Abstract
The direct detection of antigen-specific T cells using tetramers of soluble peptide-major histocompatibilty complex (pMHC) molecules is widely used in both basic and clinical immunology. However, the number of specificities that can be assessed simultaneously has been a major limitation. Here we describe and validate a method using combinations of fluorescent pMHC tetramers to simultaneously detect large numbers (≥ 15) of T cell specificities in a single human blood sample.
The highly diverse T cell receptor (TCR) repertoire of T lymphocytes plays a major role in most adaptive immune responses. Although most TCRs are highly specific for a given pMHC, the affinity of this interaction is low1 and thus multimeric pMHC staining reagents are needed for the specific detection, characterization and isolation of antigen specific cells using flow cytometry2. T cell responses to autoantigens, infectious diseases and tumor cells have all been analyzed with these reagents3-6. Recently, a number of different peptide exchange protocols have allowed rapid production of pMHC tetramers displaying, in at least one case, thousands of different peptides7-10. But there is currently no way to efficiently deploy more than a few tetramers when sample size is limited. This is especially true with clinical specimens, where one often has only one or a few million T cells per time point in a standard blood draw, enough for one, or at most a few flow-cytometrical analyses.
Here we describe and validate a method that allows the simultaneous use of large numbers of different pMHC tetramers on small numbers of cells using standard flow cytometry and reagents. This method makes use of fluorescently tagged strepavidin backbones to create pMHC tetramers that utilize all possible combinations of fluorophores, similar to an approach described for use with antibody combinations11. The number of different T cell specificities that can be detected equals 2N-1, where N is the number of different fluorescent labels. Here we use four strepavidin “colors” to detect the predicted 15 different T cell specificities, but we have also done experiments using five and six colors, suggesting that 31 and 63 specificities, respectively, are also achievable.
As a first test for this approach, we stained PBMC's from a donor with ∼0.07% CMV (Cytomegalovirus) pp65/HLA-A0201 specific CD8+ T cells with 15 different color combinations of the pp65-CMV/HLA-A0201 tetramer, using four different commercially available fluorescent streptavidin species (PE, PE-Cy5, PE-Cy7 and APC conjugates). First, the sample was split into 15 aliquots and stained separately with 15 different color combinations of this tetramer. The cells were then washed, mixed together and analyzed on an LSRII flow cytometer (BD Bioscience, Stanford Shared FACS Facility, Fig. 1a-e and see Supplementary Fig. 1a-d for the general gating scheme). All 15 possible combinations could be distinguished and the number of cells stained with each combination was approximately the same. (Fig. 1f). To test the limitations of this system, we used another CMV+ blood sample and quadruple stained it with PE, PE-Cy5, PE-Cy7, and APC labeled CMV tetramers. In separate wells, these cells were also stained with all possible combinations of PE-Cy5.5 and APC-Cy7 labeled CMV tetramers (four combinations). The cells were washed and then mixed together. The additional two tetramer colors could be differentiated even when they are used simultaneously with the four other tetramer colors used throughout this study (Fig 1g).
Figure 1.
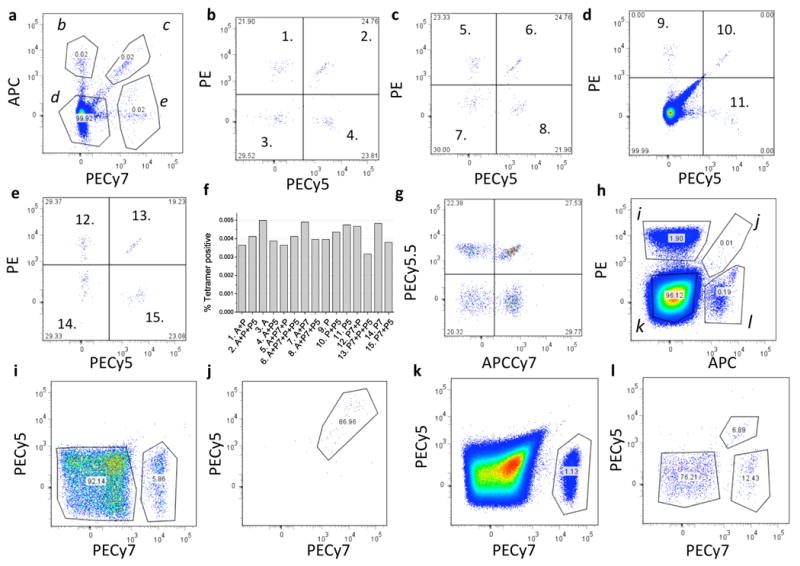
Combinatorial pMHC tetramer staining. Enriched CD8+ T cells from a CMV sero-positive donor were split into 15 aliquots and stained separately with the 15 CMV tetramer color combinations, washed, mixed together and run as a single sample. All panels show flow cytometry scatter plots. (a) Live CD8+ T cells were gated into four populations labeled b-e, based on threshold fluorescence of APC and PE-Cy7 as shown. Each population is represented in (b-e) and segregated based on PE and PE-Cy5 threshold fluorescence. The relative abundance of cells in each of the 15 tetramer-positive populations is represented in a bar chart in (f). In (g), cells from a different CMV sero-positive donor were stained with CMV tetramers labeled with PE, PE-Cy5, PE-Cy7, APC and all possible combinations of PE-Cy5.5 and APC-Cy7, washed, mixed and analyzed. A separate population was resolved for each possible color combination used. (h-l) Blood samples were processed and tetramer stained using a prepared cocktail of 15 different pMHC specificities represented by combinations of PE, PE-Cy5, PE-Cy7 and APC conjugated streptavidins (peptide scheme A, Supplementary Table 2 online). Cells were gated on PE and APC fluorescence (h) into populations labeled i-l, each of which is shown (i-l) further fractionated based on its PE-Cy5 and PE-Cy7 fluorescence.
