1. Introduction
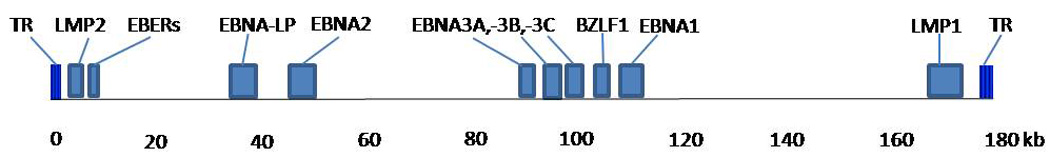
Epstein-Barr virus (EBV) is a lymphotrophic human gamma-1 herpesvirus transmitted primarily through saliva that infects over 90% of the world’s population. EBV is implicated in several benign and malignant conditions. Typical to other gamma herpesviruses, EBV establishes latent infection in B lymphocytes. The virus persists lifelong in memory B cells as episomal (circular) DNA, undergoing episodic lytic replication in B cells and in epithelial cells, essential for spread from cell to cell and from host to host. EBV has a double-stranded DNA genome comprised of approximately 170-kilobases which encode more than 85 genes (Farrell, 2005; Rickinson and Kieff, 2007; Thompson and Kurzrock, 2004). During latent infection, EBV expresses a restricted set of genes, including two EBV-encoded RNAs (EBER-1 and EBER-2), six EBV nuclear antigens (EBNA-2, EBNA-2, EBNA-3A, −3B and −3C, leader protein (EBNA-LP), and three integral membrane proteins (LMP-1, LMP-2A and −2B) (Fig. 1). During lytic infection, a large number of structural genes and regulators are expressed, including immediate early gene BZLF-1 and its promoter Zp (Delecluse et al., 2007; Farrell, 2005; IARC, 1997; Rickinson and Kieff, 2007; Thompson and Kurzrock, 2004; Thorley-Lawson and Gross, 2004).
Figure 1.
In developing countries, EBV typically occurs in early childhood and is asymptomatic; in developed countries, infection occurs in later childhood or young adulthood and can manifest as infectious mononucleosis (IM), which is self-limiting (Rickinson and Kieff, 2007). A small fraction of EBV-infected individuals develop malignancies, including Hodgkin lymphoma (HL), nasopharyngeal carcinoma (NPC), gastric adenocarcinoma, lymphoepithelioma-like carcinoma (LELC), nasal NK/T-cell lymphoma, and leiomyosarcoma (Hsu and Glaser, 2000).
Among individuals with primary immunodeficiencies (e.g, X-linked lymphoproliferative (XLP) syndrome), EBV infection frequently leads to fatal IM or malignant B cell lymphomas. Patients with acquired immunosuppression may also develop lymphoproliferative disorders. Transplant patients seronegative for EBV are at a markedly increased risk of post-transplant lymphoproliferative disease (PTLD) due to primary EBV infection (Cohen, 2003; Harrington et al., 1988; Purtilo, 1980; Purtilo, 1987). AIDS patients are at higher risk than the general population of developing both benign and malignant conditions, including oral hairy leukoplakia, immunoblastic lymphomas, Burkitt lymphomas, and Hodgkin lymphoma (Carbone, 2003; Carbone et al., 2009; Cohen, 2005).
A notable feature of EBV-associated malignancies is variation in incidence and the proportion of EBV-positive tumors in different geographic regions (Fig. 2). The incidence of NPC is higher in Southern China, Southeast Asia, and to a lesser extent the Maghrebi Arabic regions of North Africa and the Arctic, than in other regions of the world. However, the tight link between NPC and EBV is observed in all regions evaluated (Bray et al., 2008; Chang and Adami, 2006; Hsu and Glaser, 2000; IARC, 1997). Burkitt lymphoma (BL) occurs most commonly in equatorial Africa (BL in this region is recognized as the most common childhood tumor) than in other regions of the world. In contrast to NPC, the tight link between BL and EBV is only observed in Equatorial Africa, while in other regions only a fraction of BL cases are associated with EBV (Brady et al., 2008; Cardy et al., 2001; Chan et al., 1995; Gutierrez et al., 1992; Klumb et al., 2004; Mbulaiteye et al., 2009; Philip, 1985; Rochford et al., 2005). For HL, incidence is highest in Europe, in North America, and to a lesser extent in South America, and occurs less frequently in China or Africa. The fraction of HL cases that are associated with EBV have been shown to be high in Africa and S. America, but intermediate in China, N. America, and Europe (Dinand and Arya, 2006; Ferlay et al., 2004; Mueller and Grufferman, 2006; Weiss, 2000).
Figure 2.
The reasons for the restricted geographic pattern of EBV-associated malignancies are poorly understood, but variation in human host genetic factors, environmental factors, or viral factors have been suspected. Substantial genetic sequence variation in EBV has been demonstrated among EBV isolates at multiple loci on the EBV genome, but the role of genetic variations has yet to be elucidated.
Here we review the literature on EBV genetic variation in healthy and diseased individuals from different geographical regions. First, we review the two main EBV genetic types (EBV type 1 and 2) and strains based on ebnotyping and restriction fragment length polymorphisms (RFLP). Second, we examine in more depth studies focused on DNA sequence variation within the LMP-1, EBNA-1 and BZLF-1 genes of EBV. Although understanding functional differences associated with genetic variations in EBV that affect disease risk will ultimately be important to fully understand the underlying molecular mechanisms for EBV-disease associations, these are not the topic of this review.
We summarize the state of the knowledge on EBV variation, identify gaps in our understanding, and suggest areas for further study. We hope our review will highlight the benefit of closer collaboration among virologists, molecular biologists, clinicians, epidemiologists, and others to untangle the mystery of how and why EBV, a ubiquitous virus, contributes to distinct malignancies in different populations.
2. EBV type 1 and 2
EBV is classified as EBV type 1 and 2, based on sequence variation in EBNA genes. Initial classification was based on differences in EBNA-2 identified in EBV isolates from lymphoblastoid cell lines (LCLs). EBV isolated from a North American IM case, B95-8 (GenBank: V01555.2), classified as a type 1, was found to have a slightly longer EBNA-2 reading frame than that of AG876, a type 2 virus isolated from a Burkitt lymphoma case in Central Africa (Dambaugh et al., 1984). Additional differences between type 1 and 2 have been reported, including mismatched base pairs in genes encoding EBNA3A, 3B and 3C, differing in 10%, 12%, and 19% of base pairs, respectively (Sample et al., 1990).
Previous studies examining the distribution of EBV subtypes indicated predominance of EBV type 1 strains (Zimber et al., 1986). More recent data, however, suggests that a notable proportion of individuals are infected with type 2 and that co-infection of both type 1 and 2 is also possible. Co-infection is more commonly detected in immunocompromised individuals, suggesting that superinfection with type 2 is likely acquired during immunocompromised states or that immunity might differentially control the reactivation or persistence of EBV subtypes in an individual (Apolloni and Sculley, 1994; Yao et al., 1996b). The frequent detection of EBV type 2 in immunocompromised individuals is also reflected in the EBV subtypes associated with lymphomas in immunocompromised patients (Goldschmidts et al., 1992). The difference in the previous and recent results can be attributed to methodological changes in assessing EBV subtypes. Prior to the PCR-era, molecular epidemiological studies relied on EBV isolation by lymphocyte transformation, thereby introducing a selection bias towards transformation-efficient EBV subtypes (Rickinson et al., 1987). PCR has eliminated this bias through direct amplification and sequencing of EBV DNA.
While there is a greater appreciation that multiple EBV infections are common, EBV type 1 continues to be more frequently detected in Caucasian and Asian healthy individuals. Studies of Caucasian healthy populations, when combined, show that 74% of individuals tested are infected with type 1, 19% with type 2, and 7% are infected with both EBV types (Apolloni and Sculley, 1994; Correa et al., 2004; Klemenc et al., 2006; Sixbey et al., 1989; Trimeche et al., 2007). PCR studies of healthy Asian populations demonstrated an even stronger predominance of type 1: 85% of individuals tested are infected with type 1, 4% with type 2 and 11% with both types (Srivastava et al., 2000; Tiwawech et al., 2008; Zhou et al., 2001). Although the distribution of EBV type 1 and type 2 in other populations is less well understood, type 2 EBV was detected in 24% of individuals in Kenya, suggesting that this type might indeed be more prevalent in Africa (Young et al., 1987).
Although biological differences attributable to EBV types 1 and 2 provide rationale for classification of EBV into these two broad groups does, it is clear that these do not fully account for the natural diversity of EBV. First, intertypic recombinants, in which a type 1 EBNA-2 allele is paired with type 2 EBNA3A, −3B, and/or −3C alleles, have been found in both immunocompetent and immunocompromised populations in different geographic regions (Aguirre and Robertson, 1999; Burrows et al., 1996; Gorzer et al., 2006; Kim et al., 2006; Midgley et al., 2000; Yao et al., 1996a). Additionally, studies have shown that variations observed in the EBNA-2 and EBNA3A–C genes often do not correlate with variations observed in other regions of the EBV genome, including LMP-1 and EBNA-1 (Fassone et al., 2002; Gutierrez et al., 2000; Klemenc et al., 2006; Srivastava et al., 2000). Thus, a higher resolution of EBV strain classification based on sequence variation of multiple gene regions is likely to be informative.
3. RFLPs and Ebnotyping
Both restriction fragment length polymorphisms (RFLPs) and ebnotyping have been previously discussed in a review by Gratama and Ernberg (1995). RFLPs define variation due to loss or gain of restriction sites which are identified by comparing the size of EBV DNA fragment size of DNA digested by restriction enzymes. Ebnotyping involves assessing the variation in the molecular weight of EBNA-1, −2, −3a, −3b, and −3c proteins, related to the number of EBNA gene internal repeats or hydrophobicity/hydrophilicity, which may differ between strains (Gratama and Ernberg, 1995).
For example, one of the most extensively studied RFLP polymorphisms is the “f” variant that contains an extra BamHI site in the BamHI F region when compared with the F prototype (B95-8). Studies have reported a higher frequency of the “f” variant in NPC cases from Asia (142/209, 68%) compared with controls from this same region (14/49, 29%) (Abdel-Hamid et al., 1992; Lung and Chang, 1992; Lung et al., 1994; Lung et al., 1990; Lung et al., 1991; Tan et al., 2003; Wu et al., 1996). In contrast, the “f” variant is rarely observed in NPC (2.5% of 202 cases), NHL (0% of 31 cases), and healthy or nonmalignant controls (1.3% of 151 controls) from Europe, N. America, and Africa (Ayadi et al., 2007; Bouzid et al., 1994; Bouzid et al., 1998; Henry et al., 2001; Higa et al., 2002; Khanim et al., 1996; Klemenc et al., 2006; Tamura et al., 1993). These findings suggest that the “f” variant is more common in Asia than other regions of the world. Results also suggest an association between the “f” variant and NPC risk, but the small number of individuals studied precludes any definitive conclusions. Another notable RFLP polymorphism, the XhoI loss, will be discussed in the next section due to its location within the LMP-1 gene.
Classification of EBV variants based on RFLP and ebnotyping have several limitations. First, RFLP-based tests would miss variation occurring outside of the restriction sites as well as miss single nucleotide substitutions that would not change fragment size. Similarly, ebnotyping-based tests are restricted to EBNA proteins as well as would miss changes that do not affect the overall size or polarity of the fragment being tested. Finally, ebnotyping is problematic because of its reliance on LCLs: there may be bias towards variants better able to transform LCLs and the in-vitro propagation of LCLs may introduce mutations that could be erroneously classified as real variants (Jenkins and Farrell, 1996; Munch, 1998; Walling et al., 1999).
4. DNA sequencing
DNA sequencing detects single base pair changes that permit the identification of a wider range of diversity. This review summarizes EBV genetic diversity identified in three gene regions: LMP-1, EBNA-1, and BZLF-1. These three regions are the focus of this review because they are the most frequently studied regions to date. In particular, LMP-1 has been studied extensively because it is thought to play a significant role in tumorigenesis; EBNA-1 has been the focus of previous evaluations because it is required for maintenance of EBV in its latent form and because it is expressed in all EBV-associated tumors; and BZLF-1 has been targeted for evaluation in the past because of the important role it plays in the switch from latent to lytic EBV infection.
