Abstract
Background
Human infection by the pork tapeworm Taenia solium affects more than 50 million people worldwide, particularly in underdeveloped and developing countries. Cysticercosis which arises from larval encystation can be life threatening and difficult to treat. Here, we investigate for the first time the transcriptome of the clinically relevant cysticerci larval form.
Results
Using Expressed Sequence Tags (ESTs) produced by the ORESTES method, a total of 1,520 high quality ESTs were generated from 20 ORESTES cDNA mini-libraries and its analysis revealed fragments of genes with promising applications including 51 ESTs matching antigens previously described in other species, as well as 113 sequences representing proteins with potential extracellular localization, with obvious applications for immune-diagnosis or vaccine development.
Conclusion
The set of sequences described here will contribute to deciphering the expression profile of this important parasite and will be informative for the genome assembly and annotation, as well as for studies of intra- and inter-specific sequence variability. Genes of interest for developing new diagnostic and therapeutic tools are described and discussed.
Background
Taenia solium, the pork tapeworm, infects around 50 million people worldwide and is one of the foremost public health problems in developing countries [1,2]. The high influx and immigration of people coming from endemic areas to more industrialized nations has produced a complex spreading pattern for cysticercosis which is now a world-wide issue [2].
Cystercercosis arises from the development of T. solium cysticerci in soft tissues as a result of ingesting T. solium eggs [3-5]. Neurocysticercosis; which can cause epileptiform attacks, headaches, learning difficulties and convulsions; is considered the primary cause of acquired epilepsy and its clinical/therapeutic management is difficult, highlighting the importance of search for new drug targets [6-8]. In this work, we investigate for the first time the gene expression profile of T. solium in the larval form responsible – the cysticerci.
High throughput sequencing for gene discovery and gene expression profiling using traditional [9] or alternative 'Expressed Sequence Tags' (ESTs) such as 'Open Reading Frame ESTs' (ORESTES) [10,11] has greatly increased our knowledge of the set of expressed genes of some important helminthic parasites, notably Schistosoma mansoni [12,13] and its intermediate vector Biomphalaria glabrata [14], S. japonicum [15] and the cestodes Echinococcus granulosus [16], E. multilocularis [17] and Mesocestoides corti [18].
Recently, Aguilar-Díaz et al. [1] described the T. solium genome initiative designed to unravel the parasite's complete genome. The availability of transcribed sequences, such as those presented here, will be key to the facilitate genome annotation and gene discovery in T. solium.
Results
Here we present the sequencing and analysis of 2,857 ORESTES derived from T. solium cysticerci, revealing a fraction of the parasite transcriptome. A total of 1,520 high-quality ORESTES generated here were deposited in dbEST database of GenBank http://www.ncbi.nlm.nih.gov/dbEST, being 1,180 annotated as from T. solium [GenBank:EX150322 to EX151133 and GenBank:FD661301 to FD661668] and 340 corresponding to pig-derived sequences [GenBank:EX151134 to EX151473]. These sequences are also available at the STINGRAY system on the BiowebDB consortium website http://stingray.biowebdb.org/, together with relevant annotations and additional files. A list of the T. solium ORESTES and their respective GenBank accession numbers is presented on the Additional file 1.
General Features
A general overview of the T. solium ESTs generated here is presented in Table 1. More detailed analysis of the parasite transcriptome, such as codon usage and G+C content, can be obtained online at the STINGRAY system http://stingray.biowebdb.org/index.cgi?project=TS.
Table 1.
General features of Taenia solium cysticerci cDNA sequences.
| Total number of valid ORESTES* | 1,180 |
| Total number of non-redundant sequences | 812 |
| Average length of non-redundant sequences | 355 bp |
| Clusters | 185 |
| Singlets | 627 |
| Non-redundant sequences G+C average content | 49% |
| Number of non-redundant sequences hits on/with: | |
| - Blast | 424 (52.1%) |
| - RPSBlast | 150 (18.4%) |
| - InteProScan | 74 (9.0%) |
| - HMMER | 97 (12.0%) |
| - T. solium Genome ESTs | 107 (13.2%) |
| Sequences with no hits (blast/Interpro/HMMER) | 350 (43.0%) |
| Number of manually validated CDS | 191 (23.5%) |
| CDS G+C content average | 53% |
| Number of annotated sequences | 191 (23.5%) |
* After trimming by quality, length and rRNA/mtRNA, as described in the text.
A total of 2,857 clones were sequenced and, after removal of poor quality (Phred<15 and/or less than 100 bp) and less informative sequences (typically rRNA and mtRNA), the remaining 1,520 ORESTES were used for sequence assembly following detailed analysis by the STINGRAY. After assembling, sequences were arranged into distinct sets named 'Cysticerci' and 'Cysticerci PIGS', which are available at STINGRAY.
The 'Cysticerci' project http://stingray.biowebdb.org/index.cgi?project=TS corresponds to the parasite-derived transcriptome and contains a total of 1,180 ESTs clustered in 812 non-redundant sequences (185 clusters + 627 singlets), with an average size of 355 nt, totaling 288,496 nt.
The 'Cysticerci PIGS' dataset http://stingray.biowebdb.org/index.cgi?project=TP was determined on the basis of blast similarity analysis with high scores against genomic sequences of S. scrofa. It is composed of 340 non-redundant singlets with an average size of 390 nt and about 132,000 nt in total. The stringency criterion used here warrants that most of this subset is certainly composed of the host transcripts, which may include transcripts relevant for the host-parasite interaction.
The parasite transcriptome
Among the parasite's 812 non-redundant sequences (627 singlets + 185 clusters), 462 yielded significant hits with at least one of the databases used for comparative analysis http://stingray.biowebdb.org/index.cgi?project=TS. From these sequences with significant hits, 204 showed similarity with sequences of parasitic metazoan species, including sequences from the T. solium Genome Project (84), from other Taenia species such as T. saginata (2), T. crassiceps (2), T. asiatica (1), as well from other parasitic cestodes as E. granulosus (7) and E. multilocularis (4). Hits were also found against sequences from platyhelminths such as S. mansoni (88), S. japonicum (11), Clonorchis sinensis (1) and Fasciola hepatica (6). The remaining 263 sequences showed similarity with other metazoan species (see Additional file 2). For 350 sequences no hits were obtained on Blast, InterProScan or HMMER analyses.
After automated and manual annotation of all 812 non-redundant sequences, 191 were validated as coding sequences (CDS) (Table 1), of which 60 were considered hypothetical proteins or hypothetical conserved proteins. The number of ORESTES sequences according to their annotation identifiers is given in Additional file 3. As expected, this dataset enriched for coding sequences and showed a higher G+C content (53%) as compared to the total dataset (49%) (Table 1).
Analysis of the 191 annotated sequences using Gene Ontology (GO) allowed the categorization of 96 sequences, among which 84 were classified according to molecular function, 65 to biological processes and 48 to cellular component, several with multiple categories (Fig. 1). From the 65 sequences with biological processes annotation, the most frequent GO sub-categories were proteins related to cellular processes (40), followed by metabolic processes (10), biological regulation (4) and adhesion (4) (Fig. 2A). Among the GO molecular function sub-categories, binding (34), catalytic activity (24), structural molecule activity (14) and motor activity (7) were the most frequent (Fig. 2B). It is noteworthy that a relevant fraction of the transcripts revealed here appear to be related to structural aspects (such as adhesion, binding or structural molecule activity) that might be involved with the solid constitution of the cysts and their establishment on host tissues (see Additional file 4). A detailed description of each GO sub-category can be found on the annotated database available at the STINGRAY system http://stingray.biowebdb.org/index.cgi?project=TS.
