Summary
Apolipophorin III (apoLp-III) from Locustamigratoria was used as a model to investigate apolipoprotein lipid binding interactions. ApoLp-III contains eight lysine residues, of which seven are located on one side of the protein. To investigate the role of positive charges on lipid binding, lysine residues were acetylated by acetic anhydride. The degree of acetylation was analyzed by SDS-PAGE and MALDI-TOF, indicating a maximum of eight acetyl additions. Modified apoLp-III remained α-helical, but displayed a decreased α-helical content (from 78 to 54%). Acetylation resulted in a slightly increase in protein stability, as indicated by a change in the midpoint of guanidine-HCl induced denaturation from 0.55 (unmodified) to 0.65 M (acetylated apoLp-III). Lipid bound apoLp-III, either acetylated or unmodified, displayed similar increases in helical content and midpoint of guanidine-HCl-induced denaturation of ~4 M. The ability to solubilize vesicles of dimyristoylphosphatidylcholine remained unchanged. However, the rate to solubilize dimyristoylphosphatidylglycerol vesicles was reduced two-fold. In addition, a decreased ability to stabilize diacylglycerol-enriched low density lipoproteins was observed. This indicated that lysine residues are not critical for the protein’s ability to bind to zwitterionic phospholipids. Since binding interactions with ionic phospholipids and lipoproteins were affected by acetylation, lysine side chains may play a modulating role in the interaction with more complex lipid surfaces encountered in vivo.
Keywords: apolipoprotein, apolipophorin, lipoprotein, lipid, DMPC, DMPG
1. Introduction
Apolipophorin III (apoLp-III) is a key component of a remarkable lipid transport system, present in a number of insect species. During flight, low density lipophorin is employed to shuttle large amounts of diacylglycerol (DG) from fat body triacylglycerol (TG) reserves to the energy consuming flight muscles [1]. In this process, DG associates with existing lipophorin particles, which are stabilized by association of apoLp-III. Upon DG delivery and hydrolysis, apoLp-III dissociates from the lipophorin surface and can be reused. This transport system enables the insect to mobilize large amounts of stored fat reserves during strenuous exercise such as prolonged flight. ApoLp-III is found in several insect orders, most notably in Lepidoptera and Orthoptera [2]. The globular protein, approximately 18 kDa in size, shares important structural features with mammalian apolipoproteins [3–5], and is comprised of a bundle of amphipathic α-helices. In the lipid-free state, as shown by X-ray and NMR solution structures, the protein’s five α-helices are folded in an up-and-down topology [6–8]. Hydrophobic residues are buried inside the protein interior, while polar and charged residues are located on the protein surface. Upon lipid binding, the flexible protein undergoes a large conformational change. During this process the interior is exposed, allowing hydrophobic amino acid side-chains to gain direct access to the lipid surface [9–11]. The relative low protein stability of apoLp-III favors helix bundle opening and exposure of the hydrophobic interior, thereby facilitating lipid binding [7, 8, 12]. On the other hand, polar and charged amino acid residues reside on the protein surface and are exposed to the aqueous solvent and maintain apolipoprotein solubility and integrity. Potentially, these residues may be involved in binding interactions with the non-exchangeable apoLp-I/-II found in lipophorin or cell surface binding sites as shown for the N-terminal domain of human apoE [13]. Moreover, apolipoprotein-lipid binding interactions may require charged residues, as superficial insertion in the phospholipid surface potentially involves contacts with phosphatidyl headgroups. To address this, positively charged ε-amino groups of lysine side-chains were targeted for acetylation. A previous study showed that modification of the 23 lysine residues present in Manduca sexta apoLp-III (166 residues total) resulted in a complete loss of secondary structure, preventing functional analysis [14]. However, apoLp-III from Locusta migratoria bears only eight lysine residues, and may therefore be a more suitable candidate to investigate the role of lysine residues in apolipoprotein-lipid binding. In the present study, L. migratoria apoLp-III was acetylated, and analyzed for secondary structure. Subsequently, the lipid binding properties were investigated using model bilayer vesicles of either zwitterionic or negatively charged phospholipids. In addition, lipoprotein binding was analyzed using DG-enriched low density lipoproteins.
2. Materials and Methods
2.1. Protein expression, purification and modification
Recombinant apoLp-III was expressed in E. coli BL21 (DE3+) cells in minimal medium as previously reported, and lacks the carbohydrate chains normally present in native apoLp-III [15]. Protein expression was induced with isopropyl-β-D-thiogalactopyranoside (1 mM final concentration) after the optical density of the bacterial culture reached a value of 0.6. Four hours later, cells were removed by centrifugation, and the cell-free supernatant concentrated to 20 mL using a 10k tangential flow concentrator. ApoLp-III was isolated using a two-step chromatography procedure. First, the apoLp-III was separated from other proteins and cell debris using a G-75 gel filtration column (60 × 2.5 cm). Then the peak containing apoLp-III was further purified by reversed-phase HPLC (Beckman Gold) using a linear gradient of water and acetonitrile in 0.05% trifluorethanol, and a Zorbax C8 semi-preparative column (Agilent technologies, Santa Clara, CA). Solvent was removed by freeze-drying and protein samples were stored at −20 °C until further use. The purity of the preparations was verified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using precast 4–20 % Tris-glycine gels (Invitrogen, Carlsbad, CA). Proteins were fixed and stained with 0.5 % (w/v) naphthol blue black (Sigma-Aldrich, St. Louis, MO). In addition, proteins were visualized using 2,2,2,-trichloroethanol (TCE, Sigma-Aldrich). Gels were incubated with 10% TCE (v/v) in water:methanol (1:1, v/v) for 10 min, and visualized using a UV transilluminator (Ladner et al., 2004). ApoLp-III (0.5 mg/mL) was acetylated with acetic anhydride in 2 mL of a saturated sodium acetate solution [16]. The modified apoLp-III samples were subsequently dialyzed in a 0.15 M NaCl containing 0.1 % EDTA (w/v), followed by dialysis in phosphate buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4). The extent of the lysine modification was verified by MALDI-TOF (Applied Biosystems 4800) using sinapinic acid as the matrix.
