INTRODUCTION
Histidine kinase receptors are elements of the two-component signal transduction systems commonly found in bacteria and lower eukaryotes, where they are crucial for environmental adaptation through the coupling of extracellular changes to intracellular responses1,2. The typical two-component system consists of a membrane-spanning histidine kinase sensor and a cytoplasmic response regulator. In the classic system, extracellular signals such as small molecule ligands and ions are detected by the periplasmic sensor domain of the histidine kinase receptor which modulates the catalytic activity of the cytoplasmic histidine kinase domain and promotes ATP-dependent autophosphorylation of a conserved histidine residue. The phosphate is subsequently transferred to a conserved aspartate of the cognate response regulator through a phosphotransfer mechanism, and the activity of the response regulator is modulated in turn3,4. The response regulator often controls gene transcription or flagellar rotation, and thus allows cellular adaptation to an environmental signal.
The nature of histidine kinase sensor proteins is such that the catalytic domain and other cytoplasmic elements are conserved whereas the sensor domains are modular and highly variable in sequence. Thousands of histidine kinases have been identified through genome sequencing projects, but many of these remain biochemically uncharacterized. As part of a structural genomics project aimed at understanding the structural relationships between all known sensor domains, we have solved the structure of a previously uncharacterized sensor domain from a histidine kinase protein encoded by the Geobacter sulfurreducens GSU2755 gene, originally identified and annotated through genome sequencing of the organism5. We use the name HK29s to for this putative sensor domain. The structure reveals a previously uncharacterized histidine kinase sensor domain fold.
MATERIALS AND METHODS
Cloning
Using appropriately designed 5’ and 3’ primers, we amplified segment of G. sulfurreducens genomic DNA (ATCC Bioproducts) that corresponds to residues 48 to 286 of the GSU2755 gene product, indentified here as HK29s. The amplified DNA was then ligated into the ampicilin-selectable pGEX-4T-2 expression vector (Amersham Pharmacia Biotech) between the BamH1 and Xho1 restriction sites. This vector construct allowed for isopropyl-β-D-thiogalactopyranoside (IPTG)-inducible cytoplasmic expression of HK29s as an N-terminal glutathione S-transferase (GST)-fusion containing an internal thrombin cleavage site.
Expression and Purification
The GST-fusion protein was expressed from Novagen E. coli BL21(DE3) cells, induced by the addition of IPTG to a final concentration of 1mM in 4L of bacterial culture in Luria-Bertani (LB) (100µg/ml ampicilin) media at 30°C for 3 hours. The culture was originally started as a 1:100 inoculation from an overnight culture grown in LB (100µg/ml ampicilin) at 37°C, and induction began when the optical density (OD) reached 0.6. The selenomethioninyl (SeMet) protein was produced in the same manner except that 4L of minimal media containing SeMet was used instead of LB, and the overnight culture was grown in minimal media containing methionine instead of SeMet. Induced cells were harvested by centrifugation and then resuspended in 40ml of phosphate buffered saline (PBS) pH 7.4 and 5mM dithiolreitol (DTT). The cells were lysed by sonication and the suspension was then cleared by centrifugation.
The GST-fusion protein was purified by affinity chromatography. Cell supernatants containing soluble GST-HK29s was passed through a hand-poured 8ml glutathione Sepharose™ 4B (Pharmacia) gravity column. After washing of the column with phosphate-buffered saline (PBS) pH 7.4, the fusion protein was eluted with 0.1M Tris-HCl pH 8.0, 150mM NaCl, 1mM EDTA, 1mM DTT, and 20mM glutathione. Appropriate fractions containing the purified fusion protein were pooled together and dialyzed for 16 hours at 4°C into 25 volumes of a buffer containing 0.1M Tris-HCl, 150mM NaCl, 1mM EDTA, and 1mM DTT. After dialysis, thrombin (Sigma) was added to a concentration of 2U/mg of protein and cleavage was allowed to proceed for 16 hours at 20°C. The cleaved protein was then further purified by gel filtration on a Superdex 75 26/60 (Pharmacia) column equilibrated with 20mM Tris-HCl, 20mM NaCl, and1mM EDTA. HK29S appeared to be monomeric by gel filtration and was homogenous when analyzed by SDS-PAGE and native-PAGE. The appropriate fractions were pooled and concentrated to 13mg/ml .
