: 10.1007/s002320010066. [DOI] [PubMed] [Google Scholar]
- Burgoyne RD, Weiss JL. The neuronal calcium sensor family of Ca2+-binding proteins. Biochem J. 2001;353:1–12. [PMC free article] [PubMed] [Google Scholar]
- Cai X, Liang CW, Muralidharan S, Kao JP, Tang CM, Thompson SM. Unique roles of SK and Kv4.2 potassium channels in dendritic integration. Neuron. 2004;44:351–364. doi: 10.1016/j.neuron.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Chen X, Yuan LL, Zhao C, Birnbaum SG, Frick A, Jung WE, Schwarz TL, Sweatt JD, Johnston D. Deletion of Kv4.2 gene eliminates dendritic Atype K+ current and enhances induction of long-term potentiation in hippocampal CA1 pyramidal neurons. J Neurosci. 2006;26:12143–12151. doi: 10.1523/JNEUROSCI.2667-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouinard SW, Wilson GF, Schlimgen AK, Ganetzky B. A potassium channel beta subunit related to the aldo-keto reductase superfamily is encoded by the Drosophila hyperkinetic locus. Proc Natl Acad Sci U S A. 1995;92:6763–6767. doi: 10.1073/pnas.92.15.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarone VC, Polayes D, Luckow VA. E. coli Using Baculovirus Shuttle Vector. Totowa, NJ: Humana Press, Inc; 1997. Generation of Recombinant Baculovirus DNA. [DOI] [PubMed] [Google Scholar]
- Delcour AH, Lipscombe D, Tsien RW. Multiple modes of N-type calcium channel activity distinguished by differences in gating kinetics. J Neurosci. 1993;13:181–194. doi: 10.1523/JNEUROSCI.13-01-00181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett GL, Santi CM, Butler A, Harris T, Covarrubias M, Salkoff L. Mutant analysis of the Shal (Kv4) voltage-gated fast transient K+ channel in Caenorhabditis elegans. J Biol Chem. 2006;281:30725–30735. doi: 10.1074/jbc.M605814200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaymard F, Cerutti M, Horeau C, Lemaillet G, Urbach S, Ravallec M, Devauchelle G, Sentenac H, Thibaud JB. The baculovirus/insect cell system as an alternative to Xenopus oocytes. First characterization of the AKT1 K+ channel from Arabidopsis thaliana. J Biol Chem. 1996;271:22863–22870. doi: 10.1074/jbc.271.37.22863. [DOI] [PubMed] [Google Scholar]
- Gubitosi-Klug RA, Gross RW. Fatty acid ethyl esters, nonoxidative metabolites of ethanol, accelerate the kinetics of activation of the human brain delayed rectifier K+ channel, Kv1.1. J Biol Chem. 1996;271:32519–32522. doi: 10.1074/jbc.271.51.32519. [DOI] [PubMed] [Google Scholar]
- Hatano N, Ohya S, Muraki K, Clark RB, Giles WR, Imaizumi Y. Two arginines in the cytoplasmic C-terminal domain are essential for voltage-dependent regulation of A-type K+ current in the Kv4 channel subfamily. J Biol Chem. 2004;279:5450–5459. doi: 10.1074/jbc.M302034200. [DOI] [PubMed] [Google Scholar]
- Hoffman DA, Johnston D. Downregulation of transient K+ channels in dendrites of hippocampal CA1 pyramidal neurons by activation of PKA and PKC. J Neurosci. 1998;18:3521–3528. doi: 10.1523/JNEUROSCI.18-10-03521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isbrandt D, Leicher T, Waldschutz R, Zhu X, Luhmann U, Michel U, Sauter K, Pongs O. Gene structures and expression profiles of three human KCND (Kv4) potassium channels mediating A-type currents I(TO) and I(SA) Genomics. 2000;64:144–154. doi: 10.1006/geno.2000.6117. [DOI] [PubMed] [Google Scholar]
- Jan LY, Jan YN. Antibodies to horseradish peroxidase as specific neuronal markers in Drosophila and in grasshopper embryos. Proc Natl Acad Sci U S A. 1982;79:2700–2704. doi: 10.1073/pnas.79.8.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerng HH, Covarrubias M. K+ channel inactivation mediated by the concerted action of the cytoplasmic N- and C-terminal domains. Biophys J. 1997;72:163–174. doi: 10.1016/S0006-3495(97)78655-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerng HH, Pfaffinger PJ, Covarrubias M. Molecular physiology and modulation of somatodendritic A-type potassium channels. Mol Cell Neurosci. 2004;27:343–369. doi: 10.1016/j.mcn.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Kaplan WD, Trout WE., 3rd The behavior of four neurological mutants of Drosophila. Genetics. 1969;61:399–409. doi: 10.1093/genetics/61.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CA, Bowie JU. SAM domains: uniform structure, diversity of function. Trends Biochem Sci. 2003;28:625–628. doi: 10.1016/j.tibs.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Kim J, Jung SC, Clemens AM, Petralia RS, Hoffman DA. Regulation of dendritic excitability by activity-dependent trafficking of the A-type K+ channel subunit Kv4.2 in hippocampal neurons. Neuron. 2007;54:933–947. doi: 10.1016/j.neuron.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Wei DS, Hoffman DA. Kv4 potassium channel subunits control action potential repolarization and frequency-dependent broadening in rat hippocampal CA1 pyramidal neurones. J Physiol. 2005;569:41–57. doi: 10.1113/jphysiol.2005.095042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaiber K, Williams N, Roberts TM, Papazian DM, Jan LY, Miller C. Functional expression of Shaker K+ channels in a baculovirus-infected insect cell line. Neuron. 1990;5:221–226. doi: 10.1016/0896-6273(90)90311-3. [DOI] [PubMed] [Google Scholar]