We then tested the approach further by making a mixture of pMHC tetramers using HLA-A0201 bound to 15 different peptides. Using a premixed cocktail of tetramers is convenient because it reduces pipeting error and allows for accurate comparisons of various cell samples. The tetramers were mixed together and used to stain the cells simultaneously (see Methods for details of tetramer preparation). In all cases, a Hepatitis C virus (HCV) peptide was used as a negative-control or “dump” peptide for preparing the tetramer labeled with all four fluorophores (See Supplementary Tables 1 and 2 online). In this way, any cells that are non-specifically stained with all four colors would be eliminated from subsequent analysis. An HCV peptide was appropriate for this purpose because all samples used were HCV-sero-negative (as determined by the Stanford Blood Center).
For each staining experiment a different set and order of peptide epitopes were used (combinatorial “schemes”). For this example, the order of the 15 epitopes are listed in Supplementary Table 2 online using scheme A. For this particular donor blood sample, we detected six specificities of T cells with a frequency above 0.01% of CD8+ cells. These are CMV pp65, three different EBV epitopes, Influenza M1 50-58, and the characterized mutant Mart1a epitope (Supplementary Table 2 online, Fig. 1h-l).
As with all pMHC-tetramer staining experiments, it is critical to define appropriate criteria for setting positive staining thresholds. This can be complicated by the use of multiple fluorophores each with variable staining intensities making it necessary to choose arbitrary thresholds. Therefore, we used multiple approaches to verify the identities of tetramer-stained cells and thus to validate the specificity of our staining. In one approach, an aliquot of the same sample was stained with the same set of pMHC tetramers using an alternate color-coding scheme. (Supplementary Fig. 2 online, scheme B). The two staining schemes gave very similar results (Fig. 2a. In another approach, we tested for the disappearance of each T cell specificity caused by antigen dependent TCR down-regulation in response to peptide-specific interaction with pulsed antigen presenting cells. We pulsed HLA-A0201+ T2 antigen presenting cells in 15 separate wells with a single peptide overnight, then added T cells for a 5 hour incubation. We then stained with the combinatorial cocktail of pMHC tetramers. Each peptide caused a loss of signal for the appropriate peptide specificity (Fig. 2b). An additional control for non-specific tetramer staining is to use a “sham” pMHC tetramer cocktail containing only the negative-control epitope pMHC species made in parallel and used on a separate aliquot of the same sample (see Supplementary Fig. 3 online, which shows minimal staining in the sham stained cells).
Figure 2.

Specificity of combinatorial pMHC tetramer staining. (a) The percentage of cells labeled with the six different pMHC tetramers using two different staining schemes (Supplementary Table 2 online). (b) The percentage of cells stained with the indicated pMHC tetramers in cells co-cultured with T2 cells in the presence of a control peptide (HCV peptide, black bars) or the peptide corresponding to the pMHC tetramer reagent (grey bars); representative of three independent experiments. (c) Tetramer staining from four donor samples, each stained separately with 15 different PE-labeled pMHC tetramers (scheme C; Supplementary Table 2 online) is plotted on the x axis. Staining of the same sample with three separate preparations of combinatorial pMHC tetramer cocktails (Supplementary Table 2 online) is plotted +/- SEM on the y axis (d) The percentage of cells detected for each pMHC specificity before (gray bars) and after tetramer enrichment (black bars) using peptide scheme D (Supplementary Table 2).
To compare the sensitivity of the combinatorial approach to the classical approach, we used samples from 4 donors and split each of them into 18 aliquots. In the first 15, we stained each T cell specificity separately using the classical approach (a separate PE tetramer for each of the 15 peptides tested and a PE-Cy7 HIV-peptide negative-control tetramer). In the other three aliquots, we used identical but separately prepared combinatorial tetramer cocktails (all using peptide scheme C, Supplementary Table 2 online). We calculated the % tetramer positive CD8+ T cells based on by-eye thresholds and compared the values obtained using the two approaches (Fig. 2c); we found that the two approaches gave very comparable results above a 0.01% frequency threshold.
