Abstract
SC35M is a mouse-adapted variant of the highly pathogenic avian influenza virus SC35. We have previously shown that interspecies adaptation is mediated by mutations in the viral polymerase and that it is paralleled by the acquisition of high pathogenicity for mice. In the present study, we have compared virus spread and organ tropism of SC35 and SC35M in mice. We show that SC35 virus causes mild bronchiolitis in these animals, whereas infection with the mouse-adapted SC35M virus leads to severe hemorrhagic pneumonia with dissemination to other organs, including the brain. In SC35M-infected animals, viral RNA and viral antigen were detected in monocytes and macrophages, and SC35M, unlike SC35, replicated in lymphocyte and macrophage cultures in vitro. SC35M did not induce an adequate cytokine response but, unlike SC35, caused severe lymphopenia in mice. These observations suggest that the high efficiency of the SC35M polymerase is responsible for infection and depletion of lymphocytes and other white blood cells, which results in immune suppression and systemic virus spread.
Highly pathogenic avian influenza viruses (HPAIV) have raised concern in recent years as potential human pathogens. Since 1997, H5N1 viruses have spread over large parts of Asia, Europe, and Africa and have occasionally caused disease in humans with an excessive case-fatality rate.1,2 Human infection with HPAIV of the H5N1 subtype initially seemed to be restricted to the lung,3,4 but recent reports5,6,7 show that it can disseminate to organs beyond the respiratory tract with occasional involvement of the central nervous system. Cytokine dysfunction and high viral loads observed in H5N1-infected patients have been reported to contribute to H5N1 pathogenesis in humans.3,5 In 2004, a HPAIV of subtype H7N7 has caused a human outbreak. In most instances disease was mild, but one case was fatal.8 Although, all of the H5N1 and H7N7 isolates obtained up to now are still avian viruses, mutations have been observed that are thought to promote adaptation to humans. Most of these adaptive mutations are located in the hemagglutinin,1,2,8,9,10,11 the NS1 protein,12,13,14,15,16 and the polymerase proteins.17,18
Recent studies from our laboratory on strain SC35, an HPAIV of subtype H7N7, and its mouse-adapted variant SC35M have highlighted the prominent role of the viral polymerase in interspecies transmission. We have identified seven mutations in the polymerase complex that were responsible for enhanced transcription and replication efficiency of SC35M in mammalian cells and thus for adaptation to mice,17 and we found that adaptation of the polymerase proteins to the nuclear import machinery was a crucial mechanism in this process.19 It was also clear from these studies that the mutations in the polymerase were not only responsible for mouse adaptation but also for increased mouse pathogenicity of SC35M.
In the present study, we have compared SC35 and SC35M for spread of infection and tissue tropism in mice. We found that infection with SC35 is restricted to the lung with mild symptoms of disease. In contrast, infection with SC35M is characterized by an always lethal outcome with severe pneumonia and systemic spread that is presumably mediated by virus replication in T cells and monocytes and a concominant lymphocyte depletion.
Materials and Methods
Cells and Viruses
EL-4 (murine T lymphocyte) and J774 (murine macrophage) cell lines were provided by H. Garn (Philipps University Marburg, Marburg, Germany). Cells were grown in RPMI 1640 medium (PAA) supplemented with 10% fetal calf serum (Invitrogen, San Diego, CA). In addition, J774 cells were supplemented with 1% nonessential amino acids (PAA). Murine lung cells (LA-4) were grown in Ham’s F-12 (Invitrogen) supplemented with 15% fetal calf serum (Invitrogen) and 1% nonessential amino acids (PAA).