4.1. LMP-1
LMP-1 consists of a 25 amino acid (AA) cytoplasmic amino-terminus (N-terminus), six predicted transmembrane domains (AA: 26–196), and a cytoplasmic carboxy-terminus (C-terminus) (AA: 197–386) (Fig. 1 & Fig. 3) (Baer et al., 1984; Fennewald et al., 1984). LMP-1 is an integral membrane protein that is expressed in NPC, HL, and nasal NK/T-cell lymphoma, but surprisingly absent in BL (Tao et al., 2006). The C-terminus interacts with cellular proteins through the C-Terminal Activation Region 1 (CTAR1) and CTAR2 and activates several signaling pathways, including the transcriptional nuclear factor-kB (NF-kB) whose activation is linked to the inhibition of apoptosis (Hatzivassiliou and Mosialos, 2002; Izumi and Kieff, 1997)
Figure 3.
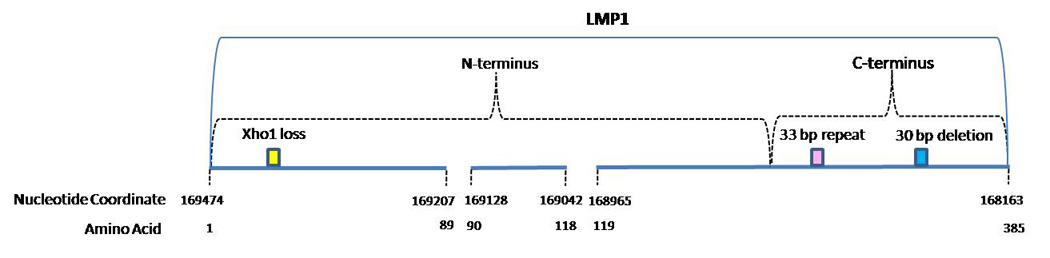
The two commonly reported LMP-1 gene polymorphisms include a 30 bp deletion in the C-terminus and the loss of restriction site XhoI in the N-terminus of the gene (Edwards et al., 1999; Nagamine et al., 2007; Saechan et al., 2006; Sung et al., 1998). The 30 bp deletion (168256–168287) causing the loss of 10 AA (343 to 352) was detected in EBV isolated from cell lines derived from NPC patients from Southern China (Miller et al., 1994). Hu et al. (1991) was the first to describe the loss of the XhoI restriction site from exon 1 of the LMP-1 gene in the CAO cell line derived from a Chinese NPC case (Hu et al., 1991). This change is caused by a single base substitution from G to T at the nucleotide position 169425. The 30 bp deletion and XhoI loss do not discriminate among variants defined by signature AA changes in the C-terminus of LMP-1 that are discussed in the next paragraph (Edwards et al., 1999).
Studies that have analyzed DNA-based variability in the C-terminus of LMP-1 have largely been evaluated based on AA changes rather than nucleotide changes. Therefore, we summarize findings from these studies at the AA level as well. At least two systems have been used to classify LMP-1 C-terminus variants (Edwards et al., 1999; Walling et al., 1999). The first categorized variants into classes defined based on signature AA changes relative to the prototypic LMP-1 (B95-8) in the C-terminus (AA 189–377), and named these classes based on the geographic region from which the initial isolate was derived: Alaskan, China 1, China 2, China 3, Mediterranean + (Med +), Med -, and North Carolina (NC) (Table 1) (Edwards et al., 1999; Mainou and Raab-Traub, 2006; Miller et al., 1994; Sung et al., 1998). Using the Edwards classification system, phylogenetic tree analysis suggested that the China 1, Med and China 3 groups were closely related. The NC and Alaskan groups formed a second cluster with which the China 2 group was also related (Edwards et al., 1999). Subsequent to the initial report using this classification system, other studies (largely from Southeast Asian) have identified additional variants closely related to one or more of the 7 major LMP-1 C-terminus variant classes (Nguyen-Van et al., 2008; Saechan et al., 2006).
Table 1.
LMP-1 Carboxy Terminus Changes
| Nucleotide Position | Nucleotide Change | Codon | AA Change | China 1 | Med+ | Med | China 3 | NC | Alaskan | China 2 |
|---|---|---|---|---|---|---|---|---|---|---|
| 168694 | T to C | 209 | no change | x | x | x | x | |||
| 168687 | G to A | 212 | Gly to Ser | x | x | x | x | |||
| 168640 | C to T | 227 | no change | x | ||||||
| 168635 | G to C | 229 | Ser to Thr | x | x | |||||
| 168631 | A to G | 230 | no change | x | x | |||||
| 168626 | G to C | 232 | Gly to Ala | x | ||||||
| 168587 | C to A | 245 | Pro to His | x | ||||||
| 33 bp repeat | 250–298 | |||||||||
| 168566 | G to A | 252 | Gly to Asp | x | ||||||
| 168404 | T to A | 306 | Leu to Gln | x | ||||||
| 168395 | G to A | 309 | Ser to Asn | x | x | x | x | x | x | |
| 168387 | G to A | 312 | Asp to Asn | x | ||||||
| 168384 | T to G | 313 | Ser to Ala | x | ||||||
| 168384 | T to C | 313 | Ser to Pro | x | ||||||
| 168361 | T to G | 320 | no change | x | ||||||
| 168357 | C to A | 322 | Gln to Asn/Lys | x | x | x | ||||
| 168357 | C to G | 322 | Gln to Lys | x | x | |||||
| 168356 | A to C | 322 | Gln to Thr | x | ||||||
| 168355 | A to T | 322 | Gln to Lys | x | ||||||
| 168330 | G to C | 331 | Gly to Gln | x | x | |||||
| 168329 | G to A | 331 | Gly to Gln | x | x | |||||
| 168329 | G to C | 331 | Gly to Ala | x | ||||||
| 168320 | A to G | 334 | Gln to Arg | x | x | x | ||||
| 168309 | T to C | 338 | Leu to Pro | x | x | x | ||||
| 168308 | T to C | 338 | Leu to Ser | x | x | x | x | x | x | x |
| 168295 | A to T | 342 | no change | x | x | x | x | x | x | x |
| 168290 | G to A | 344 | Gly to Asp | x | ||||||
| 168288 | G to A | 345 | Gly to Ser | x | ||||||
| 168287-168256 | 30 bp del | 343–352 | x | x | x | |||||
| 168267 | C to A | 352 | His to Asn | x | ||||||
| 168266 | A to G | 352 | His to Arg | x | x | |||||
| 168260 | G to A | 354 | Gly to Asp | x | ||||||
| 168257 | G to T | 355 | Gly to Val | x | x | |||||
| 168248 | A to C | 358 | His to Pro | x | ||||||
| 168238 | G to A/C | 361 | no change | x | x | |||||
| 168225 | T to A/G | 366 | Ser to Thr | x | x | x | x | x |
The second main classification system used to date characterized variants using a smaller number of AA changes in shorter segments on either side of the 33 bp repeat, while the LMP-1 variants described above were defined by changes across the entire span of the C-terminus region of LMP-1 (including the 33 bp repeat region - AA 250–298). Walling et al. (1999; 2003) identified 25 LMP-1 C-terminus variants including 14 variants with AA changes between AA 306 and 366 (Patterns 1, 2a, 2b, 2c, 3a, 3c, 4, 5, 6, 7, 8, 9) downstream of the 33 bp repeat region and 11 variants (Patterns A, B, C, D, E, F1, F2, F3, G, H) with changes between AA 209 and AA 245 or upstream of the 33 bp repeat region (Walling et al., 2003; Walling et al., 1999). The differences between the classification systems used by Edwards and Walling make comparisons across studies difficult at best and highlight the need for a consensus nomenclature so that future studies can be more directly compared.
The distributions of the 30 bp deletion, XhoI loss, and C-terminus sequence variants of LMP-1 based on the current literature are pooled in Table 2 and Table 3. These results must be interpreted cautiously due to the nonrandom sampling of individuals and the small number of individuals included in studies. It should be noted that studies published before 1995 were described in other reviews and were therefore not included here (Gratama et al., 1990; Jenkins and Farrell, 1996; Munch, 1998). Also, the geographic regions and health conditions included were those most commonly reported across studies. Finally, the varied types of specimens (e.g. blood, saliva, tissue) tested across studies were not considered when pooling results across studies.
Table 2.
| Table 2a. Prevalence (%) of 30 Base Pair Deletion in the LMP-1 gene by Clinical Condition and Geographic Region (total cases) | ||||||
|---|---|---|---|---|---|---|
| Clinical condition | China | Other Asian | Africa | South America | North America | Europe |
| NPC | 83.8% (857) | 55.1% (301) | 11.1% (18) | 27.8% (18) | ||
| BL | 38.1% (21) | 80.0% (15) | ||||
| HL | 83.1% (65) | 60.2% (88) | 48.1% (27) | 33.9% (277) | ||
| AIDS lymphoma | 47.4% (19) | 53.9% (167) | ||||
| Nonmalignant/healthy | 75.1% (189) | 34.0% (47) | 25.6% (39) | 17.1% (175) | 47.9% (48) | 33.6% (295) |
| Table 2b. Prevalence (%) of XhoI Loss in the LMP-1 Gene by Clinical Condition and Geographic Region (total cases) | ||||||
|---|---|---|---|---|---|---|
| Clinical condition | China | Other Asian | Africa | South America | North America | Europe |
| NPC | 94.8% (269) | 74.4% (117) | 0% (9) | |||
| BL | 0% (11) | |||||
| HL | 89.6% (67) | 20.0% (15) | 6.9% (29) | |||
| AIDS lymphoma | ||||||
| Nonmalignant/healthy | 79.6% (162) | 0% (22) | 5.1% (39) | 46.2% (68) | ||
(Berger et al., 1999; Chang et al., 2006; Chang et al., 1995; Chen et al., 1996b; Cheung et al., 1998; Cheung et al., 1996; Chiang et al., 1999; Correa et al., 2004; D’Addario and Chauvin, 2000; Dolcetti et al., 1997; Fassone et al., 2002; Guiretti et al., 2007; Gutierrez et al., 2000; Hayashi et al., 1997; Itakura et al., 1996; Jeon et al., 2004; Khanim et al., 1996; Lin et al., 2004; Lindsey et al., 2008; Nguyen-Van et al., 2008; Plaza et al., 2003; Sandvej et al., 1997; Santon and Bellas, 2001; Santon et al., 1995; See et al., 2008; Sung et al., 1998; Tan et al., 2003; Tang et al., 2008; Tao et al., 1998; Tiwawech et al., 2008; Trimeche et al., 2007; Vallat-Decouvelaere et al., 2002; Zhang et al., 2002; Zhou et al., 2001)
Table 3.
Prevalence (%) of Variants Based on the Carboxy Terminus of LMP-1 by Clinical Condition and Geographic Region
| Clinical Condition | LMP-1 Genotype | Asia | Africa | North America | Europe |
|---|---|---|---|---|---|
| NPC | China 1 | 82.5% | 66.0% | 0.0% | |
| China 2 | 9.7% | 0.0% | 0.0% | ||
| China 3 | 9.0% | 0.0% | 0.0% | ||
| Med | 0.0% | 0.0% | 89.0% | ||
| Alaskan | 0.0% | 33.0% | 0.0% | ||
| NC | 0.0% | 0.0% | 0.0% | ||
| B95-8 | 5.1% | 0.0% | 11.0% | ||
| Other | 1.8%a | 0.0% | 0.0% | ||
| total cases | 217 | 0 | 3 | 9 | |
| BL | China 1 | 0.0% | 0.0% | ||
| China 2 | 0.0% | 0.0% | |||
| China 3 | 0.0% | 0.0% | |||
| Med | 100.0% | 100.0% | |||
| Alaskan | 0.0% | 0.0% | |||
| NC | 0.0% | 0.0% | |||
| B95-8 | 0.0% | 0.0% | |||
| total cases | 0 | 1 | 1 | 0 | |
| HL | China 1 | 100.0% | 56.0% | ||
| China 2 | 0.0% | 0.0% | |||
| China 3 | 0.0% | 0.0% | |||
| Med | 0.0% | 12.0% | |||
| Alaskan | 0.0% | 0.0% | |||
| NC | 0.0% | 3.0% | |||
| B95-8 | 0.0% | 29.0% | |||
| total cases | 1 | 0 | 0 | 34 | |
| Nonmalignant/healthy | China 1 | 70.0% | 33.0% | 27.0% | |
| China 2 | 0.0% | 0.0% | 0.0% | ||
| China 3 | 0.0% | 0.0% | 0.0% | ||
| Med | 13.3% | 33.0% | 12.0% | ||
| Alaskan | 0.0% | 0.0% | 0.0% | ||
| NC | 0.0% | 0.0% | 18.0% | ||
| B95-8 | 10.0% | 33.0% | 42.0% | ||
| Other | 6.7%a | 0.0% | 0.0% | ||
| total cases | 60 | 0 | 3 | 33 | |
The distribution of the 30 bp deletion by geography and disease is summarized in Table 2a. The prevalence of the 30 bp deletion among controls appears relatively higher in samples from China than in samples from other areas. Within geographic regions, the 30 bp deletion was relatively more frequently detected among cancer cases than among controls. However, the interpretation of these findings in the literature is limited, given the heterogeneity in study designs and the sparsity of data for many of the geographic and disease subgroups evaluated.