- Mikhailov MV, Proks P, Ashcroft FM, Ashcroft SJ. Expression of functionally active ATP-sensitive K-channels in insect cells using baculovirus. FEBS Lett. 1998;429:390–394. doi: 10.1016/s0014-5793(98)00640-1. [DOI] [PubMed] [Google Scholar]
- Moorman JR, Kirsch GE, Vandongen AM, Joho RH, Brown AM. Fast and slow gating of sodium channels encoded by a single mRNA. Neuron. 1990;4:243–252. doi: 10.1016/0896-6273(90)90099-2. [DOI] [PubMed] [Google Scholar]
- Nadal MS, Ozaita A, Amarillo Y, Vega-Saenz De Miera E, Ma Y, Mo W, Goldberg EM, Misumi Y, Ikehara Y, Neubert TA, Rudy B. The CD26-related dipeptidyl aminopeptidase-like protein DPPX is a critical component of neuronal A-type K+ channels. Neuron. 2003;37:449–461. doi: 10.1016/s0896-6273(02)01185-6. [DOI] [PubMed] [Google Scholar]
- Nakahira K, Shi G, Rhodes KJ, Trimmer JS. Selective interaction of voltage-gated K+ channel beta-subunits with alpha-subunits. J Biol Chem. 1996;271:7084–7089. doi: 10.1074/jbc.271.12.7084. [DOI] [PubMed] [Google Scholar]
- Nehrke K, Begenisich T, Pilato J, Melvin JE. Into ion channel and transporter function. Caenorhabditis elegans ClC-type chloride channels: novel variants and functional expression. Am J Physiol Cell Physiol. 2000;279:C2052–2066. doi: 10.1152/ajpcell.2000.279.6.C2052. [DOI] [PubMed] [Google Scholar]
- Pak MD, Baker K, Covarrubias M, Butler A, Ratcliffe A, Salkoff L. mShal, a subfamily of A-type K+ channel cloned from mammalian brain. Proc Natl Acad Sci U S A. 1991;88:4386–4390. doi: 10.1073/pnas.88.10.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Ramakers GM, Storm JF. A postsynaptic transient K(+) current modulated by arachidonic acid regulates synaptic integration and threshold for LTP induction in hippocampal pyramidal cells. Proc Natl Acad Sci U S A. 2002;99:10144–10149. doi: 10.1073/pnas.152620399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettig J, Heinemann SH, Wunder F, Lorra C, Parcej DN, Dolly JO, Pongs O. Inactivation properties of voltage-gated K+ channels altered by presence of beta-subunit. Nature. 1994;369:289–294. doi: 10.1038/369289a0. [DOI] [PubMed] [Google Scholar]
- Salkoff L, Baker K, Butler A, Covarrubias M, Pak MD, Wei A. An essential ‘set’ of K+ channels conserved in flies, mice and humans. Trends Neurosci. 1992;15:161–166. doi: 10.1016/0166-2236(92)90165-5. [DOI] [PubMed] [Google Scholar]
- Salvador-Recatala V, Gallin WJ, Abbruzzese J, Ruben PC, Spencer AN. A potassium channel (Kv4) cloned from the heart of the tunicate Ciona intestinalis and its modulation by a KChIP subunit. J Exp Biol. 2006;209:731–747. doi: 10.1242/jeb.02032. [DOI] [PubMed] [Google Scholar]
- Sergeant GP, Ohya S, Reihill JA, Perrino BA, Amberg GC, Imaizumi Y, Horowitz B, Sanders KM, Koh SD. Regulation of Kv4.3 currents by Ca2+/calmodulin-dependent protein kinase II. Am J Physiol Cell Physiol. 2005;288:C304–313. doi: 10.1152/ajpcell.00293.2004. [DOI] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D, Killpack K, Laufer A, Mazzotta J, Smith RD, Stevens LM, Stuber C, Tan LR, Ventura R, Woo A, Zakrajsek I, Zhao L, Chen F, Swimmer C, Kopczynski C, Duyk G, Winberg ML, Margolis J. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- Tsunoda S, Salkoff L. Genetic analysis of Drosophila neurons: Shal, Shaw, and Shab encode most embryonic potassium currents. Journal of Neuroscience. 1995a;15:1741–1754. doi: 10.1523/JNEUROSCI.15-03-01741.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda S, Salkoff L. The major delayed rectifier in both Drosophila neurons and muscle is encoded by Shab. Journal of Neuroscience. 1995b;15:5209–5221. doi: 10.1523/JNEUROSCI.15-07-05209.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacher H, Mohapatra DP, Trimmer JS. Localization and targeting of voltage-dependent ion channels in mammalian central neurons. Physiol Rev. 2008;88:1407–1447. doi: 10.1152/physrev.00002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Wu CF. In vivo functional role of the Drosophila hyperkinetic beta subunit in gating and inactivation of Shaker K+ channels. Biophys J. 1996;71:3167–3176. doi: 10.1016/S0006-3495(96)79510-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Maclean JN, An WF, Lanning CC, Harris-Warrick RM. KChIP1 and frequenin modify shal-evoked potassium currents in pyloric neurons in the lobster stomatogastric ganglion. J Neurophysiol. 2003;89:1902–1909. doi: 10.1152/jn.00837.2002. [DOI] [PubMed] [Google Scholar]