Influenza A viruses were propagated in 11-day-old embryonated chicken eggs. The recombinant viruses SC35 and SC35M were described before.17 We ascertained the identity of the recombinant viruses by sequencing amplicons of each viral gene segment by using RT-PCR as described previously.17
Analysis of vRNA, mRNA, and cRNA by Primer Extension Assay
Murine lung cells (LA-4) were infected using multiplicity of infection 2 with recombinant virus. At 6 hours p.i., cells were harvested, and total RNA was isolated using TRIzol reagent (Invitrogen). After spectrophotometric quantification and normalization of total RNA, primer extensions were performed as described elsewhere.20,21 Briefly, 1 μg of total RNA was mixed with 1 pmol DNA primer labeled at its 5′-end with [γ-32P]ATP and T4 polynucleotide kinase in 6 μl of water and denatured at 99°C for 5 minutes. The mixture was then cooled on ice, and after addition of the reverse trancriptase SuperScriptII (Invitrogen) to the reaction buffer, primer extensions were performed at 45°C for 1 hour. Two nucleoprotein specific primers were used in the same reverse transcription reaction: 5′-GATGTGTCTTTCCAGGGGCG-3′ (for detection of vRNA), 5′-GCCTCCCTTCATAGTCGCTG-3′ (for detection of cRNA and mRNA), and 5′-TCCCAGGCGGTCTCCCATCC-3′ (for detection of 5S rRNA) for standardization. Transcription products were analyzed on 6% polyacrylamide gels containing 7 M urea in Tris-borate EDTA buffer and detected by autoradiography. Transcription products of two independent experiments were quantified using Aida Image Analyzer 3.27 software.
Growth Curves
EL-4 and J774 cells were inoculated with virus at a multiplicity of infection of 0.01. Virus inoculum was removed after 30 minutes of incubation at 37°C, and cells were washed two times with PBS. Cells were then incubated in the appropriate medium containing 0.2% bovine serum albumin at 37°C. At time points 0, 24, 48, 72, and 120 hours postinfection (p.i.) supernatants were collected and plaque forming units (PFU) determined on MDCK cells as described previously.17 The growth curves shown are the average result of two independent experiments.
Animal Experiments
The animal experiments were performed according to the guidelines of the German animal protection law. All animal protocols were approved by the relevant German authority, the Regierungspräsidium Gießen. We anesthetized 16 female BALB/c mice per group, 4–6 weeks old (purchased from Harlan Winkelmann, Borchen, Germany) with ketamine-xylazine (100 and 10 mg/kg, respectively) and inoculated intranasally with 106 PFU of SC35 or SC35M, respectively, in 50 μl diluted in PBS. For cytokine assays, three mice per group for each time point were sacrificed at 3 and 6 days p.i. Virus titers were determined in whole organ homogenates of lung, brain, and heart tissues on day 3 p.i. of three animals by plaque assay. To exclude any additional unwanted mutations in the viral genome, we have sequenced the whole cDNA from infected organ homogenisates as described before.17 The remaining animals were observed for 14 days after infection for signs of disease, and their weight was monitored daily.
In Situ Hybridization and Immunohistochemistry
For in situ hybridization and immunohistochemistry, 10 animals were infected with 106 PFU of SC35 or SC35M, respectively, as described above. Several organs, including lung, heart, brain, liver, kidney, spleen and gut of five animals per group, were removed on days 3 and 6 p.i., fixed over night in 4% buffered paraformaldehyde, and embedded in paraffin. Tissue sections (5-μm thick) were used for staining.
In Situ Hybridization
Influenza RNA in tissues was detected using single-stranded 35S-labeled RNA probes that were synthesized from a pBluescript KS+ vector containing a fragment of the NP gene (nt 1077 to 1442) of A/FPV/Rostock/34 (H7N1).22 Linearization of this plasmid with HindIII and subsequent T7 RNA polymerase transcription produced an antisense RNA probe suited to detect NP-specific RNA. Control RNA probes were obtained from the vector containing the dual-promoter plasmid of coxsackievirus B3 (pCVB3-R1).23 Pretreatment, hybridization, and washing conditions of dewaxed 5-μm paraffin tissue sections were performed as described previously.23 Slide preparations were subjected to autoradiography, exposed for 3 weeks at 4°C, and counterstained with H&E.