The distribution of XhoI loss by geography and disease is summarized in Table 2b. Although data from controls outside of Asia are limited, available data suggest that XhoI loss is more common in China than in other regions of the world. Within China and other Asian countries, XhoI loss was more frequently observed among cancer cases than controls. Additional studies are needed to fully evaluate geographic and disease patterns of XhoI loss.
The distribution of EBV variant classes defined based on AA differences in the C-terminus of LMP-1 across disease conditions and geographic regions is summarized in Table 3. Data from regions other than Asia are sparse. Within Asia, available data suggest that China1 is the most prevalent class. Additionally, the China1 variant group was slightly more frequently detected among NPC cases than controls in this region.
4.2. EBNA-1
EBNA-1 has 641 AA including a glycine-glycine-alanine (gly-gly-ala) repeat sequence thought to play a role in downregulating EBV mediated immune response by inhibiting antigen presentation (Fig. 1 & Fig. 4) (Levitskaya et al., 1995). EBNA-1 is the only latent protein that is consistently expressed in all EBV-positive malignancies including BL which express only EBNA-1 (Rowe et al., 1992). EBNA-1 is critical in maintaining EBV in the infected cells and facilitating episomal replication (Yates et al., 1984; Yates et al., 1996). In replicating latently infected EBV cells, EBNA-1 dictates equal partitioning of EBV genome, thus reducing the loss of EBV infected cells (Aiyar et al., 1998). EBNA-1 is also a transcriptional activator and activates expression of EBV transcripts via the latent C promoter (Sugden and Warren 1989).
Figure 4.
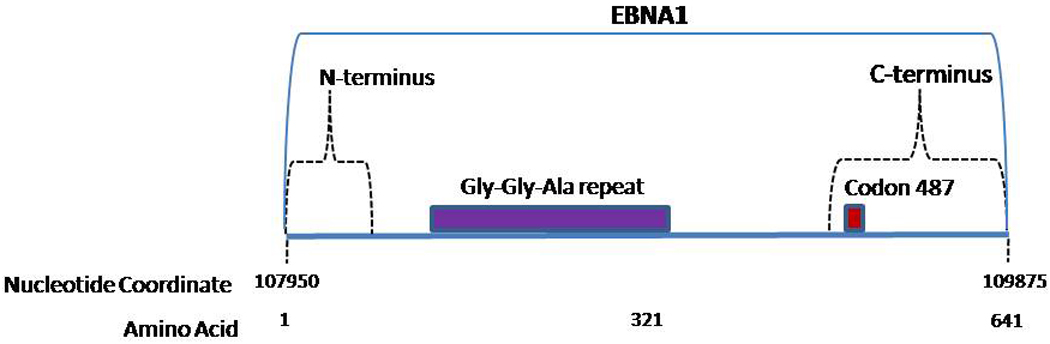
EBNA-1 variants were first identified based on the C-terminus region of EBNA-1. Two broad EBV strains based on EBNA-1 were identified. The prototype (P) strain was similar to EBNA-1 from B95-8, differing from it by only 3 AA substitutions. The second strain differed from B95.8 by 15 AA substitutions and was therefore named the variant (V) strain. Because codon 487 was found to be a hot spot for variation, P and V subtypes were further classified based on the AA present at codon 487: P-alanine (P-ala), P-threonine (P-thr), V-proline (V-pro), V-leucine (V-leu), and V-valine (V-val) (Table 4). It is of note that V-alanine (V-ala), which contains the prototype amino acid (alanine) at residue 487, exhibits differences from the prototype in other codons, including codons 499, 500, 502, 517, 520, and 524. Subvariants of P-ala, P-thr, and V-val have more recently been characterized (listed in footnote to Table 4) (Sandvej et al., 2000; Wang et al., 2003).
Table 4.
EBNA-1 Carboxy Terminus Changes
| Prototype (B95-8) | Prototype-like | Variant | |||||
|---|---|---|---|---|---|---|---|
| P-alaa | P-thrb | V-valc | V-pro | V-leu | V-ala | ||
| AA # | Codon | AA | |||||
| 471 | CAA | Gln | nc | nc | Glu | Glu (GAA) | nc |
| 475 | AAC | Asn | nc | nc | nc | Ser (AGC) | nc |
| 476 | CCG | Pro | nc | nc | Gln | Gln (CAG) | nc |
| 487 | GCT | Ala | Thr (ACT) | Val (GTT) | Pro (CCT) | Leu (CTT) | nc |
| 492 | AGT | Ser | nc | nc | Cys (TGT) | Cys (TGT) | nc |
| 499 | GAC | Asp | nc | Glu (GAG) | Glu (GAG) | Glu (GAG) | Glu (GAG) |
| 500 | GAA | Glu | nc | Asp (GAT) | Asp (GAT) | Asp (GAT) | |
| 502 | ACT | Thr | nc | Asn (AAT) | Asn (AAT) | Asn (AAT) | Asn (AAT) |
| 517 | CTT | Leu | nc | nc | nc | nc | Leu (TTA) |
| 520 | CTA | Leu | nc | Leu (CTC) | nc | Leu (CTC) | Leu (CTC) |
| 524 | ACT | Thr | Ile (ATT) | Ile (ATT) | Ile (ATT) | Ile (ATT) | Ile (ATT) |
| 525 | GCC | Ala | Gly (GGC) | nc | Gly (GGC) | Gly (GGC) | nc |
| 533 | CTT | Leu | nc | Ile (ATT) | nc | nc | nc |
Subvariants of P-ala have AA substitutions at 499, 524, and 533 (Sandvej et al., 2000)
Subvariant of P-thr has a Glu (GAG) instead of Asp (GAT) at AA 499 (Sandvej et al., 2000)
Subvariants of V-val have AA substitutions at 528 and 585- (Do et al., 2008; Wang et al., 2003) nc: no change in codon
In addition to changes in the C-terminus, EBNA-1 has variation in the N-terminus at codons 16, 24 and 27 (Gutierrez et al., 1997; Habeshaw et al., 1999). EBNA-1 N-terminus changes, though generally linked to variations in the C-terminus, have revealed additional variants that were not evident by evaluating the C-terminus alone (Gutierrez et al., 1997). The N-terminus changes reinforce the need to evaluate the EBV genome more comprehensively in order to characterize the full extent of EBV genetic heterogeneity. As an example of the degree of variability observed in EBNA-1 alone, 22 distinct DNA sequences (resulting in 19 different EBNA-1 protein sequences) based on N- and C-terminus base changes were identified in 26 healthy carriers and 17 IM patients from one study in Australia (Bell et al., 2008).
Table 5 summarizes the distribution of EBNA-1 sequence variants by disease condition and geography. In all geographic regions, P-ala or P-thr was detected either as a single strain or one of several strains in a large proportion of controls. Interestingly, V-val was detected in both cases and controls almost exclusively in China. Data from other regions and other disease conditions are sparse, precluding definitive conclusions regarding geographic and disease patterns of EBNA-1 variants.
Table 5.
Prevalence (%) of Variants Based on the Carboxy Terminus of EBNA-1 by Clinical Condition and Geographic Region
| Clinical Condition | EBNA-1 Genotype | China | Africa | South America | North America | Europe |
|---|---|---|---|---|---|---|
| NPC | P-ala | 0.0% | 18.2% | |||
| P-thr | 4.0% | 72.7% | ||||
| V-val | 95.2% | 0.0% | ||||
| V-leu | 0.0% | 0.0% | ||||
| V-pro | 0.0% | 0.0% | ||||
| Multiple/Other | 0.8%a | 9.1%d | ||||
| Total Cases | 125 | 0 | 0 | 0 | 11 | |
| BL | P-ala | 2.5% | 0.0% | 60.0% | ||
| P-thr | 41.8% | 8.3% | 40.0% | |||
| V-val | 0.0% | 0.0% | 0.0% | |||
| V-leu | 54.4% | 75.0% | 0.0% | |||
| V-pro | 1.3% | 0.0% | 0.0% | |||
| Multiple/Other | 0.0% | 16.7%c | 0.0% | |||
| Total Cases | 0 | 79 | 12 | 0 | 2 | |
| HL | P-ala | 0.0% | 0.0% | 16.7% | 13.6% | |
| P-thr | 0.0% | 25.0% | 50.0% | 81.8% | ||
| V-val | 100% | 0.0% | 0.0% | 0.0% | ||
| V-leu | 0.0% | 75.0% | 33.3% | 4.5% | ||
| V-pro | 0.0% | 0.0% | 0.0% | 0.0% | ||
| Multiple/Other | 0.0% | 0.0% | 0.0% | 0.0% | ||
| Total Cases | 2 | 0 | 20 | 12 | 22 | |
| AIDS lymphoma | P-ala | 20.0% | 15.8% | 36.7% | ||
| P-thr | 40.0% | 31.6% | 38.8% | |||
| V-val | 0.0% | 0.0% | 2.0% | |||
| V-leu | 20.0% | 26.3% | 8.2% | |||
| V-pro | 0.0% | 0.0% | 0.0% | |||
| Multiple/Other | 20.0%3 | 26.3%d | 14.3%f | |||
| Total Cases | 0 | 0 | 5 | 19 | 49 | |
| Healthy/nonmalignant | P-ala | 16.2% | 5.3% | 16.7% | 46.4% | 40.0% |
| P-thr | 2.7% | 36.8% | 33.3% | 17.9% | 55.0% | |
| V-val | 48.6% | 0.0% | 0.0% | 0.0% | 0.0% | |
| V-leu | 0.0% | 47.4% | 50% | 0.0% | 2.5% | |
| V-pro | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| Multiple/Other | 32.4%a | 10.5%b | 0.0% | 35.7%e | 2.5%d | |
| Total Cases | 37 | 38 | 6 | 28 | 8 | |
(Bhatia et al., 1996; Chang et al., 1999; Do et al., 2008; Fassone et al., 2002; Gutierrez et al., 2000; Gutierrez et al., 1997; MacKenzie et al., 1999; Sandvej et al., 2000; Wang et al., 2003; Wang et al., 2002; Zhang et al., 2004)
V-val + P-ala;
P-ala/thr + V-pro;
V-ala;
Not specified;
P-ala + P-thr/V-val
P-ala + P-thr
4.3. BZLF-1
BZLF-1 consists of 3 exons: the first exon encodes a transactivating domain, the second encodes a strong basic domain that directs the ZEBRA to the nucleus, and the third exon encodes a domain thought to play a role in regulating the switch from latency to lytic infection (Fig. 1 & Fig. 5) (IARC, 1997). Antibodies for the product of the BZLF1 gene, transcription factor BamHI Z EBV Replication Activator (ZEBRA), have been frequently detected in NPC and BL cases, indicating an increased production of the protein (Joab et al., 1991; Tedeschi et al., 2007; Yoshizaki et al., 2000). ZEBRA (also known as Z, ZTA, EB1) activation triggers the virus to switch from latent to lytic infection. BZLF1 is a master regulator for expression of several early viral genes critical for productive replication of EBV and alone can activate the entire lytic cascade (Countryman and Miller, 1985; Rooney et al., 1989).
Figure 5.