Moon et al.12 have shown that anti-PE coated magnetic beads can very efficiently enrich for pMHC tetramer+ cells to allow the detection of exceedingly rare T cell populations, including naïve T cell populations8,12,13. To demonstrate that our combinatorial staining approach can be used in conjunction with this type of enrichment, we first stained cells with the 15 specificity combinatorial cocktail and then used a mixture of anti-PE and anti-APC coated magnetic particles (MACS) to enrich the sample for tetramer stained T cells, as described12. Using this method, we saw a ∼100 fold increase in percentages of tetramer positive cells compared to a pre-enrichment sample, which is consistent with our previous experience using single or double tetramer stains. Although independent verification of the identities of these populations is beyond the scope of this study, this procedure also revealed several populations of T cells specific for one EBV epitope (EBV4) and three Influenza epitopes (Flu2, 3 and 5) that were below the threshold of detection without enrichment (Fig. 2d, see Supplementary Figs. 4 and 5 online for raw data). Using mixing experiments we also verified the accuracy of the enrichment protocol for all fluorophore combinations (Supplementary Figs. 6 and 7 online).
In summary, we describe here a method which uses all possible combinations of four different pMHC labels to detect 15 different αβ T lymphocyte specificities. This method is readily extended to encompass more colors and many more combinations of colors. So far, we have had success staining cells simultaneously with six colors using PE-Cy5.5 and APC-Cy7 streptavidins (See Fig. 1h, Supplementary Fig. 1 online), which would allow the detection of up to 63 possible specificities. We had much less success with other commercially available streptavidins tested, which varied significantly from lot-to-lot in staining quality. These problems could be caused by variations in the number of biotin binding sites14. Use of a strepavidin mutant that allows site specific fluorophore labeling has been reported to remedy this problem, and should allow the use of several more colors in the combinatorial approach described here14. One limitation of this method is the dilution of staining signal by the number of different fluorophores used to stain the same cell. Although we have seen only a 2-3 fold reduction in signal strength per color in the four color stains (Supplementary Fig. 3a online), this will certainly become more of a problem as more colors are used. Coupling streptavidin with molecular conjugates containing various combinations of fluorophores and/or quantum dots could solve this problem.
In addition to the utitilty of this approach for probing many peptide specificities bound to a single MHC allele (as demonstrated for HLA-A2), it is also well suited for probing mixtures of several different MHC Class I and Class II alleles, each bound to various peptide epitopes. This is particularly useful for non-HLA selected human samples, where there are many different possible alleles. Apropos of this, photocleavable peptides for several other MHC Class I alleles have recently been characterized15.
Methods
Preparation of 15 different streptavidin color combinations
To discern 15 different pMHC tetramer specificities, four fluorescently labeled streptavidins were purchased from eBioscience (PE-SAv “P”, PECy5-SAv “P5”, PECy7-SAv “P7” and APC-SAv “A”) and mixed to create the 15 different staining combinations. These streptavidins were mixed and adjusted with PBS to make a final volume of 95 uL (as listed in Supplementary Table 1 online). For the lots used in this study, the relative ratios of each color were empirically determined. For instance, a higher APC-SAv molar ratio was used because it generally had a lower fluorescent signal than the other dyes when used at equal molar concentration.
Biotinylated HLA-A2 protein peptide exchange
To produce the various biotinylated pMHC molecules, HLA-A2 was refolded with a UV-cleavable peptide, biotinylated, and purified as described7. After purification, the protein stock was stored in 50% glycerol at -20°C. For each peptide specificity, peptide exchange reactions were set up in 100 μL volumes each containing 10 μM peptide and 100 μg/ml HLA-A2 protein in PBS. After a 20 minute exposure to 365 nm UV irradiation using a Stratagene UV Stratalinker 2400, the protein was stored at 4°C overnight to complete the exchange. To remove aggregates, the protein was then centrifuged at 5000xg for 5 minutes and 90 μL of protein was transferred to a new plate.