Immunohistochemical Stainings
Before immunohistochemical stainings of deparaffinated lung tissue with a monoclonal ferret antibody recognizing influenza A/Vic/3/75 (H3N2) (provided by the World Health Organization, Geneva, Switzerland) (1:500) slides were pretreated with 0.1 M citrate buffer, pH 6.0. A biotinylated anti-ferret antibody (1:200; Rockland) was used followed by the Zytochem-Plus horseradish peroxidase (Zytomed) kit and AEC as substrate under the conditions described by the manufacturer. Controls using normal rat serum were run to exclude nonspecific staining.
Automated Blood Count
Blood samples of animals infected with 106 PFU of SC35 or SC35M, respectively, were collected on days 3 and 6 p.i. in K3EDTA tubes. For the automated blood cells count, 500 μl of blood was analyzed with the Animal Blood Counter ABC Vet scil (Vet Novations). Frequencies of white blood cells, red blood cells, platelets, as well as hemoglobin and hematocrit, were measured. Recommended settings and calibrations for mouse hematology were set according to the manufacturer’s operation manual. The data presented are average results from three to five infected mice for each time point.
FACS Analysis
Blood (100 μl) from either naive C57BL/6 mice or C57BL/6 mice infected with influenza virus (see above) for 3 days was diluted in 5 ml of FACS buffer (PBS 2% fetal calf serum, EDTA) and centrifuged at 300 × g for 5 minutes at 4°C. The pellet was incubated with a 1:100 dilution of anti-CD3 FITC, anti-CD8 CyChrome, and anti-CD11b PE (BD Biosciences) in 100 μl of FACS buffer for 30 minutes at 4°C. Thereafter the cells were washed once. Before FACS analysis, erythrocytes were lysed, and cells were fixed with FACS lysing solution (BD Biosciences). Flow cytometry was performed on a dual-laser FACSCalibur and analyzed with CellQuest software (BD Biosciences).
Cytokine Assay
A mouse cytokine antibody array (RayBiotech, Inc.) was used according to the manufacturer’s protocols to detect a panel of cytokines and chemokines from the lung and spleen homogenates of influenza infected mice. Organs were homogenized in 1 ml of PBS, centrifuged at 1000 rpm for 10 minutes, and the supernatant was aliquoted and frozen at −80°C. Samples were not thawed more than once for this assay. The array tests for the following cytokines and chemokines: granulocyte colony-stimulating factor, granulocyte-macrophage colony-stimulating factor, interleukin (IL)-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL12-p40p70, IL12-p70, IL-13, IL-17, interferon-γ, monocyte chemoattractant protein (MCP)-1, MCP-5, regulated on activation normal T cell expressed and secreted, stem cell factor, soluble tumor necrosis factor α receptor I (sTNFRI), TNF-α, and vascular endothelial growth factor. Briefly, membranes with bound cytokine antibodies were incubated with 200 μg of lung or spleen homogenate. After incubation, each membrane was washed three times and incubated with biotin-conjugated anti-cytokine antibody and then with horseradish peroxidase-conjugated streptavidin. Cytokine levels were then detected by chemiluminescence and revealed on X-ray film. Intensity of signals was quantified by TINA 2.0 software. Background signals comparable with PBS infected animals were set 1. Each column represents three independent assays.