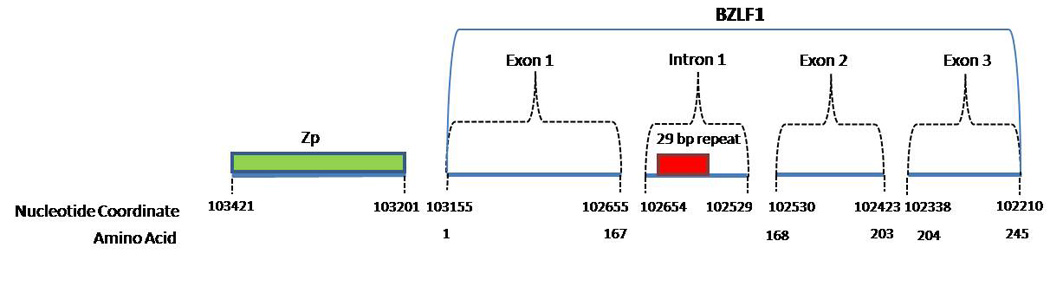
BZLF-1 variants have been defined using different regions of the gene, making comparisons across studies difficult (Chen et al., 1996a; Fassone et al., 2002; Grunewald et al., 1998; Ji et al., 2008; Sacaze et al., 2001). For example, in one study that compared the full BZLF-1 gene from 3 different cell lines, Packham et al. (1993) found that the BZLF-1 genes from the Akata and P3HR1 cell lines contained three copies of a 29 bp repeat (Fig. 5) in contrast to B95-8 which contained two copies of the repeat in the first intron (nucleotide position 102,647), an untranslated region of BZLF-1 (Packham et al., 1993). In another example, using the coding regions of BZLF-1 to define variations, Ji et al. (2008) identified five BZLF-1 variants based on the mRNA sequence changes when those were compared to the published sequence of BZLF-1 in B95-8 (Ji et al., 2008).
Based on the limited data on BZLF-1 variant distribution by geography and disease state (summarized in Table 6), there appear to be geographic differences in the distribution of variants defined based on the 29 bp repeat but no disease associations were apparent.
Table 6.
Prevalence (%) of Variants Based on the 29 Base Pair Repeat in the 1st Intron of BZLF-1 by Clinical Condition and Geographic Region
| Clinical condition | BZLF-1 | China | North Africa and Europe |
|---|---|---|---|
| NPC | B95-8 (2 copies) | 18.7% | 93.5% |
| P3HRI/Akata (3 copies) | 81.3% | 6.5% | |
| total | 75 | 35 | |
| Healthy | B95-8 (2 copies) | 21.2% | 80.0% |
| P3HRI/Akata (3 copies) | 81.8% | 20.0% | |
| total | 33 | 75 | |
4.4. Other variants
The BZLF1 promoter, Zp, is a 220 bp element located immediately upstream of the BZLF1 coding sequence (Fig. 5) containing a variety of regulatory domains that control BZLF1 transcription in response to multiple signals (Shimizu and Takada, 1993). Several variants of Zp have been identified: Zp-P which resembles the B95-8 Zp sequence, Zp-V3, Zp-V4, and most recently Zp-PV (Gutierrez et al., 2002; Martini et al., 2007). Table 8 summarizes the distribution of Zp variants across geographic regions and health conditions (Gutierrez et al., 2002; Martini et al., 2007; Tong et al., 2003). Among controls studied, the prototype Zp-P was observed more commonly in the Americas and Europe than in China, where an almost even split was observed in the prevalence of the Zp-P and Zp-V3 variants. Zp-V3 was generally more commonly observed in cancer cases than in controls across geographic regions. Larger, systematic studies are needed before firm conclusions can be made regarding geographic and/or disease associations related to variants in Zp.
Table 8.
Prevalence (%) of Variants Based on the BZLF-1 Promoter (Zp) by Clinical Condition and Geographic Region
| Health condition | Zp | China | Africa | N./ S. America | Europe |
|---|---|---|---|---|---|
| NPC | Zp-P | 13.0% | |||
| Zp-V3 | 82.6% | ||||
| Zp-V4 | 0.0% | ||||
| Zp-PV | 0.0% | ||||
| Multiple/Other | 4.3% | ||||
| Total | 46 | 0 | 0 | 0 | |
| NHL | Zp-P | 100% | 71.4% | 45.8% | |
| Zp-V3 | 0.0% | 28.6% | 54.2% | ||
| Zp-V4 | 0.0% | 0.0% | 0.0% | ||
| Zp-PV | 0.0% | 0.0% | 0.0% | ||
| Multiple/Other | 0.0% | 0.0% | 0.0% | ||
| Total | 3 | 7 | 24 | 0 | |
| AIDS lymphoma | Zp-P | 54.0% | |||
| Zp-V3 | 30.2% | ||||
| Zp-V4 | 0.0% | ||||
| Zp-PV | 11.1% | ||||
| Multiple/Other | 4.8% | ||||
| Total | 0 | 0 | 0 | 63 | |
| Non-malignant/healthy | Zp-P | 47.5% | 81.3% | 70.0% | |
| Zp-V3 | 50.0% | 0.0% | 0.0% | ||
| Zp-V4 | 0.0% | 18.8% | 0.0% | ||
| Zp-PV | 0.0% | 0.0% | 0.0% | ||
| Multiple/Other | 2.5% | 0.0% | 30.0%a | ||
| Total | 40 | 0 | 16 | 20 | |
Zp-P + Zp-V3
One of the first viral genes expressed after infection, the Epstein-Barr virus nuclear antigen-2 (EBNA2) plays an important role in B cell transformation. EBNA-2 initiates and maintains proliferation of transformed cells and prevents apoptosis of the transformed B cells. The structure of the EBNA-2 protein is characterized by a poly-proline (polyP) and poly-arginine-glycine (RG) stretch and 9 conserved regions throughout the gene region (Kempkes, 2005). A number of studies have identified heterogeneity in EBNA-2 of the type 1 virus (Aitken et al., 1994; Al-Homsi et al., 1998; Cohen et al., 1991; Schuster et al., 1996a; Schuster et al., 1996b; Sheng et al., 2004; Shim et al., 1998; Walling et al., 1994). Among the most notable nucleotide changes in EBNA-2 relative to B95-8, a triplet insertion of CTC (Leucine) at nucleotide 49136 was found in samples from German, US and African patients with a variety of non-malignant and malignant disorders (Aitken et al., 1994; Al-Homsi et al., 1998; Schuster et al., 1996a) and a 51 bp deletion at nucleotide 49102 was detected in LCLs from New Guinea as well as two German patients with fatal lymphoproliferative disease (Aitken et al., 1994; Schuster et al., 1996b). Additionally, the deletion of the entire EBNA-2 gene has been identified in a subset of African BL cell lines, but whether these recombined EBV genomes with gene-wide deletions/duplications occur in “normal” lymphocytes remains to be established (Kelly et al., 2002; Kelly et al., 2005). The findings for EBNA-2 are intriguing, but larger studies are needed to define sequence variation patterns and to evaluate associations with disease and geography.
5. Issues that should be considered when planning future studies to evaluate EBV genetic diversity and its geographic and disease patterns
5.1. Testing approach and variant classification
As summarized in the preceding sections, studies conducted to date have shown extensive diversity in EBV. Efforts that have targeted specific gene regions of the virus have identified substantial genetic heterogeneity in EBV and have demonstrated that as many as 80–100% of infected individuals carry multiple EBV variants (Bell et al., 2008; Bhatia et al., 1996; Edwards et al., 1999; Gutierrez et al., 2002; Gutierrez et al., 1997; Ji et al., 2008; Sitki-Green et al., 2003; Sitki-Green et al., 2002; Tierney et al., 2006; Walling et al., 2003; Walling et al., 1999).
Furthermore, the few studies that have evaluated diversity in more than a single region of the genome have suggested that there is incomplete linkage of genetic variation observed across these regions. As an example, Gutierrez et al. (2000) observed a larger number of unique variants when variability across EBNA-1, EBNA-2, and LMP-1 was examined concurrently than when any one of these regions was evaluated in isolation (Gutierrez et al., 2000). Thus, the full extent of EBV heterogeneity is likely to be greater than currently recognized.
Studies conducted to date have focused on different regions of EBV, making comparisons across studies difficult and limiting our ability to define the full spectrum of diversity existent within the EBV genome. Even within studies that have focused on the same gene regions, different approaches have often been used to group EBV variants, further complicating comparisons across studies. A more comprehensive exploration of EBV diversity across its entire genome might allow for the development of a consensus classification system of EBV variants into variant classes that could guide subsequent studies aimed at understanding EBV geographic and disease associations. McGeoch and Gatherer (2007) recently made genome-wide comparisons of nucleotide sequences among 3 different strains, B95-8, GD1 and AG876A (McGeoch and Gatherer, 2007). Such efforts will pave the way for future, larger studies aiming to assess EBV variation more comprehensively.
5.2. Study populations and specimen types
Studies evaluating EBV genetic diversity have often been small in size and have relied on convenience sampling of cases and controls, making estimates of the distribution of EBV variants by geography and disease status unstable and of questionable generalizability. For example, the majority of studies conducted to date have focused on China and on NPC with limited data available from other geographic regions or disease states. To address these limitations, future studies should use careful sampling techniques to ensure that the distribution of EBV variants observed in those studies is reflective of the distribution in the broader populations of interests. Studies should be large enough to provide robust epidemiologic estimates of EBV variant distribution and should include study groups representing various geographic regions and disease states (including healthy controls) to allow for a more comprehensive and systematic assessment of the distribution of EBV by geography and the possible link between specific EBV variants and disease.
The biological specimens in which EBV is detected need to be considered carefully in future studies. The wide variety of specimen types used in studies to date have included malignant and nonmalignant tissue, peripheral blood lymphocytes, and throat washings. Different EBV variants have been found in different specimen types from the same individual with indications that some variants preferentially infect certain sites or compartments (Chen et al., 1996a; Edwards et al., 2004; Gutierrez et al., 1997; Sacaze et al., 2001; Triantos et al., 1998; Walling et al., 2003). For example, in a study of EBNA-1 carboxyl sequence variants in matched blood and saliva samples of 21 immunocompetent carriers, V-pro was detected in their peripheral blood lymphocytes only while V-val was detected only in their saliva (Gutierrez et al., 1997). These observations may reflect the adaptation of EBV to particular niches, resulting in the preponderance of some variants in certain compartments. Therefore, future studies should be mindful of the specimens used for testing, and at a minimum should ensure that the same specimen type is used for the case and control groups compared. More extensive evaluation of EBV variants at multiple sites within individuals is greatly needed to clarify differences in patterns by site.
6. Conclusions
Why EBV is associated with different malignancies in different geographic regions remains a mystery and may be related in part to EBV genotypic variability through specific disease and geographic associations. We have summarized the available literature on sequence variation in EBV genes including LMP-1, EBNA-1, and BZLF-1. Given the limitations of current studies, definitive statements regarding the link between EBV genotypes, disease and geography are not possible. We conclude that the true extent of EBV genetic diversity is likely to be greater than currently recognized. Future studies conducted in carefully selected populations, sufficiently powered to provide robust epidemiologic estimates and that utilize high-throughput testing approaches that permit full characterization of viral diversity, are needed to further our understanding of patterns of EBV genetic variation and their association with malignancies in different regions.
Table 7.