Figure 1.
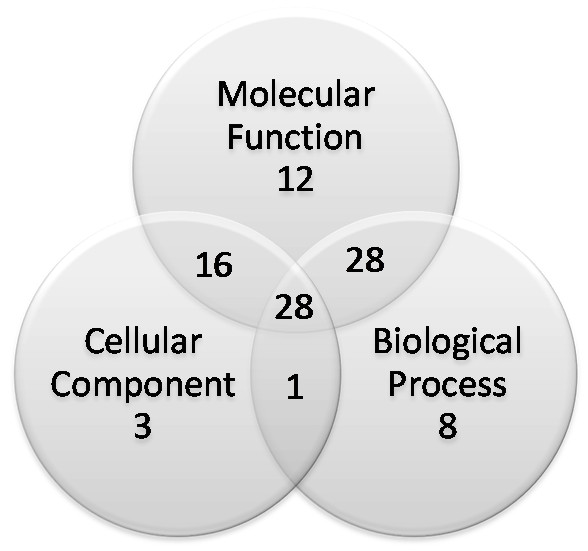
Curated Gene Ontology categorization of the Taenia solium transcriptomic sequences. Graphs shows the number of T. solium ORESTES sequences with curated Gene Ontology categorization of the distinct ontologies, Biological Process, Molecular Function and Cellular Component.
Figure 2.
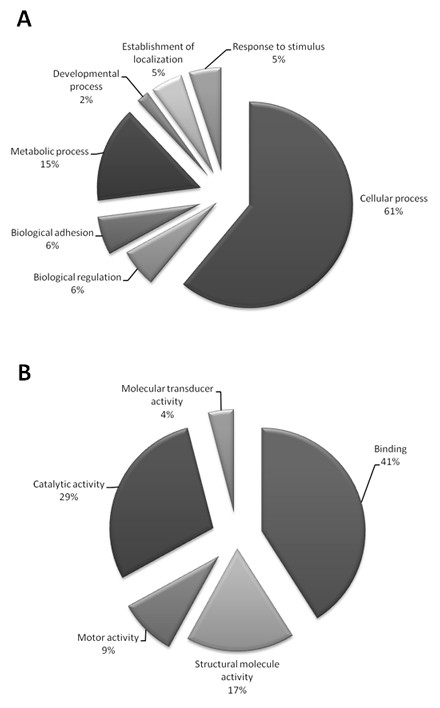
(A) Distribution of the Taenia solium sequences according to the Gene Ontology sub-categorization concerning Biological Process. The predominant category of the Biological Process ontology was Cellular Process in T. solium ORESTES sequences. (B) Distribution of the Taenia solium sequences according to Gene Ontology sub-categorization Molecular Function. Among the Molecular Function Ontology it was possible to attribute the Binding and Catalytic activity function terms to sets of sequences, indicative of a close relationship between host and parasite gene regulation during the process of cyst establishment and the host response to infection.
The search for predictive sub-cellular localization of the products related to each annotated CDS was performed using the Wolf-PSORT software [19] and returned 113 hits. Among the 13 sequences with predictions of having extracellular localization with high scores (>25), only seven have a putative function assigned by GO. Among these, three are probably not extracellular ('40S Ribosomal protein S19' [GenBank:EX150987], 'Deoxyribonuclease I' [GenBank:EX151091], and 'Sec61-like protein' [GenBank:EX150487]), two may have extracellular localization ('WD40 repeat' – [GenBank:EX150998] and 'Heat-shock protein' [GenBank:EX151058]), while the 'TolA protein' [GenBank:EX150587] and the 'Cadherin family member (cdh-4)' [GenBank: EX150991] are probably extracellular (see Additional file 5). Considering the Wolf-PSORT limitations in predicting cellular localization based on short sequences such as ESTs and the fact that none of the 113 proteins predicted as extracellular were annotated as antigens, even though most Taenia sp. proteins already reported in the literature have precisely that description, further analysis using full length sequences are necessary to confirm these results.
Conserved Domains and Motifs
The search for protein motifs among the parasite sequences was performed by similarity searches using InterProScan and RPSBlast using all databases available on the STINGRAY system and pointed out 64 distinct motifs distributed in 79 non-redundant sequences (see Additional files 6, 7 and 8). Among these, the 'pistil-specific extensin-like protein motif' [GenBank:IPR003882] was observed in 16 sequences, the 'vinculin/alpha-catenin' [GenBank:IPR006077] in 12, the 'glutelin' [GenBank:IPR000480] in six, 'fibronectin type III-like fold' [GenBank:IPR008957] in five and several others with four or less hits. Detailed categorization of the T. solium sequences according to the Eukaryotic Orthologous Groups (KOG) categories is shown on the Additional file 9.
A sensitive search for protein family recognition using multiple alignments was carried out with HMMER software and revealed 92 sequences of our parasite dataset generating at least one hit with the Pfam HMM profiles library. A domain with a still unknown function (DUF1787) was found in 12 sequences, the 'PT-PT repeat' in another nine sequences, the 'Hsp20/alpha crystallin family' (HSP20) and the 'I-set-immunoglobulin' (I-set domains) in four, the 'spectrin repeat' (SPECTRIN) and 'EGF-EGF-like domains' in another three sequences.
Comparisons with taeniid sequences
Only 117 of the 812 T. solium cysticerci clustered sequences described in the present study revealed similarity with the T. solium Genome Project ESTs available at GenBank. Among these 117 sequences, 107 showed similarity on tblastx and 100 on blastn analysis with ESTs of the T. solium Genome Project, 39 with exclusive hits to the larval stage sequence, 11 with the adult stage and 67 with genes expressed in both life-cycle stages (see Additional file 10).
Except for nine sequences from Taenia sp. or Echinococcus sp., the remaining cestode-related sequences presenting high score (>90) on blast against the T. solium sequences described in this study, were from Mesocestoides corti (heat shock 70 kDa protein) and from Diphyllobothrium dendriticum (actin). Further 36 low-score (<90) hits with the 28S ribosomal RNA from distinct cestode species were observed.
Comparative analysis against E. granulosus sequences from GenBank mainly revealed constitutive genes such as actin, paramyosin and others metabolic enzymes. However, two clusters [GenBank:EX151048, GenBank:EX151014] showed high similarity with genes coding for ERM family proteins (ezrin, radixin, moesin), exclusively with EST from larval stage of T. solium (see Additional files 6 and 8). Some of these proteins were characterized in Echinococcus species and received distinct names such as EM10, EG10, EM4 and antigen II/3, despite their high nucleotide similarity. In E. granulosus and E. multilocularis these antigens are basically found in the germinal layer of brood capsules and in the tegument of protoscolices, associated with larval stage. Gonzales et al. 2007 [20], showed that the TEG-Tsag gene of T. saginata is homologous to EM10 and EG10 genes of Echinococcus spp. and 97% identical to its T. solium homologue. However, alignment of this T. solium gene with the two clusters sequences described in the present study [GenBank:EX151048, GenBank:EX151014] clearly showed high sequence variability, despite the conserved blocks. The TEG molecules are characterized by an N-terminal FERM domain and a C-terminal ERM domain which are found in a number of cytoskeletal-associated proteins located at the interface between the plasma membrane and the cytoskeleton and in proteins interacting with lipid membranes. Thus TEG protein may play a role in tegument function and interaction with the host.