Crystallization
Initial crystallization conditions for native HK29s were determined by the hanging-drop vapor diffusion method using commercially available sparce-matrix screens, Crystal Screens I and II (Hampton Research), and Wizard I and II (Emerald Biosystems). Preliminary crystals appeared in condition #40 (10% isopropanol, 100mM 2-morpholinoethanesulfonic acid pH 6.0, and 200mM calcium acetate) of the Wizard I screen, and these were further optimized by systematically adjusting precipitant and salt concentrations, as well the inclusion of additives from Additive Screens I, II, and III (Hampton Research). Finally optimized native HK29S crystals were grown by the hanging drop vapor diffusion method against a reservoir buffer of composition 8% isopropanol, 2% PEG 3350, 150mM calcium acetate, and 100mM 2-morpholinoethanesulfonic acid (MES) pH 6.0 at 4°C. In each drop, 1µl of protein at a concentration of 13mg/ml was mixed with 1µl of crystallization buffer. SeMet HK29S was crystallized in the same method except that calcium acetate concentration was increased to 200mM. Long rod-shaped crystals typically appeared within a week and grew to full size over a period of 3 to 4 weeks. Prior to diffraction studies, crystals were frozen in liquid nitrogen after being briefly soaked in crystallization buffer supplemented with 25% ethylene glycol.
Data Collection and Structure Determination
Four-wavelength MAD measurements were made at the Se K-edge a frozen SeMet HK29S crystal at beamline X4A of NSLS (National Synchrotron Light Source) at Brookhaven National Laboratory. Diffraction to Bragg spacings of 2.9Å (260mm detector distance) were collected using 25s exposure times and 1° oscillations at each of the four wavelengths. Additional low-angle data at each wavelength were collected to 4.0Å spacings (425mm detector distance) using 15s exposure times and 3° oscillations. Data from a native crystal extending to spacings of 2.4Å (190mm detector distance) were collected at the peak wavelength using 60s exposure times and 1° oscillations. Additional low-angle data were also collected from the native crystal to 3.7Å spacings (400mm detector distance) using 20s exposure times and 3° oscillations. Data were indexed and merged using Denzo and Scalepack of the HKL program package6.
Three of the six potential selenium sites in the asymmetric unit were found using SOLVE7, and tests for the alternative enantiomorphs showed the correct space group to be P6522. Initial phases were improved by solvent flattening using DM8 of the CCP4 program package9. Phasing based on the three Se sites identified by gave a figure of merit 0.50. In order to obtain better phases, SHARP10 was used to identify the three additional Se site; after refinement, the figure of merit improved to 0.58. Phases were extended from 2.9Å to 2.4Å against the native data using RESOLVE11, and Arp/wArp 6.1.112 was used to automatically build a partial model containing 195 residues. Missing residues were manually built using O13, and the final model was refined against the native data to 2.4Å using CNS14. Data collection and refinement statistics are listed in Table I and Table II.
Table I.
HK29S Diffraction Data
| Data Set | dmin | Wavelength (Å) |
Number of Reflections |
Average Redundancy |
<I>/(δ) | Complete- ness (%) a |
Rmerge (%) b |
|---|---|---|---|---|---|---|---|
| HK29S | 2.4 | 0.9791 | 14718 | 5.1 | 14.7 | 100.0 | 4.6 |
| (99.9) | (41.9) | ||||||
| Se HK29S | 2.9 | 0.9870 | 15305 | 6.3 | 16.2 | 99.8 | 5.7 |
| (λ1) | (low remote) | (98.1) | (28.3) | ||||
| Se HK29S | 2.9 | 0.9795 | 15310 | 6.2 | 17.4 | 99.8 | 5.4 |
| (λ2) | (edge) | (98.6) | (24.9) | ||||
| Se HK29S | 2.9 | 0.9791 | 15226 | 6.0 | 16.6 | 99.4 | 5.5 |
| (λ3) | (peak) | (98.7) | (26.3) | ||||
| Se HK29S | 2.9 | 0.9715 | 15252 | 6.2 | 15.5 | 99.8 | 5.8 |
| (λ4) | (high remote) | (99.1) | (29.2) |
Values in outermost shell are given in parentheses.