Results
Pathogenicity of SC35 and SC35M Correlates with Viral Polymerase Activity in Mice
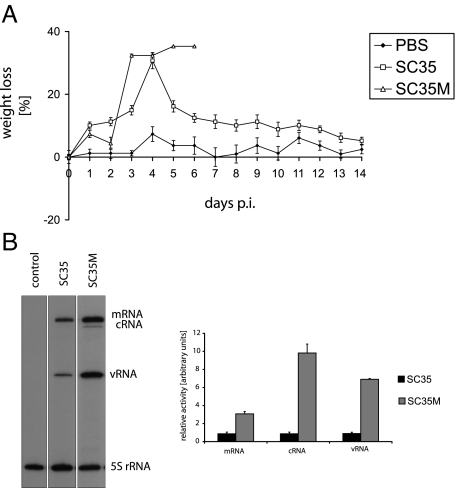
Groups of 16 mice were infected with either SC35 or SC35M virus as described in Materials and Methods. SC35-infected mice underwent a temporary weight loss of 20 to 30%, but all infected animals survived infection (log10MLD50 > 6.0 PFU). In contrast, mice infected with SC35M started to die on day 4 p.i., and all infected animals succumbed to infection by day 7 p.i. (log10MLD50 = 2.8 PFU) (Figure 1A). It has to be pointed out that SC35 and SC35M used throughout this study were recombinant viruses generated by reverse genetics.17 Nonrecombinant SC35 differed from SC35 used here by a higher pathogenicity (10 to 25% lethality at the same inoculation dose).24 Although not completely understood, this phenomenon may reflect differences in quasispecies equilibria. We have also compared the polymerase activities of SC35 and SC35M in mouse lung cells by primer extension assay. As shown in Figure 1B, the transcription and replication activity of SC35M was 3 to 10 times higher in these cells than that of SC35. These results confirm our previous observations indicating that the enhanced pathogenicity of SC35M for mice is the result of increased polymerase efficiency in murine cells.20
Figure 1.
Weight loss of mice infected with PBS, SC35, and SC35M. Groups of 10 mice were infected intranasally with 106 PFU of recombinant SC35 or SC35M, respectively. Animals were weighed daily and observed for illness for 14 days (A). Transcription and replication activity in murine lung cells (LA-4). LA-4 cells were infected with SC35 or SC35M and as a control with PBS using an multiplicity of infection 2. At 6 hours p.i., total RNA was isolated, and primer extension assays were performed as described in Materials and Methods (B). Error bars indicate SD of three independent experiments.
Tissue and Cell Tropism of SC35 and SC35M Virus in Mice
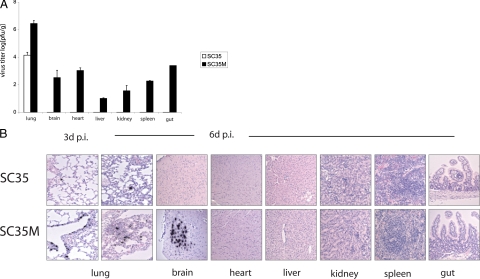
We have determined virus titers in organ homogenates of mice sacrificed at various times after infection. At 3 days p.i., SC35M was recovered from lung, brain, heart, liver, kidney, spleen, and gut (Figure 2A). In contrast, significant amounts of SC35 were only observed in the lung, but titers were about 100 times lower when compared with SC35M infection (Figure 2A). Virus was also detected at low titers (<102 PFU/ml) in the serum 3 and 6 days after infection with SC35M but not in SC35-infected animals (Table 1).
Figure 2.
Organ tropism of SC35 and SC35M in mice. Groups of five mice were infected with 106 PFU of recombinant virus. Virus titers were determined in lung, brain, heart, liver, kidney, spleen, and gut homogenates 3 days p.i. by plaque assay. Error bars indicate SD of independent experiments (A). In situ hybridization of all organs from SC35 virus-infected mice reveal virus RNA only in single alveolar cells at 3 and 6 days p.i. SC35M virus-infected mice reveal virus replication in numerous epithelial alveolar cells at days 3 and 6 p.i., and interstitial pneumonia at day 6 p.i. In addition, SC35M virus RNA was also detected in temporal lobes of the brain at day 6 p.i. Hearts or other organs were neither found to be infected with SC35 nor with SC35M virus (B).
Table 1.
Virus Titers in Serum of Infected Mice
| Virus | Virus titer 3 days p.i. (PFU/ml) | Virus titer 6 days p.i. (PFU/ml) |
|---|---|---|
| PBS | 0 | 0 |
| SC35 | 0 | 0 |
| SC35M | 30 | 60 |
Mice were infected with 106 pfu of SC35 and SC35M. As a control, PBS-inoculated mice were used. On days 3 and 6 p.i., blood was collected from three infected mice at each time point by retroocular puncture. Virus titers in serum were detected by plaque assay.