Genes of interest
A number of transcripts identified here could be of interest for further study (see Additional file 11). At least 30 genes coding for proteins potentially involved in parasite development, including transcriptional factors, component of chromatin remodelling complexes, cell adhesion-related molecules, receptors and other transducing components of signalling pathways have been identified. Moreover, putative orthologues of two proteins possibly associated to invertebrate immunity were identified for the first time in T. solium cysticerci: a 'heat shock 90 kDa protein' [GenBank:EX150676] and an 'anaphylatoxin-like domain protein' [GenBank:EX150322, GenBank:EX150873].
Fifty-one T. solium ORESTES revealed similarity with known antigens, including five previously characterized helminth antigens with potential for development of immunodiagnostic and/or vaccines. These are 'paramyosin' [GenBank:EL745866, GenBank:EL750686, GenBank:EL762552], 'major egg antigen' [GenBank:EL740635, GenBank:EL758824, GenBank:EL760346], 'cathepsin L-like cysteine proteinase' [GenBank:EL742569], 'heat shock 70 kDa protein' [GenBank:EL740975, GenBank:EL740984, GenBank:EL741400, GenBank:EL744008, GenBank:EL744338, GenBank:EL745376, GenBank:EL747548, GenBank:EL747588] and the 'H17g' or 'TEG-Tsol surface antigen' [GenBank:AJ581299], which is highly conserved among T. solium and T. saginata.
Discussion
Transcriptome investigations have greatly benefited from the recent maturation of gene expression approaches. Among these, the microarray has evolved as the most prominent high-throughput method to assess a given expression profile. However, they are still subjected to hybridization issues such as reaction kinetics and probe mismatches as former methods. Also, microarrays cannot adequately address expression profiles of samples containing mixed species, which are yielded in studies of most parasite interactions. In these situations, the use of short gene tags, such as SAGE [21] is also problematic, due to the ambiguous tag-to-gene assignment and the difficulties of gene identification, especially in situations when the genome and/or the transcriptome of one of the species is not available. By comparison, the generation of longer sequence tags, such as those derived from EST or ORESTES, can facilitate gene discovery and annotation and also provides a much less ambiguous tag-to-gene mapping.
As formerly shown, ORESTES is able to give a normalized transcriptome view, as well as to characterize sequences from the central portion of the genes, including the less-abundant transcript markers [10,11,22-24]. The normalization capability of ORESTES, together with its ability to sample the central portion of genes makes this approach complementary to traditional ESTs, more frequently used in large-scale cDNA sequencing projects. Thus, as we have shown before for other species, including humans [11], S. mansoni [12,13], Drosophila melanogaster [25] or Apis melifera [26], ORESTES provides a distinct contribution to gene discover in T. solium. The present study shows the first comparative sequence analysis of the T. solium transcriptome using ORESTES from the larval stage (cysticerci).
Comparison of the T. solium ORESTES generated in this study with all T. saginata and T. solium sequences retrieved from GenBank showed identical hits with both datasets, indicating a high level of conservation in genes like 'Tsp36 small heat shock protein'. Few hits were obtained from other taeniids (T. crassiceps and T. asiatica), which may be due to their small sequence datasets or to the higher distance from T. solium and these other species. As an example of such intra-genus variability, T. asiatica is morphologically similar to T. saginata occurring in almost all Asian countries being capable of infecting pigs and humans [27] possibly leading to cysticercosis, but unlikely neurocysticercosis [28].
Since only 119 of the T. solium sequences described in this work showed similarity to T. solium Genome Project sequences, our results significantly contribute to the knowledge of the parasite expression profile by increasing the number of sequenced transcripts and through functional annotation of several genes. Thus, the present report is complementary to the T. solium genome initiative and may be helpful on the parasite genome assembly and annotation [29], as well as on studies of intra- and inter-specific sequence variability.
Considering the overall picture of the T. solium cysticerci transcriptome presented in this work, comparative sequence analysis revealed 350 sequences (43%) producing hits with a database. Despite the small dataset, it is interesting to note that Aguilar-Diaz et al. [1] found a very similar picture in the analysis of the transcriptome of adult worms, with 40% of the genes showing no hits. A systematic, functional investigation of these unknown genes using postgenomic tools such as "gene knockout" or RNA-mediated "knockdown" is desirable.
Several protein domains related to cell structure, including cell wall organization, were found among the generated sequences. The pistil-specific extensin-like and the vinculin/alpha-catenin motifs found in this study are of special interest due their role in cell wall structure and interaction. According to Interpro, the pistil-specific extensin-like protein motif [Interpro:IPR003882] is frequently found in the cell-wall proteins of many plants, and can account for up to 20% of their dry weight. Interestingly, this motif is also found in metazoans like Brugia malayi [Interpro:A8Q5T0/A8QDB8] and C. elegans [Interpro:Q20327]. Since extensin-like proteins in plants are involved in cell wall strengthening in response to mechanical stress, such as attack by pests or plant-bending in the wind, it is reasonable to hypothesize a similar role on the T. solium cysts walls, conferring rigidity with a possible role in parasite defense.
The vinculin and/or alpha-catenin are eukaryotic actin-binding protein motifs, usually containing proline-rich motifs and several ligand-biding motifs. Vinculins are frequently used as markers for cell-cell and cell-extracellular matrix junctions, named as focal adhesions, also interacting with other structural proteins such as talin and alpha-actinins [30]. It is tempting to speculate that proteins containing these motifs may have a function on the organization of the cysticerci wall as well as on the interaction with host's tissues.
Oxidative and other types of stress are inherent to the host environment to which a parasite is exposed. Therefore, proteins that allow the cysticerci to cope with stress may be important in infection maintenance. In this study several heat shock proteins (hsp16, hsp20, hsp25, hsp70, hsp86, and hsp90) and other stress response-related proteins have been identified as being transcribed by this developmental stage. Previous studies with T. solium cysticerci showed that the expression of 70 and 80 kDa heat shock proteins was highly induced under temperature stress [31]. Recently, another heat shock protein of 35.4 kDa was described for T. solium cysticerci and points out the importance of such proteins for the parasite life cycle [32].
The host immune response to tissue parasitism is an important aspect to the establishment and development of the neurocysticercosis pathology. In this study, the 'heat shock 90 kDa' protein and the 'anaphylatoxin-like domain' (a complement-associated protein in vertebrates) – which are described for the first time for the T. solium cysticerci – have been associated to a possible immune response in invertebrates [33,34] and along with the host immunity may be involved on the host-parasite immunological interplay.
Among several genes related to the antigenic coat of the parasite, the TEG-Tsol gene is of major importance for both immune diagnostic and vaccine development, due to its high antigenicity, strong similarity (~97%) between T. solium and T. saginata paralogs, conservation among other taeniid species and reactivity to distinct animal sera [20]. TEG-Tsol was found among the ORESTES in the present study and corresponds to the major protoscolex surface antigen detected in E. granulosus (EG10) and E. multilocularis (EM10) [35], which is also expressed in the oncospheres and on adult tapeworm tegument of both T. solium and T. saginata, as well as on the tegument of the T. solium cysticerci [36-38].