2.2. Secondary structure analysis
The secondary structure of apoLp-III was analyzed with a Jasco 810 spectropolarimeter using a circular cuvet (pathlength 0.1 cm) and a protein concentration of 0.2 mg/mL in 10 mM sodium phosphate, pH 7.2. Plots are the average of three scans, measured at a scan rate of 20 nm/min. The ellipticity (θ) was converted into molar ellipticity using the equation in which MRW is the mean residue weight, θ = ellipticity in mdeg, l = pathlength in cm, and c = concentration in mg/mL:
| (1) |
The α-helical content was assessed with the following equation [17]:
| (2) |
Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco [18].
2.3. Phospholipid vesicle solubilization
Dimyristoylphosphatidylcholine (DMPC) and dimyristoylphosphatidylglycerol (DMPG) were obtained from Avanti Polar Lipids. Multilamellar vesicles were obtained by vigorously vortexing a dried lipid film with 1 ml of PBS at 30 °C [12]. Small unilamellar vesicles (SUVs) were obtained by extruding the multilamellar vesicles through a 200 nm membrane (Avanti mini extruder). To measure the rate of vesicle solubilization, SUVs were incubated at the lipid crystalline to liquid gel-state transition temperature (TM), i.e. 24 °C for DMPC and 23 °C for DMPG. To a dispersion of 250 μg DMPC vesicles, equilibrated at the TM for 10 minutes, 250 μg apoLp-III was added. In studies using DMPG vesicles, 250 μg of lipid was incubated with 25 μg of apoLp-III. The decrease in light scatter, a result from conversion of SUVs into small discoidal protein-lipid complexes, was monitored by the absorption at 325 nm in a Shimadzu spectrophotometer (UV-2401PC) equipped with a Peltier unit. Assuming first order reaction kinetics, rate constants were calculated from the initial slopes. For secondary structure and electron microscopic analysis, excess lipid-free apoLp-III was removed by Superdex-200 gel filtration chromatography. The formation of nanodisks was verified by transmission electron microscopy, using a Joel JEM 1200 EX-11 operated at 100 kV. Samples were negatively stained using phosphotungsten.
2.4. Lipoprotein binding
To measure apoLp-III binding to lipoproteins, a 200 μl solution of human low density lipoprotein (LDL, 50 μg protein; Sigma-Aldrich) in buffer (150 mM NaCl, 2 mM CaCl2, 50 mM Tris-HCl, pH 7.4) was incubated in the absence or presence of 2 mU phospholipase C (Sigma-Aldrich) [19]. PL-C-induced aggregation was measured by the absorbance at 340 nm in a microplate reader (Thermo Labsystems, Helsinki, Finland). The ability of apoLp-III to protect LDL from aggregation was measured by including μg quantities of apoLp-III in the reaction mixture, taking sample turbidity readings at the specified times.
3. Results
3.1 ApoLp-III lysine modification and characterization
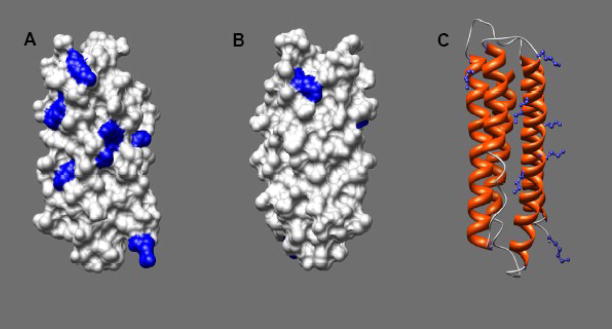
ApoLp-III contains eight lysine residues at positions 52, 54, 63, 68 (helix-2); 121 (helix 4); and 143, 145, and 161 (helix-5). The lysine residues are evenly distributed on the protein surface although seven are located on one side of the protein, more or less formed by helix 2, 3 and 5 (Fig. 1). To neutralize the ε-amino groups of the lysine side-chains, the protein was acetylated with acetic anhydride. SDS-PAGE analysis showed that the apoLp-III samples treated with acetic anhydride migrated at a slower rate compared to untreated protein. Since the naphthol blue black protein staining was significantly reduced for the acetic anhydride-treated protein samples, gels were treated with TCE to increase the staining intensity. TCE adds a formyl group to tryptophan residues, resulting into a large red-shift in the fluorescence emission maximum into the visible range [20]. As seen in Fig. 2A, apoLp-III was visualized effectively by TCE, and displayed a decreased electrophoretic mobility upon treatment with acetic anhydride compared to unmodified apoLp-III. This indicated a substantial number of the apoLp-III lysine residues were acetylated. To determine the extent of acetylation in more detail, the proteins were analyzed by mass spectrometry. The MALDI-TOF mass spectrum for acetylated (Ac)-apoLp-III displayed two major peaks with molecular masses of 17,618 and 17,661 Da (Fig 2B). Unmodified apoLp-III produced a molecular mass of 17,321 Da, close to the calculated mass of 17,327 Da based on the amino acid sequence (not shown). Thus, the mass of acetylated protein increased by 297 or 340 Da, respectively. This corresponded with the addition of seven or eight acetyl groups assuming 42 Da per lysine acetylation.
Fig. 1.
ApoLp-III high resolution structure. Surface contour of apoLp-III (PDB 1AEP) from the front (panel A) or back (panel B) indicating lysine residues in blue. Panel C: ribbon presentation (side-view) with lysine side-chains in blue. Drawings were created using Chimera [18].