Rmerge = ( Σ|Ii − < Ii>|) / Σ |Ii|, where Ii is the integrated intensity of a given reflection.
Table II.
HK29s Refinement Statistics
| Parameter | HK29S |
|---|---|
| Bragg spacings (Å) | 20 – 2.4 |
| Space group | P6522 |
| Unit cell parameters: | 122.9, 122.9, 81.7 |
| a, b, c (Å) | |
| Za | 1 |
| Solvent content (%) | 63.0 |
| R a | 0.228 |
| Rfree b | 0.285 |
| Number of Reflections | 14682 |
| Number of total atoms (non-hydrogen) |
1919 |
| Number of protein atoms | 1777 |
| Number of ligand atoms | 1 (sodium ion) |
| Number of waters | 141 |
| Average B factor (Å2) | 55.1 |
| RMS bond ideality (Å) | 0.010 |
| RMS angle ideality (°) | 1.6 |
| Rotamer Outlier (%) c | 6.1 |
| Ramachandran (favored/outlier) (%) c |
92.0/2.6 |
| PDB Code | 3H7M |
R = ( Σ ||Fo | − |Fc||) / Σ|Fo|, where Fo and Fc denote observed and calculated structure factors respectively.
Rfree was calculated using 5% of data excluded from refinement.
Molprobity analysis (http://kinemage.biochem.duke.edu).
RESULTS AND DISCUSSION
Sensor Identification
We previously performed a comparative analysis of sequences corresponding to putative histidine kinase sensor domains. These were identified from hallmark features of canonical histidine kinase receptors as being segments between two predicted transmembrane helices that precede a histidine kinase domain. We classified these protein domains into sequence families, one of which is identified as HK27 (unpublished observations). Uniquely among sensor domain families, HK27 proteins show similarity to periplasmic binding proteins.
Overall Structure and Topology
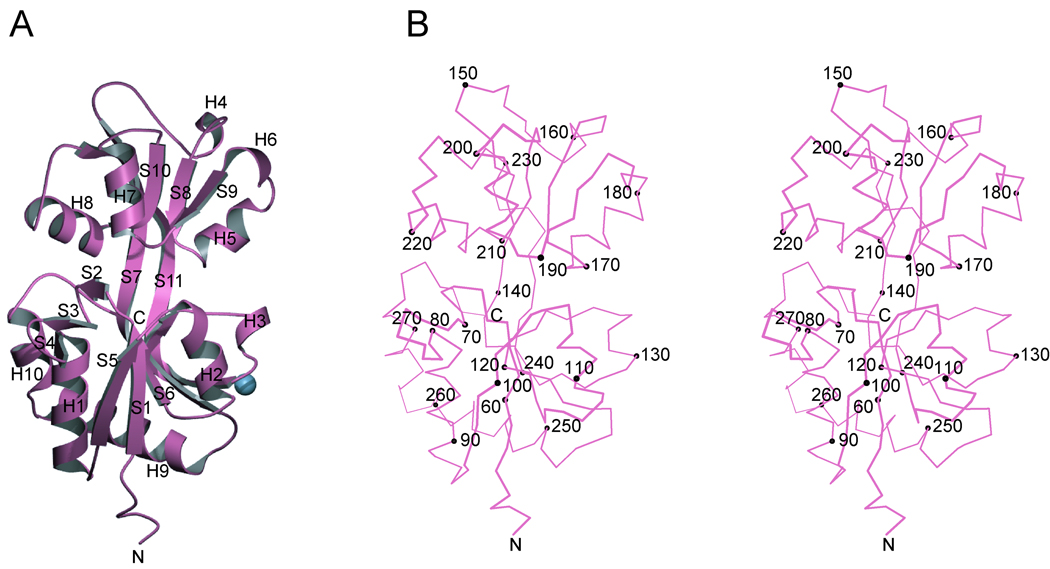
The structure of G. sulfurreducens HK29S (Figures 1A and 1B) was solved by MAD from a single crystal of SeMet HK29S (Table I). The single molecule in the asymmetric unit showed clear electron density for residues 52 to 279. A total of 228 ordered residues, 141 water molecules, and 1 sodium ion was refined against the native data to a resolution of 2.4Å with an Rwork and Rfree of 22.8% and 28.5% respectively (Table II).