Despite some encouraging results on vaccine development [39-41], several studies have pointed out intra- and inter-specific variability of taeniid species at both genotypic and phenotypic levels [26,42-50], which may represent a problem for the global-scale use of single- or multi-antigen recombinant vaccines. Thus, genome and transcriptome sequences – especially when derived from parasites collected at different endemic areas – are of major importance to address such variability and to point new vaccine and diagnostic/prognostic candidate markers. In this context, differently from genomic markers, ESTs are powerful tools not only to indicate potentially relevant candidates, but also to provide experimental evidence of expression specific developmental stages.
Conclusion
The sequencing effort presented here is complementary to the T. solium Genome Project, having described several unknown genes for this species, which may have direct and immediate applications on diagnosis, therapeutics and/or vaccine development. Furthermore, this database represents part of a key resource to understanding aspects of the cysticerci biology and host/parasite interaction. Considering the ongoing efforts to sequence the hydatid disease agents (E. granulosus and E. multilocularis) along with the T. solium Genome Project [1,28], we hope our results can contribute to the development of comparative parasitic metazoan genomics, yielding new molecular diagnosis targets [51] and new insights into the pathogenesis of cysticercosis and taeniasis.
Methods
Cysticerci collection
Taenia solium cysticerci were collected from a naturally infected, landrace, bred pig (Sus scrofa). The animal was humanely sacrificed and cysticerci, spontaneously detached from abdominal and thoracic muscles were recovered and carefully micro-dissected to remove any tissue fragments that remained attached. Cysts were extensively washed with phosphate-buffered saline and immediately stored at -80°C. The study was previously approved by the Ethics Committee on Animal Research of the Faculty of Animal Science and Food Engineering (FZEA) of Universidade de São Paulo (USP), and was carried out following the institution's guidelines for animal husbandry.
RNA extraction, RT-PCR and cDNA libraries preparation
Total RNA was obtained from cysticerci using the Trizol® (Invitrogen, Carlsbad). Messenger RNA (mRNA) was purified using the μMACs mRNA isolation kit (Miltenyi Biotec, Bergisch Gladbach), following manufacturer's directions, as described [52]. mRNA concentration was evaluated by spectrophotometry (U-3010 Hitachi, Tokyo, Japan) and 25 ng mRNA aliquots were frozen for the posterior generation of ORESTES amplification profiles as described [12]. Briefly, cDNA was synthesized and amplified with some of the oligonucleotide primers previously used in the S. mansoni transcriptome project [12]. Twenty cDNA mini-libraries were constructed using ORESTES and a set of different oligonucleotide primers (see Additional file 12). The amplification profiles were evaluated in ethidium bromide-stained agarose gels, cloned in pGEM-T-Easy plasmids (Promega Corporation, Madison, USA) and used for Escherichia coli (strain DH10β) transformation. Recombinant clones were obtained by selective growth (X-Gal, IPTG and ampicillin), screened by PCR amplification of the insert using primers pGEM-F (5'-ACG CCA AGC TAT TTA GGT GAC ACT ATA-3') and EXCEL-R (5'-GTT GTA AAA CGA CGG CCA GTG AAT-3') and stored as glycerol stocks at -80°C. For sequencing, the bacterial clones were grown in LB medium for 20 hours at 37°C, followed by plasmid DNA extraction by alkaline lysis according to standard protocols [53].
DNA sequencing and analysis
ORESTES sequencing was carried out by two laboratories located at UFSC and USP using the DYEnamic® ET Dye Terminator kit (GE Healthcare, Fairfield) or ThermoSequenase II dye terminator cycle sequencing kit (Amersham-Pharmacia Biotech) in a MegaBace 1000® DNA Analysis System (GE Healthcare) and on a ABI PRISM® 3100 Genetic Analyzer (Applied Biosystems, Foster City), respectively. Briefly, each sequencing reaction used 5 pmol of pGEM-F or EXCEL-R oligonucleotides, and PCR products [54] or plasmid DNA as templates. The labeling conditions were: 95°C/25 sec., 35 cycles of 95°C/15 sec., 50°C/20 sec. and 60°C/90 sec. The products were then precipitated (70% isopropanol), injected at 2 KV for 100 sec. and electrophoresed for 140 min. at 7 KV.
Sequence analysis was performed using the STINGRAY system (System for Integrated Genomic Resources and Analysis), an improved version of the formerly published GARSA system (Genomic Analysis Resources for Sequence Annotation) [55]. Briefly, the system workflow initially performs evaluation of the quality of the obtained chromatograms (cut-off Phred ≥ 15) following removal of vector sequences through Phred and Cross-match [56,57] and then clustering the sequences using CAP3 [58]. Following similarity searches performed by Blast (Basic Local Alignment Tool), Psi-Blast (Position-Specific Integrated Blast), RPSBlast (Reverse Position-specific Blast) [59], InterProScan [60] and HMMER (Hidden Markov Models for sequence profile analysis) [61] packages against local pre-formatted databases, blast analysis was also performed using all EST sequences from the T. solium Genome Project and the Sus scrofa genome available at GenBank. After removal of ribosomal RNA (rRNA) sequences, blast analysis against the Sus scrofa genome was used to separate parasite sequences from host sequences, creating two datasets that were evaluated separately. Functional annotation was performed using the Gene Ontology (GO) vocabulary as described by Jones et al. [62] and putative sub-cellular localization of each coding sequence was performed through the Wolf-PSORT program [63]. The G+C content of singlets and clusters was estimated by the GeeCee program (EMBOSS – European Molecular Biology Open Software Suite – package) and the tRNA sequences were predicted by tRNAscan-SE [64].
The results were then individually and manually checked during annotation, when sequences were validated as CDS when presenting i) high similarity values (e-value < = e-15 and similarity>75%) with protein databases (uniprot_swissprot, uniprot_trembl, uniref90, refseq_protein) or with protein sequences from phylogenetically related organisms (Cestoda and/or Trematoda) available on GenBank, ii) the presence of conserved domains as revealed by RPS-Blast against CDD (see Additional file 6), COG (see Additional file 7) and KOG databases (see Additional file 8); iii) the presence of protein domains as revealed by InterProScan and HMMER and iv) annotations on Gene Ontology analysis, when available. T. solium sequences having no protein domain and showing exclusive hits with high similarity values (e-value < = e-15 and similarity>75%) with 'hypothetical proteins' or 'hypothetical conserved proteins' from GenBank were annotated accordingly.
The T. solium cysticerci annotated transcripts, the host-parasite transcribed sequences, all databases used for comparative analysis as well as the additional material to this work are available online at the STINGRAY system http://stingray.biowebdb.org/index.cgi?project=TS.
Abbreviations
ESTs: Expressed Sequence Tags; ORESTES: Open Reading frame Expressed Sequence Tags; PCR: Polymerase Chain Reaction.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
CRA and PHS are the main authors. CRA, PHS, GW, AAMM, ECG and EDN have equally contributed to this work. TCMS, GR, EBS, JBR, MMS, AZ, HBF, KT and AMRD have participated on the sequence analysis. All authors have participated on the manuscript preparation.