Fig. 2.
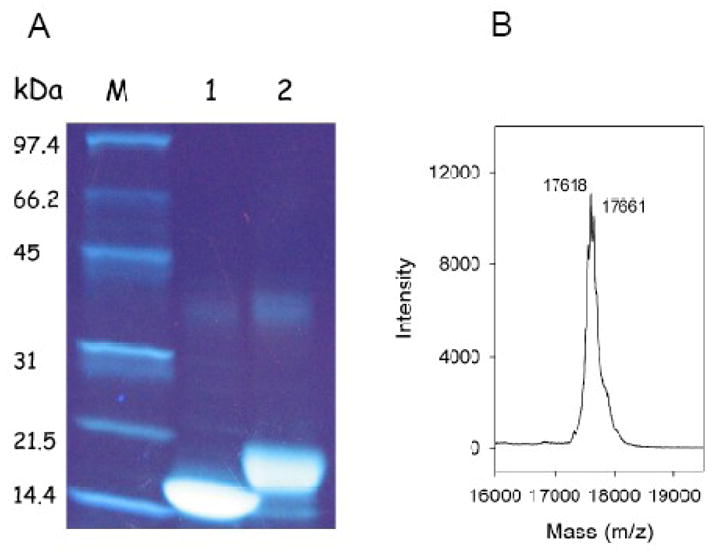
Acetylation of apoLp-III. Proteins were analyzed on 4–20% gradient SDS-PAGE gels and stained for protein with TCE (panel A). M) molecular weight standards; Lane 1) unmodified apoLp-III; lane 2) Ac-apoLp-III. Panel B: MALDI-TOF spectrum of Ac-apoLp-III.
3.2. Secondary structure analysis
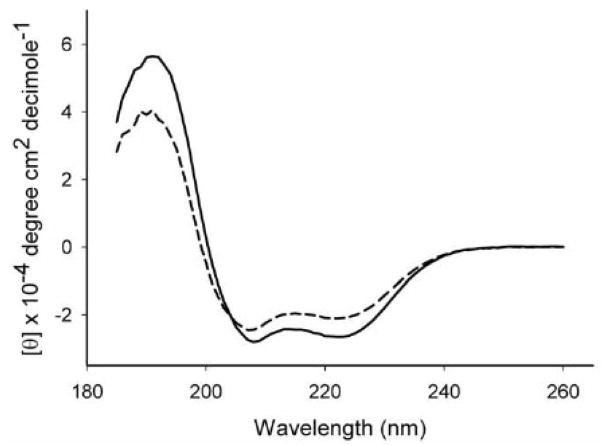
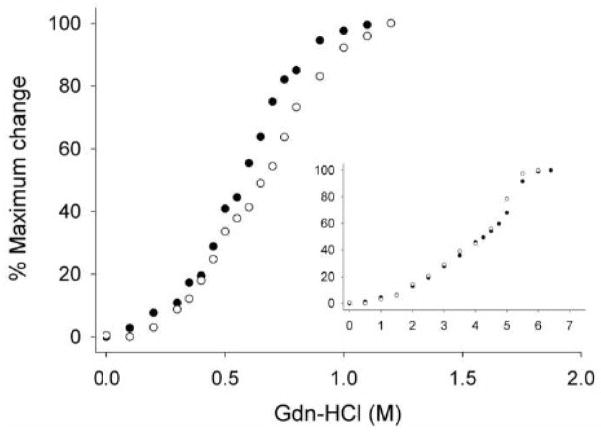
To assess potential structural changes of apoLp-III as a result of lysine acetylation, the secondary structure of Ac-apoLp-III was analyzed by circular dichroism. Far-UV CD spectra showed that the protein retained its α-helical character with troughs at 208 and 222 nm (Fig. 3). The Ac-apoLp-III spectra displayed a somewhat reduced molar ellipticity compared to unmodified apoLp-III. Secondary structure calculations revealed that the helical content was reduced from 78 % in unmodified apoLp-III to 54 % for Ac-apoLp-III. To gain insight into the protein stability properties, guanidine-HCl was employed as the denaturing agent. The denaturation profiles revealed a slight difference between the proteins. The midpoint of guanidine-HCl denaturation increased from 0.55 ± 0.02 M for unmodified apoLp-III to 0.65 ± 0.03 M guanidine-HCl for Ac-apoLp-III. Moreover, the slope of the plot was less steep for Ac-apoLp-III (Fig. 4).
Fig. 3.
Secondary structure analysis. Panel A: FAR UV CD spectra of apoLp-III (solid line) and Ac-apoLp-III (dashed line).
Fig. 4.
Gdn-HCl denaturation profiles of apoLp-III and Ac-apoLp-III. Unmodified apoLp-III (closed circles) and Ac-apoLp-III (open circles) are shown in the lipid-free state (main figure) and lipid-bound state (inset).
Since the lysine ε-amino group may be involved in ionic interactions with negatively charged moieties on phospholipid headgroups, a stabilizing effect of the protein in the lipid-bound form may be anticipated. Therefore, α-helical content and stability were also determined for apoLp-III/DMPC complexes, termed nanodisks. Far UV spectra indicated that both unmodified and Ac-apoLp-III in the nanodisks displayed a substantially increased α-helical content (Table I). As shown in Fig 4 (inset), the apoLp-III midpoint of guanidine-HCl denaturation increased greatly to values of approximately 4 M, in agreement with previous reports [21, 22]. For Ac-apoLp-III a similar increase in protein stability was observed. The denaturation plots of lipid-bound apoLp-III further indicated that the denaturation was not a simple two state process, with proteins either in a folded or unfolded state, but that unfolding intermediates may be present.