Figure 1. Structure of HK29S.
(A) Ribbon diagram. Secondary structure elements are identified with strand (S) and helix (H) labels. The blue sphere shows the location of a single bound sodium ion. (B) Stereo plot of a Cα trace of HK29S. The orientation is as in A. Every tenth Cα position is depicted as a black sphere and labeled accordingly. The figures were generated using MolScript39 and Bobscript40.
The HK29S protein is bi-lobal, comprising two α/β domains with α-helices packed against both sides of a five-stranded β-pleated sheet in each. The two domains are connected by long β-strands (S7 and S11) that extend through both central sheet cores. Domain 1 (D1) begins at the N-terminus and consists of residues 52–140 and 237–270; its central sheet is formed in the order S1-S5-S6-S7-S11. An additional β-sheet, formed from strands S2-S3-S4, is located between S1 and H1, and protrudes from the face of D1. Strand S7 is highly twisted and extends from D1 into domain 2 (D2), where it runs anti-parallel with the other four strands in the D2 sheet. D2 consists of residues 141–236, and its central sheet is formed in the order S7-S8-S9-S10-S11. Interdomain strands S7 and S11 form a two-stranded anti-parallel sheet between the two domains, and strand S11 runs anti-parallel with the other four strands of the sheet in D1. In the orientation of the molecule shown in Figure 1, the C-terminus is located behind the central sheet of D1 whereas the N-terminus is at the front. A feature of electron density within D1 was modeled as a sodium ion coordinated by two water molecules and by residues in S7, H3, and the loop between S11 and H9.
Comparisons between HK29S and Other Proteins
The fold of HK29s is similar to that observed in many periplasmic binding proteins15 which have been described as resembling a “Venus flytrap” and being adapted to bind various substrates16. In addition to being found in bacterial periplasmic binding proteins and receptors, this fold is also found in eukaryotes, notably as the ligand binding domain of ionotropic glutamate receptors17. The three highest scoring results from a DALI18 search for structural relatives of HK29S are the lysine/arginine/ornithine-binding protein LAO19 (Z score = 28.2, RMSD = 1.9Å), the cysteine transporter CjaA20 (Z score = 25.2, RMSD = 2.2Å), and the ligand binding domain of the bacterial GluR0 ion channel21 (Z score = 23.7, RMSD = 2.1Å). LAO, CjaA, and GluR0 all fold into bi-lobal structures, and similarities with HK29S are apparent in a structure-based alignment (Figure 2).