Authors' informations
GW, EBS, GR and MMS are recipients of CNPq scholarships. PHS and TCMS are recipients of CAPES scholarships. ECG is currently a CNPq Post-Doctoral Fellow at BMRC/UEA, UK. EDN is a visiting scientist at MD Anderson Cancer Center, Houston TX, USA.
Supplementary Material
Table S1. List of Taenia solium ESTs and their GenBank accession numbers.
Table S3. Distribution of the Taenia solium EST according to their characteristics and similarity analysis.
Table S4. Number of Taenia solium ORESTES according to their annotation as coding sequences (CDS).
Table S5. Distribution of Taenia solium EST hits according to the Gene Onthology (GO) classification.
Table S6. Taenia solium ESTs with predicted extra-cellular sub-localization by Wolf-PSORT.
Table S7. Number of Taenia solium EST eliciting hits with the CDD database.
Table S8. Number of Taenia solium EST eliciting hits with the COG database.
Table S9. Number of Taenia solium EST eliciting hits with the KOG database.
Picture S1. Distribution of the most frequent Eukaryotic Orthologous Groups categories observed for the Taenia solium sequences.
Table S10. List of Taenia solium EST with similarity to the parasite genome project sequences.
Table S11. List of interesting genes found in Taenia solium ESTs.
Table S2. List of cDNA mini-libraries, primers used for generation of the Taenia solium EST profiles and the number of sequences obtained in each library.
Acknowledgments
Acknowledgements
This work was supported by Associação Beneficente Alzira Denise Hertzog da Silva (ABADHS) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). The funders had no role in study design, data collection and analysis, decision to publish, or on the preparation of the manuscript.
Contributor Information
Carolina R Almeida, Email: Carolina.Almeida@appliedbiosystems.com.
Patricia H Stoco, Email: patriciastoco@gmail.com.
Glauber Wagner, Email: glauber.wagner@unoesc.edu.br.
Thaís CM Sincero, Email: thais@ccb.ufsc.br.
Gianinna Rotava, Email: gianinna_rotava@yahoo.com.br.
Ethel Bayer-Santos, Email: ethel.bs@gmail.com.
Juliana B Rodrigues, Email: juliberka@gmail.com.
Maísa M Sperandio, Email: maisamichels@hotmail.com.
Antônio AM Maia, Email: antomaia@usp.br.
Elida PB Ojopi, Email: elida@usp.br.
Arnaldo Zaha, Email: zaha@cbiot.ufrgs.br.
Henrique B Ferreira, Email: henrique@cbiot.ufrgs.br.
Kevin M Tyler, Email: k.tyler@uea.ac.uk.
Alberto MR Dávila, Email: davila@fiocruz.br.
Edmundo C Grisard, Email: grisard@ccb.ufsc.br.
Emmanuel Dias-Neto, Email: emmanuel@usp.br.
References
- Aguilar-Diaz H, Bobes RJ, Carrero JC, Camacho-Carranza R, Cervantes C, Cevallos MA, Davila G, Rodriguez-Dorantes M, Escobedo G, Fernandez JL, Fragoso G, Gaytan P, Garciarubio A, Gonzalez VM, Gonzalez L, Jose MV, Jimenez L, Laclette JP, Landa A, Larralde C, Morales-Montor J, Morett E, Ostoa-Saloma P, Sciutto E, Santamaria RI, Soberon X, De La Torre P, Valdes V, Yanez J. The genome project of Taenia solium. Parasitol Int. 2006;55:S127–130. doi: 10.1016/j.parint.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Sciutto E, Fragoso G, Fleury A, Laclette JP, Sotelo J, Aluja A, Vargas L, Larralde C. Taenia solium disease in humans and pigs: an ancient parasitosis disease rooted in developing countries and emerging as a major health problem of global dimensions. Microbes Infect. 2000;2:1875–1890. doi: 10.1016/s1286-4579(00)01336-8. [DOI] [PubMed] [Google Scholar]
- Campbell G, Garcia HH, Nakao M, Ito A, Craig PS. Genetic variation in Taenia solium. Parasitol Int. 2006;55:S121–126. doi: 10.1016/j.parint.2005.11.019. [DOI] [PubMed] [Google Scholar]
- Garcia HH, Gonzalez AE, Evans CA, Gilman RH. Taenia solium cysticercosis. Lancet. 2003;362:547–556. doi: 10.1016/S0140-6736(03)14117-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A, Nakao M, Wandra T. Human Taeniasis and cysticercosis in Asia. Lancet. 2003;362:1918–1920. doi: 10.1016/S0140-6736(03)14965-3. [DOI] [PubMed] [Google Scholar]
- Román G, Sotelo J, Del Brutto O, Flisser A, Dumas M, Wadia N, Botero D, Cruz M, Garcia H, De Bittencourt PR, Trelles L, Arriagada C, Lorenzana P, Nash TE, Spina-França A. A proposal to declare neurocysticercosis an international reportable disease. Bull World Health Organ. 2000;78:399–406. [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Sharma BS. Neurocysticercosis: A review of current status and management. J Clin Neuroscience. 2009;16:867–876. doi: 10.1016/j.jocn.2008.10.030. [DOI] [PubMed] [Google Scholar]
- Fleury A, Dessein A, Preux PM, Dumas M, Tapia G, Larralde C, Sciutto E. Symptomatic human neurocysticercosis–age, sex and exposure factors relating with disease heterogeneity. J Neurol. 2004;251:830–837. doi: 10.1007/s00415-004-0437-9. [DOI] [PubMed] [Google Scholar]
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, Kerlavage AR, Mccombie WR, Venter JC. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Dias-Neto E, Harrop R, Correa-Oliveira R, Wilson RA, Pena SD, Simpson AJ. Minilibraries constructed from cDNA generated by arbitrarily primed RT-PCR: an alternative to normalized libraries for the generation of ESTs from nanogram quantities of mRNA. Gene. 1997;186:135–142. doi: 10.1016/s0378-1119(96)00699-3. [DOI] [PubMed] [Google Scholar]
- Dias-Neto E, Correa RG, Verjovski-Almeida S, Briones MR, Nagai MA, Da Silva W, Jr, Zago MA, Bordin S, Costa FF, Goldman GH, Carvalho AF, Matsukuma A, Baia GS, Simpson DH, Brunstein A, Oliveira PS, Bucher P, Jongeneel CV, O'hare MJ, Soares F, Brentani RR, Reis LF, Souza SJ, Simpson AJ. Shotgun sequencing of the human transcriptome with ORF expressed sequence tags. Proc Natl Acad Sci USA. 2000;97:3491–3496. doi: 10.1073/pnas.97.7.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verjovski-Almeida S, Demarco R, Martins EA, Guimaraes PE, Ojopi EP, Paquola AC, Piazza JP, Nishiyama MY, Kitajima JP, Adamson RE, Ashton PD, Bonaldo MF, Coulson PS, Dillon GP, Farias LP, Gregorio SP, Ho PL, Leite RA, Malaquias LC, Marques RC, Miyasato PA, Nascimento AL, Ohlweiler FP, Reis EM, Ribeiro MA, Sa RG, Stukart GC, Soares MB, Gargioni C, Kawano T, Rodrigues V, Madeira AM, Wilson RA, Menck CF, Setubal JC, Leite LC, Dias-Neto E. Transcriptome analysis of the acoelomate human parasite Schistosoma mansoni. Nature Genet. 2003;35:148–157. doi: 10.1038/ng1237. [DOI] [PubMed] [Google Scholar]