BZLF-1 (Zp) Promoter Base Changes
| Nucleotide Position | Change | Zp-V3 | Zp-V4 | Zp-PV |
|---|---|---|---|---|
| 103322 | T to G | x | x | |
| 103316 | A to G | x | x | |
| 103281 | A to G | x | x | |
| 103233 | C to T | x | ||
| 103226 | T to C | x |
Acknowledgements
We thank Jim Goedert for his critical review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdel-Hamid M, Chen JJ, Constantine N, Massoud M, Raab-Traub N. EBV strain variation: geographical distribution and relation to disease state. Virology. 1992;190(1):168–175. doi: 10.1016/0042-6822(92)91202-6. [DOI] [PubMed] [Google Scholar]
- Aguirre AJ, Robertson ES. Characterization of intertypic recombinants of the Epstein-Barr virus from the body-cavity-based lymphomas cell lines BC-1 and BC-2. Virology. 1999;264(2):359–369. doi: 10.1006/viro.1999.0015. [DOI] [PubMed] [Google Scholar]
- Aitken C, Sengupta SK, Aedes C, Moss DJ, Sculley TB. Heterogeneity within the Epstein-Barr virus nuclear antigen 2 gene in different strains of Epstein-Barr virus. J Gen Virol. 1994;75(Pt 1):95–100. doi: 10.1099/0022-1317-75-1-95. [DOI] [PubMed] [Google Scholar]
- Aiyar A, Tyree C, Sugden B. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 1998;17(21):6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Homsi AS, Berger C, Van Baarle D, Kersten MJ, Klein MR, McQuain C, Van Oers R, Knecht H. Molecular analysis of critical sequences within the EBNA-2 type 1 gene from Epstein-Barr virus isolates from patients with infectious mononucleosis, tonsillar hyperplasia, and HIV infection. International Journal of Molecular Medicine. 1998;1(6):983–987. doi: 10.3892/ijmm.1.6.983. [DOI] [PubMed] [Google Scholar]
- Apolloni A, Sculley TB. Detection of A-type and B-type Epstein-Barr virus in throat washings and lymphocytes. Virology. 1994;202(2):978–981. doi: 10.1006/viro.1994.1422. [DOI] [PubMed] [Google Scholar]
- Ayadi W, Feki L, Khabir A, Boudawara T, Ghorbel A, Charfeddine I, Daoud J, Frikha M, Hammami A, Karray-Hakim H. Polymorphism analysis of Epstein-Barr virus isolates of nasopharyngeal carcinoma biopsies from Tunisian patients. Virus Genes. 2007;34(2):137–145. doi: 10.1007/s11262-006-0051-2. [DOI] [PubMed] [Google Scholar]
- Baer R, Bankier AT, Biggin MD, Deininger PL, Farrell PJ, Gibson TJ, Hatfull G, Hudson GS, Satchwell SC, Seguin C, et al. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Barozzi P, Luppi M, Cagossi K, Maiorana A, Marasca R, Artusi T, Poggi S, Pileri SA, Torelli G. The oncogenic 30 and 69 bp deletion variants of the EBV LMP-1 gene are common in HIV-negative lymphoproliferations, both malignant and benign. Ann Oncol. 1999;10(4):467–469. doi: 10.1023/a:1008381006612. [DOI] [PubMed] [Google Scholar]
- Bell MJ, Brennan R, Miles JJ, Moss DJ, Burrows JM, Burrows SR. Widespread sequence variation in Epstein-Barr virus nuclear antigen 1 influences the antiviral T cell response. J Infect Dis. 2008;197(11):1594–1597. doi: 10.1086/587848. [DOI] [PubMed] [Google Scholar]
- Berger C, Rothenberger S, Bachmann E, McQuain C, Nadal D, Knecht H. Sequence polymorphisms between latent membrane proteins LMP1 and LMP2A do not correlate in EBV-associated reactive and malignant lympho-proliferations. Int J Cancer. 1999;81(3):371–375. doi: 10.1002/(sici)1097-0215(19990505)81:3<371::aid-ijc10>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Bhatia K, Raj A, Guitierrez MI, Judde JG, Spangler G, Venkatesh H, Magrath IT. Variation in the sequence of Epstein Barr virus nuclear antigen 1 in normal peripheral blood lymphocytes and in Burkitt’s lymphomas. Oncogene. 1996;13(1):177–181. [PubMed] [Google Scholar]
- Bouzid M, Djennaoui D, Dubreuil J, Bouguermouh A, Ellouz D, Abdelwahab J, Decaussin G, Ooka T. Epstein-Barr virus genotypes in NPC biopsies from north Africa. Int J Cancer. 1994;56(4):468–473. doi: 10.1002/ijc.2910560403. [DOI] [PubMed] [Google Scholar]
- Bouzid M, Sheng W, Buisson M, Bouguermouh A, Morand P, Seigneurin JM, Ooka T. Different distribution of H1-H2 Epstein-Barr virus variant in oropharyngeal virus and in biopsies of Hodgkin’s disease and in nasopharyngeal carcinoma from Algeria. Int J Cancer. 1998;77(2):205–210. doi: 10.1002/(sici)1097-0215(19980717)77:2<205::aid-ijc6>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Brady G, Macarthur GJ, Farrell PJ. Epstein-Barr virus and Burkitt lymphoma. Postgrad Med J. 2008;84(993):372–377. doi: 10.1136/jcp.2007.047977. [DOI] [PubMed] [Google Scholar]
- Bray F, Haugen M, Moger TA, Tretli S, Aalen OO, Grotmol T. Age-incidence curves of nasopharyngeal carcinoma worldwide: bimodality in low-risk populations and aetiologic implications. Cancer Epidemiol Biomarkers Prev. 2008;17(9):2356–2365. doi: 10.1158/1055-9965.EPI-08-0461. [DOI] [PubMed] [Google Scholar]
- Burrows JM, Khanna R, Sculley TB, Alpers MP, Moss DJ, Burrows SR. Identification of a naturally occurring recombinant Epstein-Barr virus isolate from New Guinea that encodes both type 1 and type 2 nuclear antigen sequences. J Virol. 1996;70(7):4829–4833. doi: 10.1128/jvi.70.7.4829-4833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone A. Emerging pathways in the development of AIDS-related lymphomas. Lancet Oncol. 2003;4(1):22–29. doi: 10.1016/s1470-2045(03)00957-4. [DOI] [PubMed] [Google Scholar]
- Carbone A, Gloghini A, Serraino D, Spina M. HIV-associated Hodgkin lymphoma. Curr Opin HIV AIDS. 2009;4(1):3–10. doi: 10.1097/COH.0b013e32831a722b. [DOI] [PubMed] [Google Scholar]
- Cardy AH, Sharp L, Little J. Burkitt’s lymphoma: a review of the epidemiology. Kuwait Medical Journal. 2001;33(4):293–306. [Google Scholar]
- Chan JK, Tsang WY, Ng CS, Wong CS, Lo ES. A study of the association of Epstein-Barr virus with Burkitt’s lymphoma occurring in a Chinese population. Histopathology. 1995;(263):239–245. doi: 10.1111/j.1365-2559.1995.tb01437.x. [DOI] [PubMed] [Google Scholar]
- Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15(10):1765–1777. doi: 10.1158/1055-9965.EPI-06-0353. [DOI] [PubMed] [Google Scholar]
- Chang KL, Chen YY, Chen WG, Hayashi K, Bacchi C, Bacchi M, Weiss LM. EBNA-1 gene sequences in Brazilian and American patients with Hodgkin’s disease. Blood. 1999;94(1):244–250. [PubMed] [Google Scholar]
- Chang KP, Hao SP, Lin SY, Ueng SH, Pai PC, Tseng CK, Hsueh C, Hsieh MS, Yu JS, Tsang NM. The 30-bp deletion of Epstein-Barr virus latent membrane protein-1 gene has no effect in nasopharyngeal carcinoma. Laryngoscope. 2006;116(4):541–546. doi: 10.1097/01.mlg.0000201993.53410.40. [DOI] [PubMed] [Google Scholar]
- Chang YS, Su IJ, Chung PJ, Shu CH, Ng CK, Wu SJ, Liu ST. Detection of an Epstein-Barr-virus variant in T-cell-lymphoma tissues identical to the distinct strain observed in nasopharyngeal carcinoma in the Taiwanese population. Int J Cancer. 1995;62(6):673–677. doi: 10.1002/ijc.2910620605. [DOI] [PubMed] [Google Scholar]
- Chen HL, Lung ML, Chan KH, Griffin BE, Ng MH. Tissue distribution of Epstein-Barr virus genotypes. J Virol. 1996a;70(10):7301–7305. doi: 10.1128/jvi.70.10.7301-7305.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chen YY, Bacchi MM, Bacchi CE, Alvarenga M, Weiss LM. Genotyping of Epstein-Barr virus in Brazilian Burkitt’s lymphoma and reactive lymphoid tissue. Type A with a high prevalence of deletions within the latent membrane protein gene. Am J Pathol. 1996b;148(1):17–23. [PMC free article] [PubMed] [Google Scholar]
- Cheung ST, Leung SF, Lo KW, Chiu KW, Tam JS, Fok TF, Johnson PJ, Lee JC, Huang DP. Specific latent membrane protein 1 gene sequences in type 1 and type 2 Epstein-Barr virus from nasopharyngeal carcinoma in Hong Kong. Int J Cancer. 1998;76(3):399–406. doi: 10.1002/(sici)1097-0215(19980504)76:3<399::aid-ijc18>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Cheung ST, Lo KW, Leung SF, Chan WY, Choi PH, Johnson PJ, Lee JC, Huang DP. Prevalence of LMP1 deletion variant of Epstein-Barr virus in nasopharyngeal carcinoma and gastric tumors in Hong Kong. Int J Cancer. 1996;66(5):711–712. doi: 10.1002/(SICI)1097-0215(19960529)66:5<711::AID-IJC21>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Chiang AK, Wong KY, Liang AC, Srivastava G. Comparative analysis of Epstein-Barr virus gene polymorphisms in nasal T/NK-cell lymphomas and normal nasal tissues: implications on virus strain selection in malignancy. Int J Cancer. 1999;80(3):356–364. doi: 10.1002/(sici)1097-0215(19990129)80:3<356::aid-ijc4>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Cohen JI. Benign and malignant Epstein-Barr virus-associated B-cell lymphoproliferative diseases. Semin Hematol. 2003;40(2):116–123. doi: 10.1053/shem.2003.50018. [DOI] [PubMed] [Google Scholar]
- Cohen JI. Clinical aspects of Epstein-Barr virus infections. In: Robertson ES, editor. Epstein-Barr Virus. Norfolk: Caister Academic Press; 2005. pp. 35–54. [Google Scholar]
- Cohen JI, Wang F, Kieff E. Epstein-Barr virus nuclear protein 2 mutations define essential domains for transformation and transactivation. J Virol. 1991;65(5):2545–2554. doi: 10.1128/jvi.65.5.2545-2554.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa RM, Fellner MD, Alonio LV, Durand K, Teyssie AR, Picconi MA. Epstein-barr virus (EBV) in healthy carriers: Distribution of genotypes and 30 bp deletion in latent membrane protein-1 (LMP-1) oncogene. J Med Virol. 2004;73(4):583–588. doi: 10.1002/jmv.20129. [DOI] [PubMed] [Google Scholar]
- Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci U S A. 1985;82(12):4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Addario M, Chauvin P. Ethnic differences in the expression of Epstein-Barr virus latent membrane protein-1 mutations in nasopharyngeal carcinoma. Mutat Res. 2000;457(1–2):69–78. doi: 10.1016/s0027-5107(00)00129-9. [DOI] [PubMed] [Google Scholar]
- Dambaugh T, Hennessy K, Chamnankit L, Kieff E. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr nuclear antigen 2. Proc Natl Acad Sci U S A . 1984;81(23):7632–7636. doi: 10.1073/pnas.81.23.7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delecluse HJ, Feederle R, O’Sullivan B, Taniere P. Epstein Barr virus-associated tumours: an update for the attention of the working pathologist. J Clin Pathol. 2007;60(12):1358–1364. doi: 10.1136/jcp.2006.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinand V, Arya LS. Epidemiology? of childhood Hodgkins disease: is it different in developing countries? Indian Pediatr. 2006;43(2):141–147. [PubMed] [Google Scholar]