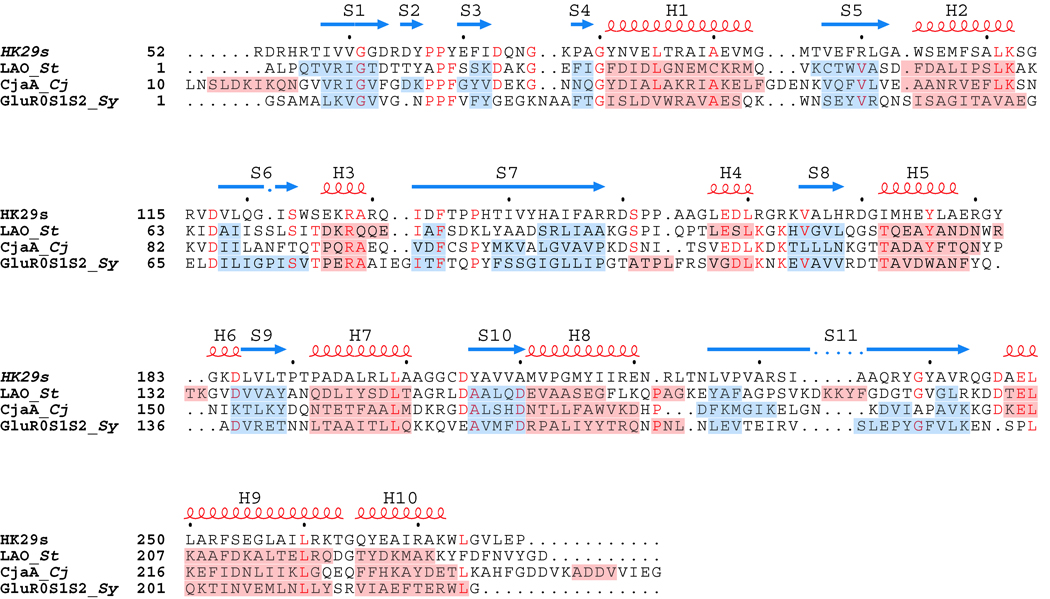
Figure 2. Structure-based Sequence Alignment.
The sequences of HK29s and structurally similar proteins LAO, CjaA, and GluR0 are aligned based on structural elements. The secondary structure assignments are labeled and displayed above the sequences as red loops (helices) and blue arrows (strands). The secondary assignments for the other structurally similar proteins are shown as red (helices) and blue (strands) highlights across the sequences themselves. Structures are considered aligned for segments that at least three consecutive Cα positions within 3.0Å of each other. Organism names are abbreviated in italics (St for Salmonella typhimurium, Cj for Campylobacter jejuni, and Sy for Synechocystis PCC 6803 ).
Pairwise sequence identities between HK29S and LAO, CjaA, and GluR0 range from 18.0% to 21.8%. We define structure-based alignments for segments having at least three consecutive Cα atoms within 3.0Å of each other. A structural alignment of HK29s D1 with the corresponding domains of LAO, CjaA, and GluR0 yields RMSD values ranging from 0.94Å to 1.49Å over a range of 112 to 123 Cα positions, and the corresponding structural alignment with D2 of HK29S yields RMSD values ranging from 1.45Å to 1.55Å over a range of 79 to 83 Cα positions. HK29S shows slightly greater structural similarity to the other three proteins in D1 than in D2.
The main differences between HK29S and LAO, CjaA, and GluR0 lie in the regions that link the two domains. In HK29S, residues 132 to 148 and residues 226 to 243 form continuous anti-parallel β-strands S7 and S11, respectively, according to DSSP22 assignments. In the cases of LAO and GluR0, the segments corresponding to S7 and S11 are broken apart into non-continuous sheets, and in CjaA this region contains three separate β-strand doublets (Figure 2). The structure of LAO also contains a four-residue helical insertion in the region corresponding to S11 of HK29S. There are also additional differences such as the insertion of short three or four residue helices in D2 of LAO and GluR0 relative to HK29S, but other structural elements are quite well conserved. The feature of continuous inter-subunit β-strands appears to be unique to HK29S as in both Type I and Type II periplasmic binding proteins, the two domains are joined by flexible linker regions15,23, or by a rigid α-helix in the case of BtuF24.
Differences between HK29s and Other Sensor Domains
HK29s is structurally very different from any of the previously solved sensor domain structures such as the all-helical NarX sensor domain25, or the PDC-type sensors such as PhoQ26,27, DcuS28,29, CitA 30,31, LuxQ32,33 and DctB28,34. The structure of HK29S is the first representative of the periplasmic-binding-protein fold in bacterial histidine kinase sensor domains, revealing a new structural class of histidine kinase sensor domain.
In comparison to other periplasmic sensor domain structures, HK29S is unique in the absence of a long N-terminal helix that is observed in all other cases. The C-terminus of HK29S is also non-helical and it is located quite far from the N-terminus. The relative location and structure of the N- and C-terminal ends of HK29s do not follow the paradigm observed in other sensor domain structures, where both the N- and C-termini form helices located near one another and poised for connection into the putative membrane-spanning helices. The structure of HK29S yields little clue about the means by which the sensor domain connects into the membrane-spanning segments in the native full-length molecule. The nature of the dimer interface and the orientation of the sensor domain with respect to the transmembrane helices in the context of a native homodimeric receptor also remain unclear.