- Verjovski-Almeida S, Leite LC, Dias-Neto E, Menck CF, Wilson RA. Schistosome transcriptome: insights and perspectives for functional genomics. Trends Parasitol. 2004;20:304–308. doi: 10.1016/j.pt.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Lockyer AE, Spinks JN, Walker AJ, Kane RA, Noble LR, Rollinson D, Dias-Neto E, Jones CS. Biomphalaria glabrata transcriptome: identification of cell-signalling, transcriptional control and immune-related genes from open reading frame expressed sequence tags (ORESTES) Developmental and Comparative Immunology. 2007;31:763–782. doi: 10.1016/j.dci.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Yan Q, Shen DK, Liu F, Zhu ZD, Song HD, Xu XR, Wang ZJ, Rong YP, Zeng LC, Wu J, Zhang X, Wang JJ, Xu XN, Wang SY, Fu G, Zhang XL, Wang ZQ, Brindley PJ, Mcmanus DP, Xue CL, Feng Z, Chen Z, Han ZG. Evolutionary and biomedical implications of a Schistosoma japonicum complementary DNA resource. Nature Genet. 2003;35:139–147. doi: 10.1038/ng1236. [DOI] [PubMed] [Google Scholar]
- Fernandez C, Gregory WF, Loke P, Maizels RM. Full-length-enriched cDNA libraries from Echinococcus granulosus contain separate populations of oligo-capped and trans-spliced transcripts and a high level of predicted signal peptide sequences. Mol Bioch Parasitol. 2002;122:171–180. doi: 10.1016/s0166-6851(02)00098-1. [DOI] [PubMed] [Google Scholar]
- Brehm K, Wolf M, Beland H, Kroner A, Frosch M. Analysis of differential gene expression in Echinococcus multilocularis larval stages by means of spliced leader differential display. Int J Parasitol. 2003;33:1145–1159. doi: 10.1016/s0020-7519(03)00169-3. [DOI] [PubMed] [Google Scholar]
- Espinoza I, Galindo M, Bizarro CV, Ferreira HB, Zaha A, Galanti N. Early post-larval development of the endoparasitic platyhelminth Mesocestoides corti: trypsin provokes reversible tegumental damage leading to serum-induced cell proliferation and growth. J Cell Physiol. 2005;205:211–217. doi: 10.1002/jcp.20382. [DOI] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucl Acids Res. 2007;35:W585–587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez LM, Ferrer E, Spickett A, Michael LM, Vatta AF, Garate T, Harrison LJ, Parkhouse RM. The Taenia saginata homologue of the major surface antigen of Echinococcus spp. is immunogenic and 97% identical to its Taenia solium homologue. Parasitol Res. 2007;101:1541–1549. doi: 10.1007/s00436-007-0673-x. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Brentani H, Caballero OL, Camargo AA, Da Silva AM, Da Silva WA, Jr, Dias-Neto E, Grivet M, Gruber A, Guimaraes PE, Hide W, Iseli C, Jongeneel CV, Kelso J, Nagai MA, Ojopi EP, Osorio EC, Reis EM, Riggins GJ, Simpson AJ, De Souza S, Stevenson BJ, Strausberg RL, Tajara EH, Verjovski-Almeida S, Acencio ML, Bengtson MH, Bettoni F, Bodmer WF, Briones MR, Camargo LP, Cavenee W, Cerutti JM, Coelho Andrade LE, Costa dos Santos PC, Ramos Costa MC, da Silva IT, Estécio MR, Sa Ferreira K, Furnari FB, Faria M, Jr, Galante PA, Guimaraes GS, Holanda AJ, Kimura ET, Leerkes MR, Lu X, Maciel RM, Martins EA, Massirer KB, Melo AS, Mestriner CA, Miracca EC, Miranda LL, Nobrega FG, Oliveira PS, Paquola AC, Pandolfi JR, Campos Pardini MI, Passetti F, Quackenbush J, Schnabel B, Sogayar MC, Souza JE, Valentini SR, Zaiats AC, Amaral EJ, Arnaldi LA, De Araújo AG, De Bessa SA, Bicknell DC, Ribeiro De Camaro ME, Carraro DM, Carrer H, Carvalho AF, Colin C, Costa F, Curcio C, Guerreiro da Silva ID, Pereira da Silva N, Dellamano M, El-Dorry H, Espreafico EM, Scattone Ferreira AJ, Ayres Ferreira C, Fortes MA, Gama AH, Giannella-Neto D, Giannella ML, Giorgi RR, Goldman GH, Goldman MH, Hackel C, Ho PL, Kimura EM, Kowalski LP, Krieger JE, Leite LC, Lopes A, Luna AM, Mackay A, Mari SK, Marques AA, Martins WK, Montagnini A, Mourão Neto M, Nascimento AL, Neville AM, Nobrega MP, O'Hare MJ, Otsuka AY, Ruas De Melo AI, Paco-Larson ML, Guimarães Pereira G, Pereira da Silva N, Pesquero JB, Pessoa JG, Rahal P, Rainho CA, Rodrigues V, Rogatto SR, Romano CM, Romeiro JG, Rossi BM, Rusticci M, Guerra De Sá R, Sant' Anna SC, Sarmazo ML, Silva TC, Soares FA, Sonati Mde F, De Freitas Sousa J, Queiroz D, Valente V, Vettore AL, Villanova FE, Zago MA, Zalcberg H, Human Cancer Genome Project/Cancer Genome Anatomy Project Annotation Consortium; Human Cancer Genome Project Sequencing Consortium The generation and utilization of a cancer-oriented representation of the human transcriptome by using expressed sequence tags. Proc Natl Acad Sci USA. 2003;100:13418–13423. doi: 10.1073/pnas.1233632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo AA, Samaia HP, Dias-Neto E, Simão DF, Migotto IA, Briones MR, Costa FF, Nagai MA, Verjovski-Almeida S, Zago MA, Andrade LE, Carrer H, El-Dorry HF, Espreafico EM, Habr-Gama A, Giannella-Neto D, Goldman GH, Gruber A, Hackel C, Kimura ET, Maciel RM, Marie SK, Martins EA, Nobrega MP, Paco-Larson ML, Pardini MI, Pereira GG, Pesquero JB, Rodrigues V, Rogatto SR, da Silva ID, Sogayar MC, Sonati MF, Tajara EH, Valentini SR, Alberto FL, Amaral ME, Aneas I, Arnaldi LA, De Assis AM, Bengtson MH, Bergamo NA, Bombonato V, De Camargo ME, Canevari RA, Carraro DM, Cerutti JM, Correa ML, Correa RF, Costa MC, Curcio C, Hokama PO, Ferreira AJ, Furuzawa GK, Gushiken T, Ho PL, Kimura E, Krieger JE, Leite LC, Majumder P, Marins M, Marques ER, Melo AS, Melo MB, Mestriner CA, Miracca EC, Miranda DC, Nascimento AL, Nobrega FG, Ojopi EP, Pandolfi JR, Pessoa LG, Prevedel AC, Rahal P, Rainho CA, Reis EM, Ribeiro ML, da Ros N, De Sa RG, Sales MM, Sant'anna SC, dos Santos ML, da Silva AM, da Silva NP, Silva WA, Jr, da Silveira RA, Sousa JF, Stecconi D, Tsukumo F, Valente V, Soares F, Moreira ES, Nunes DN, Correa RG, Zalcberg H, Carvalho AF, Reis LF, Brentani RR, Simpson AJ, De Souza SJ. The contribution of 700,000 ORF sequence tags to the definition of the human transcriptome. Proc Natl Acad Sci USA. 2001;98:12103–12108. doi: 10.1073/pnas.201182798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis EM, Ojopi EP, Alberto FL, Rahal P, Tsukumo F, Mancini UM, Guimaraes GS, Thompson GM, Camacho C, Miracca E, Carvalho AL, Machado AA, Paquola AC, Cerutti JM, Da Silva AM, Pereira GG, Valentini SR, Nagai MA, Kowalski LP, Verjovski-Almeida S, Tajara EH, Dias-Neto E, Head, Neck Consortium Large-scale transcriptome analyses reveal new genetic marker candidates of head, neck, and thyroid cancer. Cancer Res. 2005;65:1693–1699. doi: 10.1158/0008-5472.CAN-04-3506. [DOI] [PubMed] [Google Scholar]