Table 1.
Properties of Ac- and unmodified apoLp-III
| Ac-apoLp-III | unmodified apoLp-III | |
|---|---|---|
| molecular mass | 17,661 Da | 17,327 Da |
| [Gdn-HCl]½ lipid-free | 0.65 M | 0.55 M |
| [Gdn-HCl]½ DMPC-bound | 4.2 M | 4.4 M |
| helical content | 54 % | 78 % |
| helical content DMPC bound | 92 % | 99 % |
| rate of DMPC transformation | 0.26 min−1 | 0.27 min−1 |
| rate of DMPG transformation | 0.39 min−1 | 0.91 min−1 |
| LDL protectiona | 25 μg | 10 μg |
The amount of apoLp-III required to prevent aggregation of 100 μg LDL treated with 2 mU of phospholipase-C during a 45 min incubation at 37 °C.
3.3. SUV solubilization
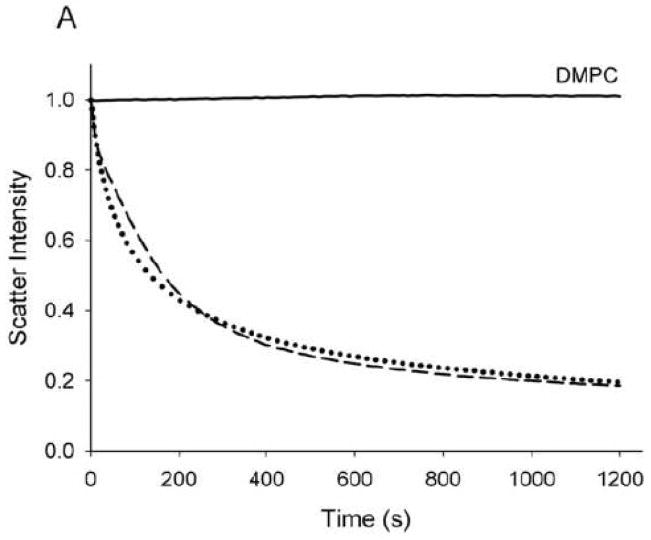
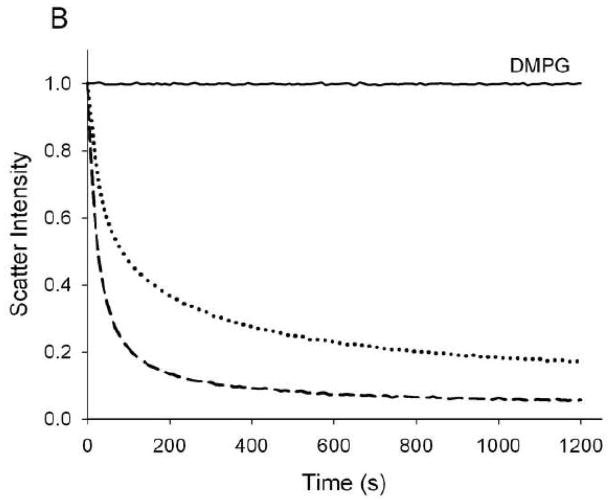
The ability of Ac-apoLp-III to interact with phospholipid bilayers was investigated using DMPC SUVs. Similar to other apolipoproteins, apoLp-III is able to spontaneously solubilize DMPC SUVs at the phospholipid TM [23]. Addition of apoLp-III to a dispersion of SUVs resulted in a steady decrease in light scatter intensity, measured spectrophotometrically at 325 nm. The change in apparent absorbance is the result of decreased light scatter intensity when SUVs (~200 nm in diameter) were transformed into much smaller nanodisks (~15 nm diameter). The DMPC SUV solubilization rate induced by Ac-apoLp-III was very similar to that of unmodified apoLp-III (Fig. 5A). Assuming a 1st order reaction, arate constant of 0.27 ± 0.05 min−1 for unmodified apoLp-III and 0.26 ± 0.06 min−1 for Ac-apoLp-III was obtained. Transmission electron microscopy analysis of the newly formed nanodisks showed no differences between Ac-apoLp-III and unmodified apoLp-III (not shown). To further analyze the effect of charge, negatively charged DMPG vesicles were employed. The solubilization rates with DMPG are an order of magnitude faster compared to DMPC, therefore a 10 fold lesser amount of apoLp-III was used to obtain measurable rates. In this case, the ability to solubilize DMPG SUVs was significantly reduced for Ac-apoLp-III (Fig. 5B). The rate constants of this transformation, 0.91 ± 0.28 min−1 for unmodified apoLp-III and 0.39 ± 0.04 min−1 for Ac-apoLp-III, showed a 2-fold decrease the acetylated protein.
Fig. 5.
SUV solubilization. Vesicles of DMPC (panel A) and DMPG (panel B) were incubated at the phospholipid transition temperature, after the addition of apoLp-III. The decrease in light scatter was monitored by the absorption at 325 nm. Solid line: no apoLp-III; dashed line: unmodified apoLp-III; dotted line: Ac-apoLp-III.