Organ tropism was also analyzed by radioactive in situ hybridization. At day 3 p.i., SC35 was only observed in single cells of the lung. Also, at day 6 p.i., a few alveolar cells were found to contain viral RNA; however, further development of the infection was not observed in the lung. Pulmonary damage and inflammation was only marginal, both at days 3 and 6 p.i. SC35 RNA, inflammation, or necrosis/apoptosis was neither detected in brain nor in heart liver, kidney, spleen, and gut tissues (Figure 2B). In contrast to mice infected with SC35, in situ hybridization revealed numerous alveolar and bronchial epithelial cells positive for viral RNA in SC35M-infected animals at days 3 (Figures 2B and 3) and 6 p.i. (Figure 2B). Furthermore, at day 6 p.i., the lungs of SC35M-infected mice revealed typical pulmonary lesions with desquamation and destruction of epithelial cells and inflammation with hemorrhages (Figure 2B). Viral RNA was not only detected in the lung but also in the brain where it was specifically located in the temporal lobes. Neither inflammatory and other histological lesions nor viral RNA could be detected in heart, liver, kidney, spleen, and gut (Figure 2B). These observations suggest that SC35M infects parenchymal cells only in the brain and that infectious virus recovered from other organs (Figure 2A) originates from blood present in these tissues.
Figure 3.
Cell tropism in the respiratory tract and in peripheral blood. Visualization of virus RNA (A and B) and protein (C–E) in lungs of SC35M-infected mice 3 days p.i. by in situ hybridization and immunohistochemistry. Groups of five mice were infected with 106 PFU of recombinant virus. Numerous bronchial (A and C) epithelial cells as well as alveolar epithelial cells (B and D) were found to be positive early in infection. Also, mononuclear immune cells in the lung (A and C (arrows)) and in the blood (E) were found to be infected. Noninfected lungs were consistently negative in immunohistochemistry (F).
The data obtained by in situ hybridization were complemented by immunohistochemical analysis of viral proteins in lung tissue. The patterns of SC35M-infected cells that were obtained with both procedures in alveolar and bronchial epithelia were similar (Figure 3, A–D). Interestingly, the viral RNA (Figure 3A, arrow) and protein (Figure 3C, arrow) were also present in mononuclear inflammatory cells in the lung. In addition, infected monocytes were detected in peripheral blood (Figure 3, E and F).
Taken together, these data show that SC35 causes a mild infection in mice restricted to the lung, whereas SC35M causes lethal hemorrhagic pneumonia with systemic virus spread and infection of the central nervous system.
SC35M Infection Leads to Lymphocyte Depletion in Mice
We have then compared blood parameters in infected mice and control animals. On days 3 and 6 p.i., blood was collected by retroocular puncture and analyzed for white blood cells, red blood cells, hemoglobin, hematocrit, platelets, lymphocytes, monocytes, and granulocytes (Table 2). In mice infected with SC35, levels of monocytes, lymphocytes, and granulocytes as well as total white blood cells increased over the period of 6 days, while platelet counts dropped transiently at day 3 p.i. On the other hand, in SC35M-infected mice, a substantial drop of hemoglobin and hematocrit were observed on day 6 p.i., which is most likely due to blood loss as the result of the hemorrhagic pneumonia (Figures 2B and 3, A–F). SC35M infection further led to a constant reduction of white blood cell numbers, which was already apparent at day 3 p.i. and further continued until day 6 p.i. The decrease was most prominent with lymphocytes. Their number dropped by >50% at day 3 p.i. and stayed at this low level throughout the period of observation. To shed light on the response of individual subsets of lymphocytes to infection with SC35 and SC35M, we performed FACS analysis of blood on day 3 p.i. using specific antibodies targeting CD3+, CD4+, and CD8+ T cells and CD11b monocytes as described in Materials and Methods. In SC35M-infected mice, about 50% of CD8+ T cells and >70% of CD4+ T cells were depleted 3 days p.i. (Figure 4, A and C), whereas SC35 infection resulted only in a moderate shift in T-cell proportions (Figure 4B). These data are compatible with the view that infection with SC35 that is confined to the lung elicits protective T-cell response in mice. In contrast, systemic infection with SC35M is characterized by a rapid and severe depletion of T cells.