- Maia RM, Valente V, Cunha MA, Sousa JF, Araujo DD, Silva W, Zago MA, Dias-Neto E, Souza SJ, Simpson AJ, Monesi N, Ramos RG, Espreafico EM, Paco-Larson ML. Identification of unannotated exons of low abundance transcripts in Drosophila melanogaster and cloning of a new serine protease gene upregulated upon injury. BMC Genomics. 2007;8:249. doi: 10.1186/1471-2164-8-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes FM, Valente V, Sousa JF, Cunha MA, Pinheiro DG, Maia RM, Araujo DD, Costa MC, Martins WK, Carvalho AF, Monesi N, Nascimento AM, Peixoto PM, Silva MF, Ramos RG, Reis LF, Dias-Neto E, Souza SJ, Simpson AJ, Zago MA, Soares AE, Bitondi MM, Espreafico EM, Espindola FS, Paco-Larson ML, Simoes ZL, Hartfelder K, Silva WA. The use of Open Reading frame ESTs (ORESTES) for analysis of the honey bee transcriptome. BMC Genomics. 2004;5:84. doi: 10.1186/1471-2164-5-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A, Yamasaki H, Nakao M, Sako Y, Okamoto M, Sato MO, Nakaya K, Margono SS, Ikejima T, Kassuku AA, Afonso SM, Ortiz WB, Plancarte A, Zoli A, Geerts S, Craig PS. Multiple genotypes of Taenia solium-ramifications for diagnosis, treatment and control. Acta Trop. 2003;87:95–101. doi: 10.1016/s0001-706x(03)00024-x. [DOI] [PubMed] [Google Scholar]
- Galan-Puchades MT, Fuentes MV. Taenia asiatica intermediate hosts. Lancet. 2004;363:660. doi: 10.1016/S0140-6736(04)15607-9. [DOI] [PubMed] [Google Scholar]
- Garciarrubio A, Bobes RJ, Carrero JC, Cevallos MA, Fragoso G, González VM, José MV, Landa A, Larralde C, Mendoza L, Morales-Montor J, Morett1 E, Sciutto E, Soberón X, Laclette JP. The Genome Project of Taenia solium. International Journal of Infectious Diseases. 2008;12:e395. [Google Scholar]
- Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Vargas-Parada L, Solis CF, Laclette JP. Heat shock and stress response of Taenia solium and T. crassiceps (Cestoda) Parasitol. 2001;122:583–588. doi: 10.1017/s0031182001007764. [DOI] [PubMed] [Google Scholar]
- Ferrer E, Gonzalez LM, Foster-Cuevas M, Cortez MM, Davila I, Rodriguez M, Sciutto E, Harrison LJ, Parkhouse RM, Garate T. Taenia solium: characterization of a small heat shock protein (Tsol-sHSP35.6) and its possible relevance to the diagnosis and pathogenesis of neurocysticercosis. Exp Parasitol. 2005;110:1–11. doi: 10.1016/j.exppara.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Nair SV, Ramsden A, Raftos DA. Ancient origins: complement in invertebrates. Invert Surv J. 2005;2:114–123. [Google Scholar]
- Robert J. Evolution of heat shock protein and immunity. Devel Comp Immunol. 2003;27:449–464. doi: 10.1016/s0145-305x(02)00160-x. [DOI] [PubMed] [Google Scholar]
- Felleisen R, Gottstein B. Echinococcus multilocularis: molecular and immunochemical characterization of diagnostic antigen II/3–10. Parasitol. 1993;107:335–342. doi: 10.1017/s0031182000079300. [DOI] [PubMed] [Google Scholar]
- Ferrer E, Gonzalez LM, Martinez-Escribano JA, Gonzalez-Barderas ME, Cortez MM, Davila I, Harrison LJ, Parkhouse RM, Garate T. Evaluation of recombinant HP6-Tsag, an 18 kDa Taenia saginata oncospheral adhesion protein, for the diagnosis of cysticercosis. Parasitol Res. 2007;101:517–525. doi: 10.1007/s00436-007-0507-x. [DOI] [PubMed] [Google Scholar]
- Rosas G, Fragoso G, Garate T, Hernandez B, Ferrero P, Foster-Cuevas M, Parkhouse RM, Harrison LJ, Briones SL, Gonzalez LM, Sciutto E. Protective immunity against Taenia crassiceps murine cysticercosis induced by DNA vaccination with a Taenia saginata tegument antigen. Microbes Infect Pasteur. 2002;4:1417–1426. doi: 10.1016/s1286-4579(02)00025-4. [DOI] [PubMed] [Google Scholar]
- Frosch PM, Mühlschlegel F, Sygulla L, Hartmann M, Frosch M. Identification of a cDNA clone from the larval stage of Echinococcus granulosus with homologies to the E. multilocularis antigen EM10-expressing cDNA clone. Parasitol Res. 1994;80:703–705. doi: 10.1007/BF00932958. [DOI] [PubMed] [Google Scholar]
- Cruz-Revilla C, Toledo A, Rosas G, Huerta M, Flores-Perez I, Pena N, Morales J, Cisneros-Quinones J, Meneses G, Diaz-Orea A, Anciart N, Goldbaum F, Aluja A, Larralde C, Fragoso G, Sciutto E. Effective protection against experimental Taenia solium tapeworm infection in hamsters by primo-infection and by vaccination with recombinant or synthetic heterologous antigens. J Parasitol. 2006;92:864–867. doi: 10.1645/GE-779R1.1. [DOI] [PubMed] [Google Scholar]
- Flisser A, Gauci CG, Zoli A, Martinez-Ocana J, Garza-Rodriguez A, Dominguez-Alpizar JL, Maravilla P, Rodriguez-Canul R, Avila G, Aguilar-Vega L, Kyngdon C, Geerts S, Lightowlers MW. Induction of protection against porcine cysticercosis by vaccination with recombinant oncosphere antigens. Infect Immun. 2004;72:5292–5297. doi: 10.1128/IAI.72.9.5292-5297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flisser A, Rodriguez-Canul R, Willingham AL., 3rd Control of the taeniosis/cysticercosis complex: future developments. Vet Parasitol. 2006;139:283–292. doi: 10.1016/j.vetpar.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Fernandez S, Costa AC, Katsuyama AM, Madeira AM, Gruber A. A survey of the inter- and intraspecific RAPD markers of Eimeria spp. of the domestic fowl and the development of reliable diagnostic tools. Parasitol Res. 2003;89:437–445. doi: 10.1007/s00436-002-0785-2. [DOI] [PubMed] [Google Scholar]