3.4. LDL protection
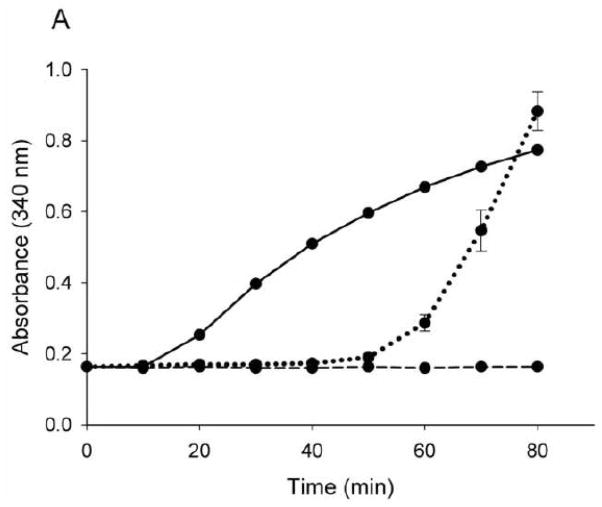
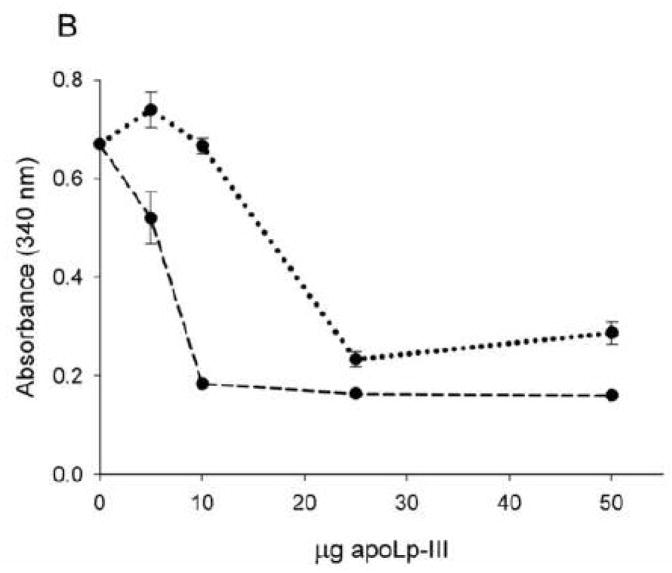
The effect of apoLp-III acetylation on lipoprotein binding interaction was analyzed using modified LDL. While apoLp-III does not bind to human LDL as such, it rapidly associates with LDL when DG emerges on the lipoprotein surface. This process mimics the association of apoLp-III with lipophorin in vivo when large amounts of DG are transferred from the fat body to insect lipoprotein during prolonged flight [24]. To create DG on the surface of LDL particles, phospholipase-C was incubated with LDL, converting phosphatidylcholine into DG [19]. As a result, modified LDL forms large aggregates, as is evident by the rapid increase in sample turbidity measured at 340 nm. However, DG-enriched LDL recruits apolipoproteins on its surface, thereby preventing lipoprotein aggregation, and the incubation mixture remains optically clear. As shown in Fig. 6A, 50 μg of either Ac- or unmodified apoLp-III incubated simultaneously with LDL and phospholipase-C prevented lipoprotein aggregation for a period of 50 minutes. However, at extended incubation times, Ac-apoLp-III lost its protective role, as is evident by the onset of sample turbidity. Using limited amounts of apoLp-III, differences between the two proteins became more pronounced. While 10 μg of unmodified apoLp-III remained effective in providing full protection for a during an incubation time of 60 minutes, a minimum of 25 μg of Ac-apoLp-III was required to prevent the onset of phospholipase-C-induced LDL aggregation (Fig. 6B). This result indicated that the ability of Ac-apoLp-III to bind to the DG-enriched LDL surface was decreased compared to the unmodified protein.
Fig. 6.
Protection of apoLp-III to LDL aggregation. Phospholipase-C was used to induce LDL aggregation, which was measured by the absorbance at 340 nm for 80 min (Panel A, solid line). Incubation of LDL and phospholipase-C in the presence of unmodified apoLp-III (dashed line) provided full protection, while the ability of Ac-apoLp-III (dotted line) to prevent LDL aggregation was reduced. Panel B: LDL and phospholipase-C were incubated with μg quantities of apoLp-III to measure the minimum amount of apoLp-III required to prevent aggregation (measured at 60 min).
4. Discussion
ApoLp-III is an exchangeable apolipoprotein with a predominantly hydrophobic interior. Few polar residues are buried, and it is thought that some of these residues have a destabilizing effect on the protein structure. In turn, the reduced stability promotes helix bundle opening, facilitating binding to lipid surfaces [7, 8]. The stability of apoLp-III is relatively low, as indicated by a midpoint of guanidine-HCl denaturation of ~ 0.6 M [22]. A mutant apoLp-III which was more stable displayed a decreased ability to solubilize DMPC SUVs [12]. Such an inverse correlation between protein stability and lipid binding has been reported for human apoE isoforms as well [3, 17]. In addition to hydrophobic forces, electrostatic interactions are expected to play a significant role in apoLp-III lipid binding interaction. Human apoA-I, apoE and sphinxmoth apoLp-III from M. sexta, Galleria mellonella and Bombyx mori, are all relatively rich in lysine and arginine residues (~15% of total amino acids). On the other hand, apoLp-III from L. migratoria bears only eight lysine and one arginine residue (~6%), and contains two N-linked complex carbohydrate chains [25]. Removal of carbohydrate does not affect the structural properties, but enhanced the vesicle solubilization rate to a level comparable with M. sexta apoLp-III [26]. The role of carbohydrate in locust apoLp-III in vivo remains unknown, although it may be possible that it maintains the protein solubility at high concentrations. In the present study, the positive charges of lysine side chains were neutralized by acetylation with acetic anhydride. MALDI-TOF analysis revealed that up to eight residues were modified. Besides a decreased electrophoretic mobility on SDS-PAGE, protein staining of Ac-apoLp-III was significantly reduced, probably because naphthol blue black binding depends in part on the presence of lysine. Soaking the protein gels with a solution containing TCE improved the visualization of Ac-apoLp-III substantially. TCE depends directly on the presence of tryptophan residues (two present in L. migratoria apoLp-III), thus lysine modification does not affect the visualization. Also, protein bands become visible after only a few minutes [20], while prolonged destaining periods are necessary when employing naphthol blue black. This protein dye is routinely employed to stain L. migratoria apoLp-III on polyacrylamide gels as the more commonly used Coomassie Brilliant Blue depends too much on the presence of positively charged residues [27].