Table 2.
Blood Parameters of Infected Mice
| Blood cell count | PBS | SC35
|
SC35 mol/L
|
||
|---|---|---|---|---|---|
| 3 days p.i. | 6 days p.i. | 3 days p.i. | 6 days p.i. | ||
| WBC (103/mm3) | 6.4 | 6.7 | 9.2 | 5.0 | 3.8 |
| ±0.7 | ±3.0 | ±0.9 | ±0.1 | ±1.2 | |
| RBC (106/mm3) | 7.9 | 9.1 | 8.5 | 10.4 | 5.8 |
| ±0.8 | ±1.3 | ±0.9 | ±0.3 | ±1.1 | |
| HGB (g/dl) | 14.1 | 16.3 | 14.5 | 17.6 | 9.6 |
| ±1.1 | ±1.8 | ±1.7 | ±0.3 | ±1.7 | |
| HCT (%) | 44.4 | 45.4 | 47.1 | 56.2 | 30.5 |
| ±3.8 | ±6.4 | ±5.3 | ±1.6 | ±6.0 | |
| PLT (103/mm3) | 1161 | 447 | 782 | 1348 | 446 |
| ±157 | ±179 | ±141 | ±68 | ±234 | |
| LYM (103/mm3) | 4.8 | 3.1 | 5.9 | 2.1 | 1.9 |
| ±0.6 | ±0.5 | ±0.4 | ±0.1 | ±0.7 | |
| MO (103/mm3) | 0.01 | 0.2 | 0.1 | 0.1 | 0.05 |
| ±0.0 | ±0.3 | ±0.0 | ±0.0 | ±0.1 | |
| GRA (103/mm3) | 1.6 | 3.9 | 3.3 | 2.8 | 1.9 |
| ±0.2 | ±0.4 | ±0.4 | ±0.3 | ±0.4 | |
Mice were infected with 106 PFU of SC35 and SC35M. Blood of three to five infected animals was collected 3 and 6 days p.i. by retroocular puncture and analyzed using an ABC blood counter for detection of white blood cells (WBC), red blood cells (RBC), hemoglobin (HGB), hematocrit (HCT), platelets (PLT), lymphocytes (LYM), monocytes (MO), and granulocytes (GRA). As a control, PBS-inoculated mice were used. SDs (±) are indicated underneath the bold measurements.
Figure 4.
Lymphopenia in mice. Groups of six mice were infected with 106 pfu of SC35 (B) and SC35M (C) or PBS control (A). At 3 days p.i., blood was collected from mice. CD4+ and CD8+ T cells and monocytes were detected using FACS analysis. Error bars indicate SD of independent experiments.
Cytokine and Chemokine Response
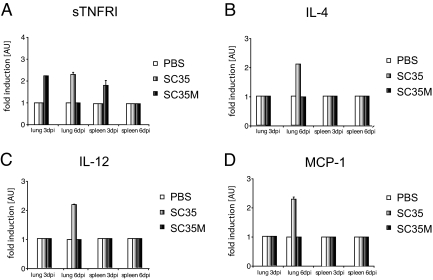
To assess the cytokine and chemokine response in mice on infection with the two different influenza viruses, we have screened lungs and spleens of infected animals using a panel of antibodies specific for 22 different cytokines and chemokines. As shown in Figure 5, A–D, four of them, sTNFRI, IL-4, IL-12, and MCP-1, were moderately elevated in lung and spleen early after infection with SC35. In contrast, SC35M-infected animals showed only increased sTNFRI levels. These data demonstrate that SC35M is unable to induce an adequate cytokine and chemokine response in mice.