- Do NV, Ingemar E, Phi PT, Jenny A, Chinh TT, Zeng Y, Hu L. A major EBNA1 variant from Asian EBV isolates shows enhanced transcriptional activity compared to prototype B95.8. Virus Res. 2008;132(1–2):15–24. doi: 10.1016/j.virusres.2007.10.020. [DOI] [PubMed] [Google Scholar]
- Dolcetti R, Zancai P, De Re V, Gloghini A, Bigoni B, Pivetta B, De Vita S, Carbone A, Boiocchi M. Epstein-Barr virus strains with latent membrane protein-1 deletions: prevalence in the Italian population and high association with human immunodeficiency virus-related Hodgkin’s disease. Blood. 1997;89(5):1723–1731. [PubMed] [Google Scholar]
- Edwards RH, Seillier-Moiseiwitsch F, Raab-Traub N. Signature amino acid changes in latent membrane protein 1 distinguish Epstein-Barr virus strains. Virology. 1999;261(1):79–95. doi: 10.1006/viro.1999.9855. [DOI] [PubMed] [Google Scholar]
- Edwards RH, Sitki-Green D, Moore DT, Raab-Traub N. Potential selection of LMP1 variants in nasopharyngeal carcinoma. J Virol. 2004;78(2):868–881. doi: 10.1128/JVI.78.2.868-881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell PJ. Epstein-Barr Virus Genome. In: Robertson ES, editor. Epstein-Barr Virus. Norfolk: Caister Academic Press; 2005. pp. 263–288. [Google Scholar]
- Fassone L, Cingolani A, Martini M, Migliaretti G, Oreste PL, Capello D, Gloghini A, Vivenza D, Dolcetti R, Carbone A, Antinori A, Gaidano G, Larocca LM. Characterization of Epstein-Barr virus genotype in AIDS-related non-Hodgkin’s lymphoma. AIDS Res Hum Retroviruses. 2002;18(1):19–26. doi: 10.1089/088922202753394682. [DOI] [PubMed] [Google Scholar]
- Fennewald S, van Santen V, Kieff E. Nucleotide sequence of an mRNA transcribed in latent growth-transforming virus infection indicates that it may encode a membrane protein. J Virol. 1984;51(2):411–419. doi: 10.1128/jvi.51.2.411-419.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Bray F, Pisani P, Parkin DM. IARC CancerBase. Vol. 5. Lyon: IARCPress; 2004. GLOBOCAN 2002: Cancer Incidence, Mortality and Prevalence Worldwide. [Google Scholar]
- Goldschmidts WL, Bhatia K, Johnson JF, Akar N, Gutierrez MI, Shibata D, Carolan M, Levine A, Magrath IT. Leukemia. 9. Vol. 6. 1992. Epstein-Barr virus genotypes in AIDS-associated lymphomas are similar to those in endemic Burkitt’s lymphomas; pp. 875–878. [PubMed] [Google Scholar]
- Gorzer I, Niesters HG, Cornelissen JJ, Puchhammer-Stockl E. Characterization of Epstein-Barr virus Type I variants based on linked polymorphism among EBNA3A, −3B, and −3C genes. Virus Res. 2006;118(1–2):105–114. doi: 10.1016/j.virusres.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Gratama JW, Ernberg I. Molecular epidemiology of Epstein-Barr virus infection. Adv Cancer Res. 1995;67:197–255. [PubMed] [Google Scholar]
- Gratama JW, Oosterveer MA, Klein G, Ernberg I. EBNA size polymorphism can be used to trace Epstein-Barr virus spread within families. J Virol. 1990;64(10):4703–4708. doi: 10.1128/jvi.64.10.4703-4708.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald V, Bonnet M, Boutin S, Yip T, Louzir H, Levrero M, Seigneurin JM, Raphael M, Touitou R, Martel-Renoir D, Cochet C, Durandy A, Andre P, Lau W, Zeng Y, Joab I. Amino-acid change in the Epstein-Barr-virus ZEBRA protein in undifferentiated nasopharyngeal carcinomas from Europe and North Africa. Int J Cancer. 1998;75(4):497–503. doi: 10.1002/(sici)1097-0215(19980209)75:4<497::aid-ijc2>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Guiretti DM, Chabay PA, Valva P, Stefanoff CG, Barros MH, De Matteo E, Renault IZ, Preciado MV, Hassan R. Structural variability of the carboxy-terminus of Epstein-Barr virus encoded latent membrane protein 1 gene in Hodgkin’s lymphomas. J Med Virol. 2007;79(11) doi: 10.1002/jmv.21020. 1730-22. [DOI] [PubMed] [Google Scholar]
- Gutierrez MI, Bhatia K, Barriga F, Diez B, Muriel FS, de Andreas ML, Epelman S, Risueno C, Magrath IT. Molecular epidemiology of Burkitt’s lymphoma from South America: differences in breakpoint location and Epstein-Barr virus association from tumors in other world regions. Blood. 1992;79(12):3261–3266. [PubMed] [Google Scholar]
- Gutierrez MI, Ibrahim MM, Dale JK, Greiner TC, Straus SE, Bhatia K. Discrete alterations in the BZLF1 promoter in tumor and non-tumor-associated Epstein-Barr virus. J Natl Cancer Inst. 2002;94(23):1757–1763. doi: 10.1093/jnci/94.23.1757. [DOI] [PubMed] [Google Scholar]
- Gutierrez MI, Kingma DW, Sorbara L, Tran M, Raffeld M, Jaffe ES, Magrath I, Bhatia K. Association of EBV strains, defined by multiple loci analyses, in non-Hodgkin lymphomas and reactive tissues from HIV positive and HIV negative patients. Leuk Lymphoma. 2000;37(3–4):425–429. doi: 10.3109/10428190009089443. [DOI] [PubMed] [Google Scholar]
- Gutierrez MI, Raj A, Spangler G, Sharma A, Hussain A, Judde JG, Tsao SW, Yuen PW, Joab I, Magrath IT, Bhatia K. Sequence variations in EBNA-1 may dictate restriction of tissue distribution of Epstein-Barr virus in normal and tumour cells. J Gen Virol. 1997;78(Pt 7):1663–1670. doi: 10.1099/0022-1317-78-7-1663. [DOI] [PubMed] [Google Scholar]
- Habeshaw G, Yao QY, Bell AI, Morton D, Rickinson AB. Epstein-barr virus nuclear antigen 1 sequences in endemic and sporadic Burkitt’s lymphoma reflect virus strains prevalent in different geographic areas. J Virol. 1999;73(2):965–975. doi: 10.1128/jvi.73.2.965-975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington DS, Weisenburger DD, Purtilo DT. Epstein-Barr virus--associated lymphoproliferative lesions. Clin Lab Med. 1988;8(1):97–118. [PubMed] [Google Scholar]
- Hatzivassiliou E, Mosialos G. Cellular signaling pathways engaged by the Epstein-Barr virus transforming protein LMP1. Front Biosci. 2002;7:d319–d329. doi: 10.2741/hatziva. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Chen WG, Chen YY, Bacchi MM, Bacchi CE, Alvarenga M, Abreu ES, Chang KL, Weiss LM. Deletion of Epstein-Barr virus latent membrane protein 1 gene in United States and Brazilian Hodgkin’s disease and reactive lymphoid tissue: high frequency of a 30-bp deletion. Hum Pathol. 1997;28(12):1408–1414. doi: 10.1016/s0046-8177(97)90231-8. [DOI] [PubMed] [Google Scholar]
- Henry S, Sacaze C, Berrajah L, Karray H, Drira M, Hammami A, Icart J, Mariame B. In nasopharyngeal carcinoma-bearing patients, tumors and lymphocytes are infected by different Epstein-Barr virus strains. Int J Cancer. 2001;91(5):698–704. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1110>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Higa M, Kinjo T, Kamiyama K, Iwamasa T, Hamada T, Iyama K. Epstein-Barr virus (EBV) subtype in EBV related oral squamous cell carcinoma in Okinawa, a subtropical island in southern Japan, compared with Kitakyushu and Kumamoto in mainland Japan. J Clin Pathol. 2002;55(6):414–423. doi: 10.1136/jcp.55.6.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JL, Glaser SL. Epstein-barr virus-associated malignancies: epidemiologic patterns and etiologic implications. Crit Rev Oncol Hematol. 2000;34(1):27–53. doi: 10.1016/s1040-8428(00)00046-9. [DOI] [PubMed] [Google Scholar]
- Hu LF, Zabarovsky ER, Chen F, Cao SL, Ernberg I, Klein G, Winberg G. Isolation and sequencing of the Epstein-Barr virus BNLF-1 gene (LMP1) from a Chinese nasopharyngeal carcinoma. J Gen Virol. 1991;72(Pt 10):2399–2409. doi: 10.1099/0022-1317-72-10-2399. [DOI] [PubMed] [Google Scholar]
- IARC. Vol. 8. Lyon: IARC; 1997. Epstein-Barr virus and Kaposi’s sarcoma herpesvirus/human herpesvirus; p. 70. [PMC free article] [PubMed] [Google Scholar]
- Itakura O, Yamada S, Narita M, Kikuta H. High prevalence of a 30-base pair deletion and single-base mutations within the carboxy terminal end of the LMP-1 oncogene of Epstein-Barr virus in the Japanese population. Oncogene. 1996;13(7):1549–1553. [PubMed] [Google Scholar]
- Izumi KM, Kieff ED. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-kappaB. Proc Natl Acad Sci U S A. 1997;94(23):12592–12597. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins PJ, Farrell PJ. Are? particular Epstein-Barr virus strains linked to disease? Semin Cancer Biol. 1996;7(4):209–215. doi: 10.1006/scbi.1996.0028. [DOI] [PubMed] [Google Scholar]
- Jeon YK, Lee BY, Kim JE, Lee SS, Kim CW. Molecular characterization of Epstein-Barr virus and oncoprotein expression in nasopharyngeal carcinoma in Korea. Head Neck. 2004;26(7):573–583. doi: 10.1002/hed.10370. [DOI] [PubMed] [Google Scholar]
- Ji KM, Li CL, Meng G, Han AD, Wu XL. New BZLF1 sequence variations in EBV-associated undifferentiated nasopharyngeal carcinoma in southern China. Arch Virol. 2008;153(10):1949–1953. doi: 10.1007/s00705-008-0195-6. [DOI] [PubMed] [Google Scholar]
- Joab I, Nicolas JC, Schwaab G, de-The G, Clausse B, Perricaudet M, Zeng Y. Detection of anti-Epstein-Barr-virus transactivator (ZEBRA) antibodies in sera from patients with nasopharyngeal carcinoma. Int J Cancer. 1991;48(5):647–649. doi: 10.1002/ijc.2910480503. [DOI] [PubMed] [Google Scholar]
- Kelly G, Bell A, Rickinson A. Epstein-Barr virus-associated Burkitt lymphomagenesis selects for downregulation of the nuclear antigen EBNA2. Nat Med. 2002;8(10):1098–1104. doi: 10.1038/nm758. [DOI] [PubMed] [Google Scholar]
- Kelly GL, Milner AE, Tierney RJ, Croom-Carter DS, Altmann M, Hammerschmidt W, Bell AI, Rickinson AB. Epstein-Barr virus nuclear antigen 2 (EBNA2) gene deletion is consistently linked with EBNA3A, −3B, and −3C expression in Burkitt’s lymphoma cells and with increased resistance to apoptosis. J Virol. 2005;79(16):10709–10717. doi: 10.1128/JVI.79.16.10709-10717.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempkes B. EBNA-2, and notch signalling. In: Robertson ES, editor. Epstein-Barr Virus. Norfolk: Caister Academic Press; 2005. pp. 463–500. [Google Scholar]
- Khanim F, Yao QY, Niedobitek G, Sihota S, Rickinson AB, Young LS. Blood. 9. Vol. 88. 1996. Analysis of Epstein-Barr virus gene polymorphisms in normal donors and in virus-associated tumors from different geographic locations; pp. 3491–3501. [PubMed] [Google Scholar]
- Kim SM, Kang SH, Lee WK. Identification of two types of naturally-occurring intertypic recombinants of Epstein-Barr virus. Mol Cells. 2006;21(2):302–307. [PubMed] [Google Scholar]
- Klemenc P, Marin J, Soba E, Gale N, Koren S, Strojan P. Distribution of Epstein-Barr virus genotypes in throat washings, sera, peripheral blood lymphocytes and in EBV positive tumor biopsies from Slovenian patients with nasopharyngeal carcinoma. J Med Virol. 2006;78(8):1083–1090. doi: 10.1002/jmv.20666. [DOI] [PubMed] [Google Scholar]