Acetylation resulted in a moderate decrease in secondary structure content, possibly by diminished ionic interactions, but overall the protein remained α-helical and soluble in buffer. In addition, DMPC binding induced a substantial increase in helical content. The stability of Ac-apoLp-III in the lipid-free state was similar to unmodified protein. Nevertheless, close inspection of the guanidine-HCl denaturation plots revealed a small, yet measurable increase in protein stability. Also, the slope of the plot was less steep for Ac-apoLp-III, indicating less cooperativity of unfolding in the absence of ε-amino groups of lysine side chains, perhaps because of the lack of charge repulsion between neighboring lysine residues. Similar results were reported for acetylated apoA-I, although the cooperative effect was much more pronounced in the latter. This may be a reflection of the 21 lysine and 16 arginine side-chains present in the 243-residue human apolipoprotein [28]. Since Ac-apoLp-III retained its helical character we were able to assess the lipid binding properties. In contrast, lysine residues carry out a significant structural role in M. sexta apoLp-III as acetylation resulted in a protein almost devoid of secondary structure. Not surprisingly, M. sexta apoLp-III lipoprotein binding and the ability to spontaneously form nanodisks at the phospholipid TM was virtually absent [14].
The ability to solubilize DMPC vesicles remained unchanged for L. migratoria Ac-apoLp-III. This indicates that neutralization of lysine side-chains has no effect on the ability of the protein to recognize potential lipid surface defects and to open its hydrophobic interior to initiate binding. In this respect, a hydrophobic patch formed by leucine residues on the apolipoprotein surface may be responsible to initiate lipid binding as suggested previously [29, 30]. It was predicted that lysine residues participated in ionic interactions with phosphate moieties of phosphatidylcholine, but the rate of nanodisk formation was essentially similar for both unmodified and Ac-apoLp-III. Furthermore, acetylation of apoLp-III did not result in changes in protein stability when complexed with DMPC and both modified and Ac-apoLp-III showed the distinctive large increase in resistance to guanidine-HCl induced denaturation in nanodisks. These findings indicate that lysine side chains are not essential for binding to zwitterionic DMPC vesicles, and are not a significant factor for nanodisk stabilization. Thus, these processes are likely dominated by hydrophobic forces, while inter-helical contacts between neighboring apoLp-III molecules may also contribute [31]. The six lysine residues that reside in the polar-nonpolar interface characterize these amphipathic α-helices as class A, while the three remaining helices are class G* [32]. It has been suggested that interfacial lysines are a crucial part of the snorkel model [33], if so acetylation may change the character of these helices. Nevertheless, it is possible that the 4-carbon aliphatic portion of lysine remains in close contact with the hydrophobic interior of the phospholipid bilayer, and instead of charged amino groups, the acetyl moieties point out in the aqueous environment. Therefore, the hydrophobic face of the helices has not been changed significantly, explaining the unaltered DMPC binding interaction. Nevertheless, the rate constant for DMPG vesicle solubilization was decreased by 2-fold upon acetylation, indicating that a potential role in the binding process of electrostatic attraction between lysines and negative glycerol headgroups. ApoLp-III association to lipoprotein surfaces was assessed with human LDL. Induced by phospholipase-C catalysis, DG appeared on the LDL surface, triggering apolipoprotein binding. This elegant assay is particularly relevant for apoLp-III because it creates DG on the LDL surface. This resembles the in vivo apoLp-III lipoprotein binding interaction in insect hemolymph, where up to 16 molecules of apoLp-III associate with DG-enriched lipophorins to maintain lipoprotein integrity [34, 35]. While well suited for apoLp-III, the assay is also highly applicable to apoA-I and apoE and illustrates the structural and functional similarity between these apolipoproteins with amphipathic α-helices as the element responsible for lipid binding [19, 36]. At conditions for which unmodified apoLp-III binds to the LDL surface providing full protection from lipoprotein aggregation, the ability of Ac-apoLp-III to prevent lipoprotein aggregation was significantly reduced. The observation that Ac-apoLp-III lost in part its protective role indicated that lysine residues interact to some extent with the LDL surface through ionic forces. Obviously, the surface of LDL is more complex compared to SUVs, and harbors apoB100, phosphatidylcholine and sphingomyelin (66 and 29% of the total lipid mass, respectively) [37]. The conversion of phosphatidylcholine into DG causes major lipid packing problems, which is resolved by superficial apoLp-III association with the LDL surface [38]. As postulated earlier, the amphipathic α-helices are able to cover the hydrophobic gaps between phosphatidylcholine headgroups, when DG intercalates in the phosphatidylcholine monolayer [39].
While electrostatic interactions were postulated to be major forces that direct apoLp-III phospholipid bilayers interaction [40], the present data with DMPC SUVs does not support this. Monolayer studies with DG also pointed out that hydrophobic forces may play a critical role [40, 41]. The present study with DMPC bilayers is in support of the view that such forces are essential for apolipoprotein lipid interaction. Nevertheless, electrostatic interactions, modulated by positively charged lysine side chains, may contribute to binding interactions in more complex lipid mixtures encountered in vivo.