Potential Ligand Binding Site in a Closed Apo State
Most histidine kinase sensors bind to small molecule ligands to initiate signal transduction. The putative ligand for HK29S is unknown, but it seems likely that the ligand-binding site would be at a position between domains as in the periplasmic binding proteins. The inter-domain cleft of HK29S does not contain a ligand; but there is a hydrogen-bonded web of ordered water molecules coordinated by residues that line a pocket between the domains. HK29S residues involved in this water coordination are Tyr66, Glu70, Asn82, Gly121, Ser123, Arg128, His168, and Tyr238, which lie within the secondary structure elements S2, S3, H1, S6, S8, and S11. Analogous residues involved in ligand binding for LAO and CjaA include a subset of these positions. In particular, residues 66, 121, 123, and 168 of HK29s are correspondent, and Tyr66 and Ser123 are conserved between HK29S and LAO, and Arg128 is conserved among HK29S, LAO and CjaA. Thus, we suggest that the water-filled pocket in the inter-domain cleft of HK29s is the putative ligand binding site for this sensor.
Typically in periplasmic binding proteins, ligand binding is accompanied by a hinging motion in the relatively flexible linker between the two domains whereby ligand binding stabilizes a closed state in which the two domains are more proximate than in the more open unliganded state15. In LAO, the lysine-liganded conformation differs from the unliganded conformation by a 52° closure about an axis through the hinge region and represents a large conformational change in the relative orientation of the two domains19. The two domains in our structure of HK29S are in an orientation more similar to that of LAO in the lysine complex than in the unliganded apo state, but slightly opened toward the apo LAO state. With D1 domains superimposed, a 10.1° rotation is required to further close D2 from HK29S into superposition with D2 of the liganded LAO structure. The axis of rotation passes through the end of S10 in D2 and runs at an angle in proximity to the inter-domain cleft. The 10.1° differences in domain closure between HK29s and ligand-bound LAO is comparable to the ~8° difference in domain closure observed between the different closed states of GluR2 when full and partial agonists bind35. It appears that the conformation observed in our crystal of HK29S may represent a closed apo state. Ligand access to the putative binding site of HK29S would be expected to require movement to a more open state, but the β-structured S7-S11 linker might preclude full opening as in LAO. Ligand binding might be expected to elicit a slightly more closed state than observed here for HK29S.
Closed apo states have been observed previously for the periplasmic glucose/galactose receptor36 and for the GluR0 ligand binding domain21. In the absence of ligand, such molecules are thought to exist in both the open and closed states which are in facile equilibrium37,38, whereby crystallization conditions might preferentially selected for one state over the other. In both GluR0 and the glucose/galactose receptor, inter-domain conformations are similar in the closed apo and the closed ligand-bound state, and, in each case, a hydrogen-bonded network of water molecules is found to mimic the bound ligand and satisfy the hydrogen bonding capacity of residues that line the binding site. If the structure of HK29S indeed represents a closed apo state, then it is likely that the hydrogen-bonding network of the ordered waters found in its inter-domain cleft also mimics the hydrogen bonding of the unknown putative ligand. Therefore at least a subset of the residues involved in the coordination of such waters may be involved with ligand binding.
Conclusion
The HK29S histidine kinase sensor domain is structurally very similar to periplasmic binding proteins, representing the first example of such a fold in sensor domains. It appears from structural comparisons that this HK29S may have adopted a closed apo conformation. A water-filled pocket at a site homologous with the ligand-binding sites in homologs serves to identify candidate residues for ligand binding by HK29 receptors. From the structured linker in this protein, inter-domain flexion may be limited as compared to that in typical periplasmic binding proteins. It is unclear how ligand binding by this sensor would be relayed into transmembrane signals for kinase activation.