Figure 5.
Chemokine and cytokine response in mice. Homogenates of the lung and spleen of six mice each group were infected with 106 PFU of SC35 and SC35M; three animals for each time point (3 and 6 days p.i.) were assessed for chemokine and cytokine response as described in Materials and Methods. Positive signals yielded with antibodies against sTNFRI (A), IL-4 (B), IL-12 (C), and MCP-1 (D) 3 and 6 days p.i. are shown as arbitrary units (AU). Each bar is representative for three independent experiments. Error bars indicate SD of independent experiments.
SC35M, unlike SC35, Replicates in Murine T Cells and Macrophages in Vitro
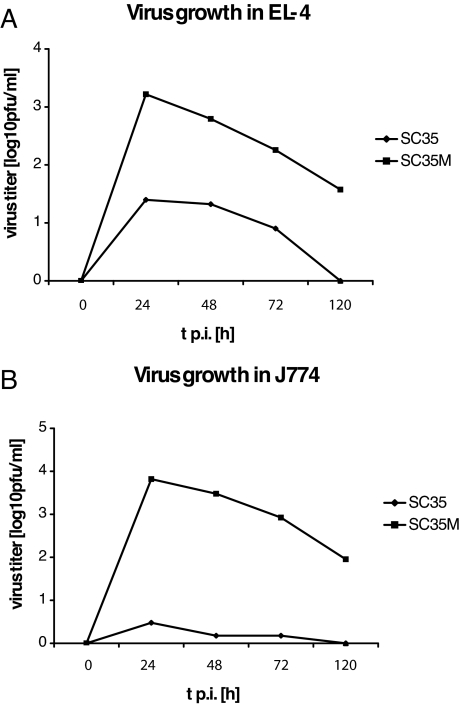
Since SC35M infects macrophages (Figure 3) and induces lymphocyte depletion in mice, it was of interest to find out if these cells allow production of infectious progeny virus. We have therefore infected cultured murine T cells (EL-4) and macrophages (J774) and determined virus titers in the supernatant of these cultures by plaque assay. The growth curves shown in Figure 6, A and B, indicate that SC35M replicates in these cells, whereas SC35 is recovered only at very low titers or not at all.
Figure 6.
Virus growth in EL-4 murine T cells (A) and J774 murine macrophages (B) after infection with SC35 and SC35M at a multiplicity of infection of 10−2. At time points 24, 48, 72, and 120 hours p.i., supernatants of infected cells were collected, and virus titers were determined using plaque assay. The figure shown is representative of two independent experiments.
Discussion
Influenza virus infection in man, in general, is localized in the respiratory tract. On some occasions, however, infection disseminates to other organs. Systemic spread is characteristic for human H5N1 infections,5,6,7,25 and involvement of the central nervous system was often observed in the 1918 outbreak of the Spanish influenza and less frequently in the pandemics of 1957 and 1968.26,27,28,29 Polymerase, hemagglutinin, and neuraminidase have previously been reported to be important determinants for these differences in the course of infection.30 In the system analyzed in the present study, the polymerase including the nucleocapsid protein is the only critical factor. We show that the polymerase of SC35 has a low efficiency in murine lung cells. As a result, the virus load is low, and infection is restricted to the respiratory tract. In contrast, the high efficiency of the polymerase of SC35M is responsible for high virus loads in the lung that allow hematogenic spread of the infection to the brain and other distant organs. These observations confirm our previous findings indicating that pathogenicity depends on the efficiency of the polymerase, which, in turn, is a host-dependent trait, since the efficiency of the SC35M polymerase is only high in mammalian cells but not in avian cells.17,20
The observation that infectious virus was recovered from all organs analyzed indicates that SC35M infection was widely disseminated. However, it was not clear whether virus replicated in each of these organs since most of them did not show tissue lesions or inflammatory infiltrations. Deposits of viral RNA and viral antigens were only observed in brain and in mononuclear cells of the lung and of the peripheral blood, and direct evidence for virus replication in monocytes and lymphocytes was also obtained when cell cultures were infected. Thus, some tissues may be better targets than others, and in addition to a high virus load flooding the entire organism, cell tropism may be another determinant of pathogenicity. Using another HPAIV of subtype H7, we have previously shown that endotheliotropism plays a crucial role in systemic infection of the chicken embryo.22,31 In the present study, there was no evidence for infection of endothelia in any of the analyzed organs. Thus, endotheliotropism can be ruled out as a mechanism promoting systemic spread of SC35M infection in mice.