Acknowledgments
This work was supported by grants of the National Institute of Health (AREA HL077135 and SCORE GM 063119). LJV was supported by scholarships from NIH-MBRS/RISE (5R25GM07638-03), and the Howard Hughes Medical Institute (52002663). The authors would like to acknowledge Dr. Ashraf Elamin, Institute for Integrated Research in Materials, Environments, and Society (IIRMES) at the California State University Long Beach for the MALDI-TOF analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Van der Horst DJ, Van Hoof D, Van Marrewijk WJA, Rodenburg KW. Alternative lipid mobilization: the insect shuttle system. Mol Cell Biochem. 2002;239:113–119. [PubMed] [Google Scholar]
- 2.Weers PMM, Ryan RO. Apolipophorin III: Role model apolipoprotein. Insect Biochem Mol Biol. 2006;36:231–240. doi: 10.1016/j.ibmb.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Jonas A, Phillips MC. Lipoprotein structure. In: Vance DE, Vance JE, editors. Biochemistry of Lipids, Lipoproteins and Membranes. Elsevier BV; Amsterdam: 2008. pp. 485–506. [Google Scholar]
- 5.Saito H, Lund-Katz S, Phillips MC. Contributions of domain structure and lipid interaction to the functionality of exchangeable human apolipoproteins. Prog Lipid Res. 2004;43:350–380. doi: 10.1016/j.plipres.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Breiter DR, Kanost MR, Benning MM, Wesenberg G, Law JH, Wells MA, Rayment I, Holden HM. Molecular structure of an apolipoprotein determined at 2.5-Å resolution. Biochemistry. 1991;30:603–608. doi: 10.1021/bi00217a002. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Sykes BD, Ryan RO. Structural basis for the conformational adaptability of apolipophorin III, a helix bundle exchangeable apolipoprotein. Proc Natl Acad Sci USA. 2002;99:1188–1193. doi: 10.1073/pnas.032565999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan D, Zheng Y, Yang D, Wang J. NMR solution structure and dynamics of an exchangeable apolipoprotein, Locustamigratoria apolipophorin III. J Biol Chem. 2003;278(21212–21220) doi: 10.1074/jbc.M208486200. [DOI] [PubMed] [Google Scholar]
- 9.Wientzek M, Kay CM, Oikawa K, Ryan RO. Binding of insect apolipophorin III to dimyristoylphosphatidylcholine vesicles. Evidence for a conformational change. J Biol Chem. 1994;269:4605–4612. [PubMed] [Google Scholar]
- 10.Sahoo D, Weers PMM, Ryan RO, Narayanaswami V. Lipid triggered molecular switch of apoLp-III helix bundle to an extended helix conformation. J Mol Biol. 2002;321:201–214. doi: 10.1016/s0022-2836(02)00618-6. [DOI] [PubMed] [Google Scholar]
- 11.Garda HA, Arrese EL, Soulages JL. Structure of apolipophorin III in discoidal lipoproteins. Interhelical distances in the lipid-bound state and conformational change upon binding to lipid. J Biol Chem. 2002;277:19773–19782. doi: 10.1074/jbc.M110089200. [DOI] [PubMed] [Google Scholar]
- 12.Weers PMM, Cabrera J, Abdullahi EW, Hsu TC. Role of buried polar residues in helix bundle stability and lipid binding of apolipophorin III: destabilization by threonine 31. Biochemistry. 2005;44:8810–8816. doi: 10.1021/bi050502v. [DOI] [PubMed] [Google Scholar]
- 13.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 14.Upadhyaya M, Oikawa K, Kay CM, Scraba DG, Bradley R, Ryan RO. Role of charged amino acid side chains in the stability and lipid binding of Manduca sexta apolipophorin. Arch Insect Biochem Physiol. 1995;30:211–223. [Google Scholar]
- 15.Weers PMM, Wang J, Van der Horst DJ, Kay CM, Sykes BD, Ryan RO. Recombinant locust apolipophorin III: characterization and NMR spectroscopy. Biochim Biophys Acta. 1998;1393:99–107. doi: 10.1016/s0005-2760(98)00063-0. [DOI] [PubMed] [Google Scholar]
- 16.Innerarity TL, Pitas RE, Mahley RW. Lipoprotein–receptor interactions. Methods Enzymol. 1986;129:542–565. doi: 10.1016/0076-6879(86)29091-6. [DOI] [PubMed] [Google Scholar]
- 17.Morrow JA, Segall ML, Lund-Katz S, Phillips MC, Knapp M, Rupp B, Weisgraber KH. Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry. 2000;39:11657–11666. doi: 10.1021/bi000099m. [DOI] [PubMed] [Google Scholar]
- 18.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - A Visualization System for Exploratory Research and Analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 19.Liu H, Scraba DG, Ryan RO. Prevention of phospholipase- C induced aggregation of low-density lipoprotein by amphipathic apolipoproteins. FEBS Lett. 1993;316:27–33. doi: 10.1016/0014-5793(93)81730-n. [DOI] [PubMed] [Google Scholar]
- 20.Ladner CL, Yang JY, Turner RJ, Edwards RA. Visible fluorescent detection of proteins in polyacrylamide gels without staining. Anal Biochem. 2004;326:13–20. doi: 10.1016/j.ab.2003.10.047. [DOI] [PubMed] [Google Scholar]
- 21.Ryan RO, Oikawa K, Kay CM. Conformational, thermodynamic and stability properties of Manduca sexta apolipophorin III. J Biol Chem. 1993;268:1525–1530. [PubMed] [Google Scholar]
- 22.Weers PMM, Kay CM, Oikawa K, Wientzek M, Van der Horst DJ, Ryan RO. Factors affecting the stability and conformation of Locusta migratoria apolipophorin III. Biochemistry. 1994;33:3617–3624. doi: 10.1021/bi00178a019. [DOI] [PubMed] [Google Scholar]