- Klumb CE, Hassan R, De Oliveira DE, De Resende LM, Carrico MK, De Almeida Dobbin J, Pombo-De-Oliveira MS, Bacchi CE, Maia RC. Geographic variation in Epstein-Barr virus-associated Burkitt’s lymphoma in children from Brazil. Int J Cancer. 2004;108(1):66–70. doi: 10.1002/ijc.11443. [DOI] [PubMed] [Google Scholar]
- Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen PM, Klein G, Kurilla MG, Masucci MG. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375(6533):685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- Lin JC, Cherng JM, Lin HJ, Tsang CW, Liu YX, Lee SP. Amino acid changes in functional domains of latent membrane protein 1 of Epstein-Barr virus in nasopharyngeal carcinoma of southern China and Taiwan: prevalence of an HLA A2-restricted ‘epitope-loss variant’. J Gen Virol. 2004;85(Pt 7):2023–2034. doi: 10.1099/vir.0.19696-0. [DOI] [PubMed] [Google Scholar]
- Lin JC, Lin SC, Luppi M, Torelli G, Mar EC. Geographic sequence variation of latent membrane protein 1 gene of Epstein-Barr virus in Hodgkin’s lymphomas. J Med Virol. 1995;45(2):183–191. doi: 10.1002/jmv.1890450213. [DOI] [PubMed] [Google Scholar]
- Lin SX, Zong YS, Wu QL, Han AJ, Liang YJ. Loss of an XhoI-site within N-terminal region of Epstein-Barr virus LMP1 gene in nasopharyngeal carcinoma. Ai Zheng. 2003;22(11):1147–1151. [PubMed] [Google Scholar]
- Lindsey JW, Patel S, Zou J. Epstein-Barr virus genotypes in multiple sclerosis. Acta Neurol Scand. 2008;117(2):141–144. doi: 10.1111/j.1600-0404.2007.00917.x. [DOI] [PubMed] [Google Scholar]
- Lung ML, Chang GC. Detection of distinct Epstein-Barr virus genotypes in NPC biopsies from southern Chinese and Caucasians. Int J Cancer. 1992;52(1):34–37. doi: 10.1002/ijc.2910520108. [DOI] [PubMed] [Google Scholar]
- Lung ML, Chang GC, Miller TR, Wara WM, Phillips TL. Genotypic analysis of Epstein-Barr virus isolates associated with nasopharyngeal carcinoma in Chinese immigrants to the United States. Int J Cancer. 1994;59(6):743–746. doi: 10.1002/ijc.2910590605. [DOI] [PubMed] [Google Scholar]
- Lung ML, Chang RS, Huang ML, Guo HY, Choy D, Sham J, Tsao SY, Cheng P, Ng MH. Epstein-Barr virus genotypes associated with nasopharyngeal carcinoma in southern China. Virology. 1990;177(1):44–53. doi: 10.1016/0042-6822(90)90458-4. [DOI] [PubMed] [Google Scholar]
- Lung ML, Lam WP, Sham J, Choy D, Yong-Sheng Z, Guo HY, Ng MH. Detection and prevalence of the “f” variant of Epstein-Barr virus in southern China. Virology. 1991;185(1):67–71. doi: 10.1016/0042-6822(91)90754-y. [DOI] [PubMed] [Google Scholar]
- MacKenzie J, Gray D, Pinto-Paes R, Barrezueta LF, Armstrong AA, Alexander FA, McGeoch DJ, Jarrett RF. Analysis of Epstein-Barr virus (EBV) nuclear antigen 1 subtypes in EBV-associated lymphomas from Brazil and the United Kingdom. J Gen Virol. 1999;80(Pt 10):2741–2745. doi: 10.1099/0022-1317-80-10-2741. [DOI] [PubMed] [Google Scholar]
- Mainou BA, Raab-Traub N. LMP1 strain variants: biological and molecular properties. J Virol. 2006;80(13):6458–6468. doi: 10.1128/JVI.00135-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini M, Capello D, Serraino D, Navarra A, Pierconti F, Cenci T, Gaidano G, Larocca LM. Characterization of variants in the promoter of EBV gene BZLF1 in normal donors, HIV-positive patients and in AIDS-related lymphomas. J Infect. 2007;54(3):298–306. doi: 10.1016/j.jinf.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Mbulaiteye SM, Biggar RJ, Bhatia K, Linet MS, Devesa SS. Sporadic childhood Burkitt lymphoma incidence in the United States during 1992–2005. Pediatr Blood Cancer. 2009 doi: 10.1002/pbc.22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ, Gatherer D. Lineage structures in the genome sequences of three Epstein-Barr virus strains. Virology. 2007;359(1):1–5. doi: 10.1016/j.virol.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Midgley RS, Blake NW, Yao QY, Croom-Carter D, Cheung ST, Leung SF, Chan AT, Johnson PJ, Huang D, Rickinson AB, Lee SP. Novel intertypic recombinants of epstein-barr virus in the chinese population. J Virol. 2000;74(3):1544–1548. doi: 10.1128/jvi.74.3.1544-1548.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WE, Edwards RH, Walling DM, Raab-Traub N. Sequence variation in the Epstein-Barr virus latent membrane protein 1. J Gen Virol. 1994;75(Pt 10):2729–2740. doi: 10.1099/0022-1317-75-10-2729. [DOI] [PubMed] [Google Scholar]
- Mueller NE, Grufferman S. Hodgkin Lymphoma. In: Schottenfeld D, Fraumeni JF, editors. Cancer Epidemiology and Prevention. New York: Oxford University Press, Inc.; 2006. pp. 872–897. [Google Scholar]
- Munch M. Epstein-Barr virus strain characterization. APMIS. 1998;106(4):425–433. doi: 10.1111/j.1699-0463.1998.tb01367.x. [DOI] [PubMed] [Google Scholar]
- Nagamine M, Takahara M, Kishibe K, Nagato T, Ishii H, Bandoh N, Ogino T, Harabuchi Y. Sequence variations of Epstein-Barr virus LMP1 gene in nasal NK/T-cell lymphoma. Virus Genes. 2007;34(1):47–54. doi: 10.1007/s11262-006-0008-5. [DOI] [PubMed] [Google Scholar]
- Nguyen-Van D, Ernberg I, Phan-Thi Phi P, Tran-Thi C, Hu L. Epstein-Barr virus genetic variation in Vietnamese patients with nasopharyngeal carcinoma: full-length analysis of LMP1. Virus Genes. 2008;37(2):273–281. doi: 10.1007/s11262-008-0262-9. [DOI] [PubMed] [Google Scholar]
- Packham G, Brimmell M, Cook D, Sinclair AJ, Farrell PJ. Strain variation in Epstein-Barr virus immediate early genes. Virology. 1993;192(2):541–550. doi: 10.1006/viro.1993.1070. [DOI] [PubMed] [Google Scholar]
- Philip T. Burkitt’s lymphoma in Europe. IARC Sci Publ. 1985;(60):107–118. [PubMed] [Google Scholar]
- Plaza G, Santon A, Vidal AM, Bellas C. Latent membrane protein-1 oncogene deletions in nasopharyngeal carcinoma in Caucasian patients. Acta Otolaryngol. 2003;123(5):664–668. [PubMed] [Google Scholar]
- Purtilo DT. Epstein-Barr-virus-induced oncogenesis in immune-deficient individuals. Lancet. 1980;1(8163):300–303. doi: 10.1016/s0140-6736(80)90792-8. [DOI] [PubMed] [Google Scholar]
- Purtilo DT. Epstein-Barr virus: the spectrum of its manifestations in human beings. South Med J. 1987;80(8):943–947. doi: 10.1097/00007611-198708000-00003. [DOI] [PubMed] [Google Scholar]
- Rickinson A, Kieff E. Epstein-Barr Virus. In: Knipe DM, Howley PM, editors. Fields Virology, Fifth. Vol. 2. 2 vols. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2655–2700. [Google Scholar]
- Rickinson AB, Young LS, Rowe M. Influence of the Epstein-Barr virus nuclear antigen EBNA 2 on the growth phenotype of virus-transformed B cells. J Virol. 1987;61(5):1310–1317. doi: 10.1128/jvi.61.5.1310-1317.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochford R, Cannon MJ, Moormann AM. Endemic Burkitt’s lymphoma: a polymicrobial disease? Nat Rev Microbiol. 2005;3(2):182–187. doi: 10.1038/nrmicro1089. [DOI] [PubMed] [Google Scholar]
- Rooney CM, Rowe DT, Ragot T, Farrell PJ. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J Virol. 1989;63(7) doi: 10.1128/jvi.63.7.3109-3116.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe M, Lear AL, Croom-Carter D, Davies AH, Rickinson AB. Three pathways of Epstein-Barr virus gene activation from EBNA1-positive latency in B lymphocytes. J Virol. 1992;66(1) doi: 10.1128/jvi.66.1.122-131.1992. 3109-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacaze C, Henry S, Icart J, Mariame B. Tissue specific distribution of Epstein-Barr virus (EBV) BZLF1 gene variants in nasopharyngeal carcinoma (NPC) bearing patients. Virus Res. 2001;81(1–2):133–142. doi: 10.1016/s0168-1702(01)00376-8. [DOI] [PubMed] [Google Scholar]
- Saechan V, Mori A, Mitarnun W, Settheetham-Ishida W, Ishida T. Analysis of LMP1 variants of EBV in Southern Thailand: evidence for strain-associated T-cell tropism and pathogenicity. J Clin Virol. 2006;36(2):119–125. doi: 10.1016/j.jcv.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Sample J, Young L, Martin B, Chatman T, Kieff E, Rickinson A. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol. 1990;64(9):4084–4092. doi: 10.1128/jvi.64.9.4084-4092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandvej K, Gratama JW, Munch M, Zhou XG, Bolhuis RL, Andresen BS, Gregersen N, Hamilton-Dutoit S. Sequence analysis of the Epstein-Barr virus (EBV) latent membrane protein-1 gene and promoter region: identification of four variants among wild-type EBV isolates. Blood. 1997;90(1):323–330. [PubMed] [Google Scholar]
- Sandvej K, Zhou XG, Hamilton-Dutoit S. EBNA-1 sequence variation in Danish and Chinese EBV-associated tumours: evidence for geographical polymorphism but not for tumour-specific subtype restriction. J Pathol. 2000;191(2):127–131. doi: 10.1002/(SICI)1096-9896(200006)191:2<127::AID-PATH614>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Santon A, Bellas C. Deletions within the epstein-barr virus latent membrane protein-1 oncogene in adult ordinary, HIV-associated and paediatric Hodgkin’s disease. Leuk Lymphoma. 2001;40(3–4):235–242. doi: 10.3109/10428190109057922. [DOI] [PubMed] [Google Scholar]
- Santon A, Manzanal AI, Campo E, Bellas C. Deletions in the Epstein-Barr virus latent membrane protein-1 oncogene in Hodgkin’s disease. Clin Mol Pathol. 1995;48(4):M184–M187. doi: 10.1136/mp.48.4.m184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster V, Ott G, Seidenspinner S, Kreth HW. Common Epstein-Barr virus (EBV) type-1 variant strains in both malignant and benign EBV-associated disorders. Blood. 1996a;87(4):1579–1585. [PubMed] [Google Scholar]
- Schuster V, Seidenspinner S, Kreth HW. Detection of a nuclear antigen 2 (EBNA2)-variant Epstein-Barr virus strain in two siblings with fatal lymphoproliferative disease. J Med Virol. 1996b;48(1):114–120. doi: 10.1002/(SICI)1096-9071(199601)48:1<114::AID-JMV18>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- See HS, Yap YY, Yip WK, Seow HF. Epstein-Barr virus latent membrane protein-1 (LMP-1) 30-bp deletion and Xho I-loss is associated with type III nasopharyngeal carcinoma in Malaysia. World J Surg Oncol. 2008;6:18. doi: 10.1186/1477-7819-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng W, Bouguermouh A, Bouzid M, Djennaoui D, Ooka T. BAMHI DNA fragment H-polymorphism of Epstein-Barr virus is associated with the mutations present in an 89 BP sequence localized in EBNA2 gene. Virus Genes. 2004;29(1):99–108. doi: 10.1023/B:VIRU.0000032793.30419.6c. [DOI] [PubMed] [Google Scholar]