Tetramerization
After UV exchange to create 15 different biotinylated pMHC monomers in separate wells, the streptavidin mixtures described above were added incrementally to achieve > 4:1 pMHC:SAv molar ratio. In all cases, the combination #15 was used as a “dump” (negative control) to remove “sticky” cells. The Hepatitis C virus epitope (HCV) was used for the dump tetramer. To improve our signal to noise ratio and prevent subsequent pMHC cross-pairing, streptavidin agarose was added to each tetramer to quench any unbound, biotinylated pMHC. This agarose was removed by filtration. Next, biotinylated agarose was added and filtered out to remove any unsaturated strepavidin molecules. Before mixing all 15 tetramer preparations together, excess free biotin was added to fully quench any unsaturated streptavidin. The tetramer mixtures were then concentrated and exchanged using a 4 ml 10 kDa cut-off Amicon Ultra (Millipore) protein concentrator. Finally, 10 μM HCV peptide was added to the mixture to prevent peptide exchange between the tetramer specificities. Mixtures were stored at 4°C for later use. For consistency purposes, the optical density at 565 nm was used to ensure similar concentrations of tetramer mixtures were used in each experiment. The tetramer mixures were diluted when added to the cells to achieve a final OD565 of 0.5 (corresponding to a final total PE concentration of ∼250 nM).
Cells and staining
Platelet donor aphaeresis lymphocytes were obtained from the Stanford Blood Center and enriched for CD8+ T cells by negative selection using the human CD8+ T cell Rosette-sep enrichment kit (StemCell Technologies). In all cases, tetramer staining was done in PBS with 2 mM EDTA, 2% Fetal calf serum, 0.1% sodium azide, and 1 μg/ml purified anti-CD16 and anti-CD32 (ebioscience) for 1 hr at room temperature. Fifteen combinations of PE, PE-Cy5, PE-Cy7, and APC streptavidin were used to make separate preparations of CMV tetramer of each color combination. The cells were subsequently stained on ice with various fluorescent markers, always including CD8 (QDot 605 or 705, Invitrogen; or PerCP-Cy5.5 anti-CD8, eBioscience) and an amine-reactive viability stain (LIVE/DEAD Fixable aqua dead cell stain kit, Invitrogen).
Supplementary Material
Acknowledgments
The authors would like to thank K. Adachi, M. Kuhns, Y.-H. Chien and D. Furman for helpful discussions. This work was supported by NIH grant #5U19AI057229, A Bill and Melinda Gates Foundation grant and The Howard Hughes Medical Institute. EWN is supported by The American Cancer Society Steven Stanley and Edward Albert Bielfelt Post-Doctoral Fellowship. LOK is supported by a National Science Foundation Graduate Research Fellowship. YW is supported by a Damon Runyon Postdoctoral Fellowship.
References
- 1.Matsui K, Boniface JJ, Reay PA, et al. Science. 1991;254(5039):1788. doi: 10.1126/science.1763329. [DOI] [PubMed] [Google Scholar]
- 2.Altman JD, Moss PA, Goulder PJ, et al. Science. 1996;274(5284):94. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 3.Danke NA, Kwok WW. J Immunol. 2003;171(6):3163. doi: 10.4049/jimmunol.171.6.3163. [DOI] [PubMed] [Google Scholar]
- 4.Serbina N, Pamer EG. Curr Opin Immunol. 2003;15(4):436. doi: 10.1016/s0952-7915(03)00071-2. [DOI] [PubMed] [Google Scholar]
- 5.Kita H, He XS, Gershwin ME. Autoimmun Rev. 2003;2(1):43. doi: 10.1016/s1568-9972(02)00124-6. [DOI] [PubMed] [Google Scholar]
- 6.Pittet MJ, Speiser DE, Valmori D, et al. Int Immunopharmacol. 2001;1(7):1235. doi: 10.1016/s1567-5769(01)00048-0. [DOI] [PubMed] [Google Scholar]
- 7.Toebes M, Coccoris M, Bins A, et al. Nat Med. 2006;12(2):246. doi: 10.1038/nm1360. [DOI] [PubMed] [Google Scholar]
- 8.Day CL, Seth NP, Lucas M, et al. J Clin Invest. 2003;112(6):831. doi: 10.1172/JCI18509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, James EA, Huston L, et al. Clin Immunol. 2006;120(1):21. doi: 10.1016/j.clim.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Grotenbreg GM, Roan NR, Guillen E, et al. Proc Natl Acad Sci U S A. 2008;105(10):3831. doi: 10.1073/pnas.0711504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horan PK, Slezak SE, Poste G. Proc Natl Acad Sci U S A. 1986;83(21):8361. doi: 10.1073/pnas.83.21.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moon JJ, Chu HH, Pepper M, et al. Immunity. 2007;27(2):203. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scriba TJ, Purbhoo M, Day CL, et al. J Immunol. 2005;175(10):6334. doi: 10.4049/jimmunol.175.10.6334. [DOI] [PubMed] [Google Scholar]
- 14.Ramachandiran V, Grigoriev V, Lan L, et al. J Immunol Methods. 2007;319(1-2):13. doi: 10.1016/j.jim.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bakker AH, Hoppes R, Linnemann C, et al. Proc Natl Acad Sci U S A. 2008;105(10):3825. doi: 10.1073/pnas.0709717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