- 23.Chromy BA, Arroyo E, Blanchette CD, Bench G, Benner H, Cappuccio JA, Coleman MA, Henderson PT, Hinz AK, Kuhn EA, Pesavento JB, Segelke BW, Sulchek TA, Tarasow T, Walsworth VL, Hoeprich PD. Different apolipoproteins impact nanolipoprotein particle formation. J Am Chem Soc. 2007;129:14348–54. doi: 10.1021/ja074753y. [DOI] [PubMed] [Google Scholar]
- 24.Ryan RO, Van der Horst DJ. Lipid transport biochemistry and role in energy production. Ann Rev Entomol. 2000;45:233–260. doi: 10.1146/annurev.ento.45.1.233. [DOI] [PubMed] [Google Scholar]
- 25.Hård K, Van Doorn JM, Thomas-Oates JE, Kamerling JP, Van der Horst DJ. Structures of the Asn-linked oligosaccharides of apolipophorin III from the insect Locusta migratoria. Carbohydrate-linked 2-aminoethylphosphonate as a constituent of a glycoprotein. Biochemistry. 1993;32:766–775. doi: 10.1021/bi00054a005. [DOI] [PubMed] [Google Scholar]
- 26.Weers PMM, Van der Horst DJ, Ryan RO. Interaction of locust apolipophorin III with lipoproteins and phospholipid vesicles: effect of glycosylation. J Lipid Res. 2000;41:416–423. [PubMed] [Google Scholar]
- 27.Tal M, Silberstein A, Nusser EJ. Why does Coomassie Brilliant Blue R interact differently with different proteins? J Biol Chem. 1985;260:9976–9980. [PubMed] [Google Scholar]
- 28.Brubaker G, Peng D-Q, Somerlot B, Abdollahian DJ, Smith JD. Apolipoprotein A-I lysine modification: effects on helical content, lipid binding and cholesterol acceptor activity. Biochim Biophys Acta. 2006;1761:64–72. doi: 10.1016/j.bbalip.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Weers PMM, Narayanaswami V, Kay CM, Ryan RO. Interaction of an exchangeable apolipoprotein with phospholipid vesicles and lipoprotein particles. Role of leucines 32, 34 and 95 in Locusta migratoria apolipophorin III. J Biol Chem. 1999;274:21804–21810. doi: 10.1074/jbc.274.31.21804. [DOI] [PubMed] [Google Scholar]
- 30.Ajees AA, Anantharamaiah GM, Mishra VK, Hussain MM, Murthy HMK. Crystal structure of human apolipoprotein A-I: Insights into its protective effect against cardiovascular diseases. Proc Natl Acad Sci USA. 2006;103:2126–2131. doi: 10.1073/pnas.0506877103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Silva RA, Huang R, Morris J, Fang J, Gracheva EO, Ren G, Kontush A, Jerome WG, Rye KA, Davidson WS. Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proc Natl Acad Sci USA. 2008;105:12176–12181. doi: 10.1073/pnas.0803626105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narayanaswami V, Weers PMM, Bogerd J, Kooiman FP, Kay CM, Scraba DG, Van der Horst DJ, Ryan RO. Spectroscopic and lipid binding studies on the amino and carboxy terminal fragments of Locusta migratoria apolipophorin III. Biochemistry. 1995;34:11822–11830. doi: 10.1021/bi00037a021. [DOI] [PubMed] [Google Scholar]
- 33.Mishra VK, Palgunachari MN. Interaction of model class A1, class A2, and class Y amphipathic helical peptides with membranes. Biochemistry. 1996;35:11210–11220. doi: 10.1021/bi960760f. [DOI] [PubMed] [Google Scholar]
- 34.Wells MA, Ryan RO, Kawooya JK, Law JH. The role of apolipophorin III in vivo lipoprotein interconversions in adult Manduca sexta. J Biol Chem. 1987;262:4172–4176. [PubMed] [Google Scholar]
- 35.Van der Horst DJ, Van Doorn JM, Voshol H, Kanost MR, Ziegler R, Th Beenakkers AM. Different isoforms of an apoprotein (apolipophorin III) associate with lipoproteins in Locusta migratoria. Eur J Biochem. 1991;196:509–517. doi: 10.1111/j.1432-1033.1991.tb15843.x. [DOI] [PubMed] [Google Scholar]
- 36.Segrest JP, Jones MK, De Loof H, Brouillette CG, Venkatachalapathi YV, Anantharamaiah GM. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J Lipid Res. 1992;33:141–166. [PubMed] [Google Scholar]
- 37.Sommer A, Prenner E, Gorges R, Stütz H, Grillhofer H, Kostner GM, Paltauf F, Hermetter A. Organization of phosphatidylcholine and sphingomyelin in the surface monolayer of low density lipoprotein and lipoprotein(a) as determined by time-resolved fluorometry. J Biol Chem. 1992;267:24217–24222. [PubMed] [Google Scholar]
- 38.Sahoo D, Narayanaswami V, Kay CM, Ryan RO. Fluorescence studies of exchangeable apolipoprotein-lipid interactions. Superficial association of apolipophorin III with lipoprotein surfaces. J Biol Chem. 1998;273:1403–8. doi: 10.1074/jbc.273.3.1403. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Liu H, Sykes BD, Ryan RO. Identification and localization of two distinct microenvironments for the diacylglycerol component of lipophorin particles by 13C NMR. Biochemistry. 1995;34:6755–6761. doi: 10.1021/bi00020a021. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Lewis RNAH, McElhaney RN, Ryan RO. Calorimetric and spectroscopic studies of the interaction of Manduca sexta apolipophorin III with zwitterionic, anionic, and nonionic lipids. Biochemistry. 1993;32:3942–52. doi: 10.1021/bi00066a014. [DOI] [PubMed] [Google Scholar]
- 41.Demel RA, Van Doorn JM, Van der Horst DJ. Insect apolipophorin III: interaction of locust apolipophorin III with diacylglycerol. Biochim Biophys Acta. 1992;1124:151–158. doi: 10.1016/0005-2760(92)90091-9. [DOI] [PubMed] [Google Scholar]