- Shim YS, Kim CW, Lee WK. Sequence variation of EBNA2 of Epstein-Barr virus isolates from Korea. Mol Cells. 1998;8(2):226–232. [PubMed] [Google Scholar]
- Shimizu N, Takada K. Analysis of the BZLF1 promoter of Epstein-Barr virus: identification of an anti-immunoglobulin response sequence. J Virol. 1993;67(6):3240–3245. doi: 10.1128/jvi.67.6.3240-3245.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu CH, Tu TY. Prevalence of the Taiwan variant of the Epstein-Barr virus in nasopharyngeal carcinoma patients and normal individuals. Zhonghua Yi Xue Za Zhi (Taipei) 2000;63(4):288–293. [PubMed] [Google Scholar]
- Sitki-Green D, Covington M, Raab-Traub N. Compartmentalization and transmission of multiple epstein-barr virus strains in asymptomatic carriers. J Virol. 2003;77(3):1840–1877. doi: 10.1128/JVI.77.3.1840-1847.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitki-Green D, Edwards RH, Webster-Cyriaque J, Raab-Traub N. Identification of Epstein-Barr virus strain variants in hairy leukoplakia and peripheral blood by use of a heteroduplex tracking assay. J Virol. 2002;76(19):9645–9656. doi: 10.1128/JVI.76.19.9645-9656.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sixbey JW, Shirley P, Chesney PJ, Buntin DM, Resnick L. Detection of a second widespread strain of Epstein-Barr virus. Lancet. 1989;2(8666):761–765. doi: 10.1016/s0140-6736(89)90829-5. [DOI] [PubMed] [Google Scholar]
- Srivastava G, Wong KY, Chiang AK, Lam KY, Tao Q. Coinfection of multiple strains of Epstein-Barr virus in immunocompetent normal individuals: reassessment of the viral carrier state. Blood. 2000;95(7):2443–2445. [PubMed] [Google Scholar]
- Sung NS, Edwards RH, Seillier-Moiseiwitsch F, Perkins AG, Zeng Y, Raab-Traub N. Epstein-Barr virus strain variation in nasopharyngeal carcinoma from the endemic and non-endemic regions of China. Int J Cancer. 1998;76(2):207–215. doi: 10.1002/(sici)1097-0215(19980413)76:2<207::aid-ijc7>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Tamura S, Kunimoto M, Tabata T, Yoshie O. Genotypic analysis of Epstein-Barr virus associated with nasopharyngeal carcinoma of Japanese patients. Jpn J Cancer Res. 1993;84(3):246–249. doi: 10.1111/j.1349-7006.1993.tb02863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EL, Peh SC, Sam CK. Analyses of Epstein-Barr virus latent membrane protein-1 in Malaysian nasopharyngeal carcinoma: high prevalence of 30-bp deletion, Xho1 polymorphism and evidence of dual infections. J Med Virol. 2003;69(2):251–257. doi: 10.1002/jmv.10282. [DOI] [PubMed] [Google Scholar]
- Tang YL, Lu JH, Cao L, Wu MH, Peng SP, Zhou HD, Huang C, Yang YX, Zhou YH, Chen Q, Li XL, Zhou M, Li GY. Genetic variations of EBV-LMP1 from nasopharyngeal carcinoma biopsies: potential loss of T cell epitopes. Braz J Med Biol Res. 2008;41(2):110–116. doi: 10.1590/s0100-879x2008000200006. [DOI] [PubMed] [Google Scholar]
- Tao Q, Robertson KD, Manns A, Hildesheim A, Ambinder RF. Epstein-Barr virus (EBV) in endemic Burkitt’s lymphoma: molecular analysis of primary tumor tissue. Blood. 1998;91(4):1373–1381. [PubMed] [Google Scholar]
- Tao Q, Young LS, Woodman CB, Murray PG. Epstein-Barr virus (EBV) and its associated human cancers--genetics, epigenetics, pathobiology and novel therapeutics. Front Biosci. 2006;11:2672–2713. doi: 10.2741/2000. [DOI] [PubMed] [Google Scholar]
- Tedeschi R, Pin E, Martorelli D, Bidoli E, Marus A, Pratesi C, Bortolin MT, Zanussi S, Vaccher E, Dolcetti R, De Paoli P. Serum antibody response to lytic and latent Epstein-Barr virus antigens in undifferentiated nasopharyngeal carcinoma patients from an area of nonendemicity. Clin Vaccine Immunol. 2007;14(4):435–441. doi: 10.1128/CVI.00466-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MP, Kurzrock R. Epstein-Barr virus and cancer. Clin Cancer Res. 2004;10(3):803–821. doi: 10.1158/1078-0432.ccr-0670-3. [DOI] [PubMed] [Google Scholar]
- Thorley-Lawson DA, Gross A. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N Engl J Med. 2004;350(13):1328–1337. doi: 10.1056/NEJMra032015. [DOI] [PubMed] [Google Scholar]
- Tierney RJ, Edwards RH, Sitki-Green D, Croom-Carter D, Roy S, Yao QY, Raab-Traub N, Rickinson AB. Multiple Epstein-Barr virus strains in patients with infectious mononucleosis: comparison of ex vivo samples with in vitro isolates by use of heteroduplex tracking assays. J Infect Dis. 2006;193(2):287–297. doi: 10.1086/498913. [DOI] [PubMed] [Google Scholar]
- Tiwawech D, Srivatanakul P, Karalak A, Ishida T. Association between EBNA2 and LMP1 subtypes of Epstein-Barr virus and nasopharyngeal carcinoma in Thais. J Clin Virol. 2008;42(1):1–6. doi: 10.1016/j.jcv.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Tong JH, Lo KW, Au FW, Huang DP, To KF. Re: Discrete alterations in the BZLF1 promoter in tumor and non-tumor-associated Epstein-Barr virus. J Natl Cancer Inst. 2003;95(13):1008–1009. doi: 10.1093/jnci/95.13.1008. author reply 1009. [DOI] [PubMed] [Google Scholar]
- Triantos D, Leao JC, Porter SR, Scully CM, Teo CG. Tissue distribution of Epstein-Barr virus genotypes in hosts coinfected by HIV. AIDS. 1998;12(16):2141–2146. doi: 10.1097/00002030-199816000-00008. [DOI] [PubMed] [Google Scholar]
- Trimeche M, Bonnet C, Korbi S, Boniver J, de Leval L. Association between Epstein-Barr virus and Hodgkin’s lymphoma in Belgium: a pathological and virological study. Leuk Lymphoma. 2007;48(7):1323–1331. doi: 10.1080/10428190701411177. [DOI] [PubMed] [Google Scholar]
- Vallat-Decouvelaere AV, Bretel MA, Vassias I, Laplanche JL, Polivka M, Wassef M, Brunet M, Thiebaut JB, Gosselin B, Morinet F, Mikol J. High frequency of a 30-bp deletion of Epstein-Barr virus latent membrane protein 1 gene in primary HIV non-Hodgkin’s brain lymphomas. Neuropathol Appl Neurobiol. 2002;28(6):471–479. doi: 10.1046/j.1365-2990.2002.t01-1-00418.x. [DOI] [PubMed] [Google Scholar]
- Walling DM, Brown AL, Etienne W, Keitel WA, Ling PD. Multiple Epstein-Barr virus infections in healthy individuals. J Virol. 2003;77(11):6546–6550. doi: 10.1128/JVI.77.11.6546-6550.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling DM, Perkins AG, Webster-Cyriaque J, Resnick L, Raab-Traub N. The Epstein-Barr virus EBNA-2 gene in oral hairy leukoplakia: strain variation, genetic recombination, and transcriptional expression. J Virol. 1994;68(12):7918–7926. doi: 10.1128/jvi.68.12.7918-7926.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling DM, Shebib N, Weaver SC, Nichols CM, Flaitz CM, Webster-Cyriaque J. The molecular epidemiology and evolution of Epstein-Barr virus: sequence variation and genetic recombination in the latent membrane protein-1 gene. J Infect Dis. 1999;179(4):763–774. doi: 10.1086/314672. [DOI] [PubMed] [Google Scholar]
- Wang JT, Sheeng TS, Su IJ, Chen JY, Chen MR. EBNA-1 sequence variations reflect active EBV replication and disease status or quiescent latency in lymphocytes. J Med Virol. 2003;69(3):417–425. doi: 10.1002/jmv.10305. [DOI] [PubMed] [Google Scholar]
- Wang WY, Chien YC, Jan JS, Chueh CM, Lin JC. Consistent sequence variation of Epstein-Barr virus nuclear antigen 1 in primary tumor and peripheral blood cells of patients with nasopharyngeal carcinoma. Clin Cancer Res. 2002;8(8):2586–2590. [PubMed] [Google Scholar]
- Weiss LM. Epstein-Barr virus and Hodgkin’s disease. Curr Oncol Rep. 2000;2(2):199–204. doi: 10.1007/s11912-000-0094-9. [DOI] [PubMed] [Google Scholar]
- Wu SJ, Lay JD, Chen CL, Chen JY, Liu MY, Su IJ. Genomic analysis of Epstein-Barr virus in nasal and peripheral T-cell lymphoma: a comparison with nasopharyngeal carcinoma in an endemic area. J Med Virol. 1996;50(4):314–321. doi: 10.1002/(SICI)1096-9071(199612)50:4<314::AID-JMV6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Yao QY, Tierney RJ, Croom-Carter D, Cooper GM, Ellis CJ, Rowe M, Rickinson AB. Isolation of intertypic recombinants of Epstein-Barr virus from T-cell-immunocompromised individuals. J Virol. 1996a;70(8):4895–4903. doi: 10.1128/jvi.70.8.4895-4903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao QY, Tierney RJ, Croom-Carter D, Dukers D, Cooper GM, Ellis CJ, Rowe M, Rickinson AB. Frequency of multiple Epstein-Barr virus infections in T-cell-immunocompromised individuals. J Virol. 1996b;70(8):4884–4894. doi: 10.1128/jvi.70.8.4884-4894.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci U S A. 1984;81(12):3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JL, Camiolo SM, Ali S, Ying A. Comparison of the EBNA1 proteins of Epstein-Barr virus and herpesvirus papio in sequence and function. Virology. 1996;222(1):1–13. doi: 10.1006/viro.1996.0392. [DOI] [PubMed] [Google Scholar]
- Yoshizaki T, Miwa H, Takeshita H, Sato H, Furukawa M. Elevation of antibody against Epstein-Barr virus genes BRLF1 and BZLF1 in nasopharyngeal carcinoma. J Cancer Res Clin Oncol. 2000;126(2):69–73. [PubMed] [Google Scholar]
- Young LS, Yao QY, Rooney CM, Sculley TB, Moss DJ, Rupani H, Laux G, Bornkamm GW, Rickinson AB. New type B isolates of Epstein-Barr virus from Burkitt’s lymphoma and from normal individuals in endemic areas. J Gen Virol. 1987;68(Pt 11):2853–2862. doi: 10.1099/0022-1317-68-11-2853. [DOI] [PubMed] [Google Scholar]
- Zhang XS, Song KH, Mai HQ, Jia WH, Feng BJ, Xia JC, Zhang RH, Huang LX, Yu XJ, Feng QS, Huang P, Chen JJ, Zeng YX. The 30-bp deletion variant: a polymorphism of latent membrane protein 1 prevalent in endemic and non-endemic areas of nasopharyngeal carcinomas in China. Cancer Lett. 2002;176(1):65–73. doi: 10.1016/s0304-3835(01)00733-9. [DOI] [PubMed] [Google Scholar]
- Zhang XS, Wang HH, Hu LF, Li A, Zhang RH, Mai HQ, Xia JC, Chen LZ, Zeng YX. V-val subtype of Epstein-Barr virus nuclear antigen 1 preferentially exists in biopsies of nasopharyngeal carcinoma. Cancer Lett. 2004;211(1):11–18. doi: 10.1016/j.canlet.2004.01.035. [DOI] [PubMed] [Google Scholar]
- Zhou XG, Sandvej K, Li PJ, Ji XL, Yan QH, Zhang XP, Da JP, Hamilton-Dutoit SJ. Epstein--Barr virus gene polymorphisms in Chinese Hodgkin’s disease cases and healthy donors: identification of three distinct virus variants. J Gen Virol. 2001;82(Pt 5):1157–1167. doi: 10.1099/0022-1317-82-5-1157. [DOI] [PubMed] [Google Scholar]
- Zimber U, Adldinger HK, Lenoir GM, Vuillaume M, Knebel-Doeberitz MV, Laux G, Desgranges C, Wittmann P, Freese UK, Schneider U, et al. Geographical prevalence of two types of Epstein-Barr virus. Virology. 1986;154(1):56–66. doi: 10.1016/0042-6822(86)90429-0. [DOI] [PubMed] [Google Scholar]