AKNOWLEDGMENTS
We thank Randy Abramowitz and John Schwanof for help with synchrotron data collection. This work was supported in part by NIH grant GM34102. Beamline X4A of the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory, a DOE facility, is supported by the New York Structural Biology Center.
Footnotes
ACCESSION NUMBERS
Atomic coordinates and structure factors (code 3H7M) have been deposited in the Protein Data Bank (http://www/rcsb/pdb).
References
- 1.West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 2.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 3.Dutta R, Qin L, Inouye M. Histidine kinases: diversity of domain organization. Mol. Microbio. 1999;34:633–640. doi: 10.1046/j.1365-2958.1999.01646.x. [DOI] [PubMed] [Google Scholar]
- 4.Robinson VL, Buckler DR, Stock AM. A tale of two components: a novel kinase and a regulatory switch. Nat. Struct. Biol. 2000;7:626–633. doi: 10.1038/77915. [DOI] [PubMed] [Google Scholar]
- 5.Methe BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, Heidelberg JF, Wu D, Wu M, Ward N, Beanan MJ, Dodson RJ, Madupu R, Brinkac LM, Daugherty SC, DeBoy RT, Durkin AS, Gwinn M, Kolonay JF, Sullivan SA, Haft DH, Selengut J, Davidsen TM, Zafar N, White O, Tran B, Romero C, Forberger HA, Weidman J, Khouri H, Feldblyum TV, Utterback TR, Van Aken SE, Lovley DR, Fraser CM. Genome of Geobacter sulfurreducens: Metal reduction in subsurface environments. Science. 2003;302:1967–1969. doi: 10.1126/science.1088727. [DOI] [PubMed] [Google Scholar]
- 6.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 7.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D. Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowtan K. CCP4/ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 9.Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallogr. D. Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 10.de la Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 11.Terwilliger TC. Reciprocal-space solvent flattening. Acta Crystallogr. D. Biol. Crystallogr. 1999;55:1863–1871. doi: 10.1107/S0907444999010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 13.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved Methods for Building Protein Models in Electron-Density Maps and the Location of Errors in These Models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 14.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 15.Quiocho FA, Ledvina PS. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: Variation of common themes. Mol. Microbiol. 1996;20:17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- 16.Felder CB, Graul RC, Lee AY, Merkle HP, Sadee W. The venus flytrap of periplasmic binding proteins: An ancient protein module present in multiple drug receptors. Aaps Pharmsci. 1999;1:E2. doi: 10.1208/ps010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- 18.Holm L, Sander C. Protein-Structure Comparison by Alignment of Distance Matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 19.Oh BH, Pandit J, Kang CH, Nikaido K, Gokcen S, Ames GFL, Kim SH. 3-Dimensional Structures of the Periplasmic Lysine Arginine Ornithine-Binding Protein with and without a Ligand. J. Biol. Chem. 1993;268:17648–17649. [PubMed] [Google Scholar]
- 20.Muller A, Thomas GH, Horler R, Brannigan JA, Blagova E, Levdikov VM, Fogg MJ, Wilson KS, Wilkinson AJ. An ATP-binding cassette-type cysteine transporter in Campylobacter jejuni inferred from the structure of an extracytoplasmic solute receptor protein. Mol. Microbiol. 2005;57:143–155. doi: 10.1111/j.1365-2958.2005.04691.x. [DOI] [PubMed] [Google Scholar]
- 21.Mayer ML, Olson R, Gouaux E. Mechanisms for ligand binding to GluR0 ion channels: Crystal structures of the glutamate and serine complexes and a closed Apo state. J. Mol. Biol. 2001;311:815–836. doi: 10.1006/jmbi.2001.4884. [DOI] [PubMed] [Google Scholar]