- Ferrer E, Bonay P, Foster-Cuevas M, Gonzalez LM, Davila I, Cortez MM, Harrison LJ, Parkhouse RM, Garate T. Molecular cloning and characterisation of Ts8B1, Ts8B2 and Ts8B3, three new members of the Taenia solium metacestode 8 kDa diagnostic antigen family. Mol Bioch Parasitol. 2007;152:90–100. doi: 10.1016/j.molbiopara.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Gonzalez LM, Bonay P, Benitez L, Ferrer E, Harrison LJ, Parkhouse R, Garate T. Molecular and functional characterization of a Taenia adhesion gene family (TAF) encoding potential protective antigens of Taenia saginata oncospheres. Parasitol Res. 2007;100:519–528. doi: 10.1007/s00436-006-0297-6. [DOI] [PubMed] [Google Scholar]
- Gonzalez LM, Montero E, Sciutto E, Harrison LJ, Parkhouse RM, Garate T. Differential diagnosis of Taenia saginata and Taenia solium infections: from DNA probes to polymerase chain reaction. Transac Roy Soc Trop Med Hyg. 2002;96:S243–250. doi: 10.1016/s0035-9203(02)90083-0. [DOI] [PubMed] [Google Scholar]
- Jeon HK, Eom KS. Taenia asiatica and Taenia saginata: genetic divergence estimated from their mitochondrial genomes. Exp Parasitol. 2006;113:58–61. doi: 10.1016/j.exppara.2005.11.018. [DOI] [PubMed] [Google Scholar]
- Maravilla P, Souza V, Valera A, Romero-Valdovinos M, Lopez-Vidal Y, Dominguez-Alpizar JL, Ambrosio J, Kawa S, Flisser A. Detection of genetic variation in Taenia solium. J Parasitol. 2003;89:1250–1254. doi: 10.1645/GE-2786RN. [DOI] [PubMed] [Google Scholar]
- Maravilla P, Valera A, Souza V, Martinez-Gordillo M, Flisser A. Isozyme analysis of Taenia solium isolates from Mexico and Colombia. Mem Inst O Cruz. 2003;98:1049–1050. doi: 10.1590/s0074-02762003000800013. [DOI] [PubMed] [Google Scholar]
- Sciutto E, Rosas G, Hernandez M, Morales J, Cruz-Revilla C, Toledo A, Manoutcharian K, Gevorkian G, Blancas A, Acero G, Hernandez B, Cervantes J, Bobes RJ, Goldbaum FA, Huerta M, Diaz-Orea A, Fleury A, De Aluja AS, Cabrera-Ponce JL, Herrera-Estrella L, Fragoso G, Larralde C. Improvement of the synthetic tri-peptide vaccine (S3Pvac) against porcine Taenia solium cysticercosis in search of a more effective, inexpensive and manageable vaccine. Vaccine. 2007;25:1368–1378. doi: 10.1016/j.vaccine.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Vega R, Pinero D, Ramanankandrasana B, Dumas M, Bouteille B, Fleury A, Sciutto E, Larralde C, Fragoso G. Population genetic structure of Taenia solium from Madagascar and Mexico: implications for clinical profile diversity and immunological technology. Int J Parasitol. 2003;33:1479–1485. doi: 10.1016/s0020-7519(03)00206-6. [DOI] [PubMed] [Google Scholar]
- Almeida CR, Ojopi EP, Nunes CM, Machado LR, Takayanagui OM, Livramento JA, Abraham R, Gattaz WF, Vaz AJ, Dias-Neto E. Taenia solium DNA is present in the cerebrospinal fluid of neurocysticercosis patients and can be used for diagnosis. Eur Arch Psych Clin Neurosc. 2006;256:307–310. doi: 10.1007/s00406-006-0612-3. [DOI] [PubMed] [Google Scholar]
- Ojopi EP, Oliveira PS, Nunes DN, Paquola A, Demarco R, Gregorio SP, Aires KA, Menck CF, Leite LC, Verjovski-Almeida S, Dias-Neto E. A quantitative view of the transcriptome of Schistosoma mansoni adult-worms using SAGE. BMC Genomics. 2007;8:186. doi: 10.1186/1471-2164-8-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel D. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Silva WA, Costa MC, Valente V, Sousa JF, Paco-Larson ML, Espreafico EM, Camargo SS, Monteiro E, Holanda AJ, Zago MA, Simpson AJ, Dias-Neto E. PCR template preparation for capillary DNA sequencing. Biotechniques. 2001;30:537–542. doi: 10.2144/01303st05. [DOI] [PubMed] [Google Scholar]
- Davila AM, Lorenzini DM, Mendes PN, Satake TS, Sousa GR, Campos LM, Mazzoni CJ, Wagner G, Pires PF, Grisard EC, Cavalcanti MC, Campos ML. GARSA: genomic analysis resources for sequence annotation. Bioinformatics. 2005;21:4302–4303. doi: 10.1093/bioinformatics/bti705. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder N, Apweiler R. InterPro and InterProScan: tools for protein sequence classification and comparison. Meth Mol Biol. 2007;396:59–70. doi: 10.1007/978-1-59745-515-2_5. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Finn RD, Sonnhammer EL. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucl Acids Res. 1999;27:260–262. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CE, Baumann U, Brown AL. Automated Methods of predicting the function of biological sequences using GO and BLAST. BMC Bioinformat. 2005;6:272. doi: 10.1186/1471-2105-6-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucl Acids Res. 2007;35:W585–587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucl Acids Res. 2005;33:W686–689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of Taenia solium ESTs and their GenBank accession numbers.
Table S3. Distribution of the Taenia solium EST according to their characteristics and similarity analysis.
Table S4. Number of Taenia solium ORESTES according to their annotation as coding sequences (CDS).
Table S5. Distribution of Taenia solium EST hits according to the Gene Onthology (GO) classification.
Table S6. Taenia solium ESTs with predicted extra-cellular sub-localization by Wolf-PSORT.
Table S7. Number of Taenia solium EST eliciting hits with the CDD database.
Table S8. Number of Taenia solium EST eliciting hits with the COG database.
Table S9. Number of Taenia solium EST eliciting hits with the KOG database.
Picture S1. Distribution of the most frequent Eukaryotic Orthologous Groups categories observed for the Taenia solium sequences.
Table S10. List of Taenia solium EST with similarity to the parasite genome project sequences.
Table S11. List of interesting genes found in Taenia solium ESTs.
Table S2. List of cDNA mini-libraries, primers used for generation of the Taenia solium EST profiles and the number of sequences obtained in each library.