- 22.Kabsch W, Sander C. Dictionary of Protein Secondary Structure - Pattern-Recognition of Hydrogen-Bonded and Geometrical Features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 23.Fukami-Kobayashi K, Tateno Y, Nishikawa K. Domain dislocation: a change of core structure in periplasmic binding proteins in their evolutionary history. J. Mol. Biol. 1999;286:279–290. doi: 10.1006/jmbi.1998.2454. [DOI] [PubMed] [Google Scholar]
- 24.Karpowich NK, Huang HH, Smith PC, Hunt JF. Crystal structures of the BtuF periplasmic-binding protein for vitamin B12 suggest a functionally important reduction in protein mobility upon ligand binding. J. Biol. Chem. 2003;278:8429–8434. doi: 10.1074/jbc.M212239200. [DOI] [PubMed] [Google Scholar]
- 25.Cheung J, Hendrickson WA. Structural Analysis of Ligand Stimulation of the Histidine Kinase NarX. Structure. 2009;17:190–201. doi: 10.1016/j.str.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung J, Bingman CA, Reyngold M, Hendrickson WA, Waldburger CD. Crystal structure of a functional dimer of the PhoQ sensor domainq. J. Biol. Chem. 2008;283:13762–13770. doi: 10.1074/jbc.M710592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho US, Bader MW, Amaya MF, Daley ME, Klevit RE, Miller SI, Xu WQ. Metal bridges between the PhoQ sensor domain and the membrane regulate transmembrane signaling. J. Mol. Biol. 2006;356:1193–1206. doi: 10.1016/j.jmb.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 28.Cheung J, Hendrickson WA. Crystal Structures of C-4-Dicarboxylate Ligand Complexes with Sensor Domains of Histidine Kinases DcuS and DctB. J. Biol. Chem. 2008;283:30256–30265. doi: 10.1074/jbc.M805253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pappalardo L, Janausch IG, Vijayan V, Zientz E, Junker J, Peti W, Zweckstetter M, Unden G, Griesinger C. The NMR structure of the sensory domain of the membranous two-component fumarate sensor (histidine protein kinase) DcuS of Escherichia coli. J. Biol. Chem. 2003;278:39185–39188. doi: 10.1074/jbc.C300344200. [DOI] [PubMed] [Google Scholar]
- 30.Reinelt S, Hofmann E, Gerharz T, Bott M, Madden DR. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J. Biol. Chem. 2003;278:39189–39196. doi: 10.1074/jbc.M305864200. [DOI] [PubMed] [Google Scholar]
- 31.Sevvana M, Vijayan V, Zweckstetter M, Reinelt S, Madden DR, Herbst-Irmer R, Sheldrick GM, Bott M, Griesinger C, Becker S. A ligand-induced switch in the periplasmic domain of sensor histidine kinase CitA. J. Mol. Biol. 2008;377:512–523. doi: 10.1016/j.jmb.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 32.Neiditch MB, Federle MJ, Miller ST, Bassler BL, Hughson FM. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol Cell. 2005;18:507–518. doi: 10.1016/j.molcel.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 33.Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou YF, Nan BY, Nan J, Ma QJ, Panjikar S, Liang YH, Wang YP, Su XD. C-4-Dicarboxylates Sensing Mechanism Revealed by the Crystal Structures of DctB Sensor Domain. J. Mol. Biol. 2008;383:49–61. doi: 10.1016/j.jmb.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 35.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-Sensitive glutamate receptor: Crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 36.Flocco MM, Mowbray SL. The 1.9 Ångstrom X-Ray Structure of a Closed Unliganded Form of the Periplasmic Glucose/Galactose Receptor from Salmonella typhimurium. J. Biol. Chem. 1994;269:8931–8936. [PubMed] [Google Scholar]
- 37.Careaga CL, Sutherland J, Sabeti J, Falke JJ. Large-Amplitude Twisting Motions of an Interdomain Hinge - a Disulfide Trapping Study of the Galactose-Glucose Binding-Protein. Biochemistry. 1995;34:3048–3055. doi: 10.1021/bi00009a036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolf A, Shaw EW, Nikaido K, Ames GFL. The Histidine-Binding Protein Undergoes Conformational-Changes in the Absence of Ligand as Analyzed with Conformation-Specific Monoclonal-Antibodies. J. Biol. Chem. 1994;269:23051–23058. [PubMed] [Google Scholar]
- 39.Kraulis PJ. Molscript - a Program to Produce both Detailed and Schematic Plots of Protein Structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 40.Esnouf RM. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]