It is clear from our data, however, that infection of lymphocytes and monocytes in the blood plays a crucial role in the spread of SC35M in mice. Mononuclear inflammatory cells infected with SC35M were also detected in the lungs of infected animals by in situ hybridization and immunohistochemistry, suggesting that not only circulating immune cells may function as carriers for influenza virus. Similar observations were made in other studies. Autopsy studies of H5N1-infected patients revealed viral RNA and viral antigens in T lymphocytes present in lymph nodes and cerebral neurons and in fetal macrophages of the placenta.6 It was also reported that the H5N1 virus as well as the pandemic 1918 influenza virus infect immune cells like macrophages and dendritic cells.32,33 Taken together, these findings underline the important role of immune cells in virus dissemination.
Another important factor in pathogenesis is lymphocyte depletion. In mice infected with SC35M, we observed a severe drop in the number of circulating lymphocytes as well as lymphocytes resident in the lung. Both CD4+ and CD8+ T cells were affected. Infection with SC35 did not induce lymphocyte depletion. Infection of mice with the HPAIV of the H5N1 subtype has also been associated with severe lymphopenia, whereas white blood cell counts were not altered after infection with a low pathogenic variant.34 Furthermore, autopsies of two H5N1-infected patients revealed extensive lymphopenia in spleen, lymph nodes, and mucosal tissue of the gastrointestinal tract.6 These observations indicate that SC35M as well as H5N1 viruses use immunosuppression resulting from lymphocyte depletion as a mechanism to fuel the infection process.
It has been reported that infection with HPAIV and 1918 virus causes a “cytokine storm” that significantly contributes to pathogenicity.5,35 With SC35M, we did not observe this phenomenon. There was a cytokine response to SC35 infection yet at a low level. Interestingly, Tumpey et al34 made similar observations with H5N1 viruses. They compared the mouse-lethal A/HK/483/97 isolate with the nonlethal A/HK/486/97 isolate for their effects on the murine immune system and found a reduction of CD4+ and CD8+ T cells and a reduced synthesis of γ-interferon and macrophage inflammatory protein in lung and lymphoid tissue of A/HK/483/97-infected mice.34 Their data and our results presented here suggest that the inability of the immune system to combat infection with these viruses is, in part, due to poor cytokine response.
The cytokines and chemokines induced by low pathogenic SC35 virus were IL-12p70, IL-4, and MCP-1. IL-12p70 is important for the differentiation of T helper cells and cell-mediated immunity.36 Indeed, IL-12 was previously shown to be important for resistance to influenza virus infection in mice.37 IL-4 has an important role in regulating antibody production, inflammation, and the development of effector T-cell responses.38 Furthermore, IL-4 stimulates the production of the MCP-1, which recruits macrophages to eliminate infected cells.39 The only chemokine found to be up-regulated early in SC35M infection was sTNFRI. Although the exact role of sTNFR is still a matter of speculation, soluble forms of the TNF receptor released from monocytes were shown to inhibit TNF-α action in vitro and in vivo and to down-regulate the inflammatory response.40,41 The soluble form of the TNF receptor also down-regulates TNF-α activity during lung response.42 Whether the ability of HPAIV to modulate shedding of sTNFR from the surface of monocytes interferes with TNF-α action, and thus contributes to pathogenesis, remains to be seen.
In conclusion, we have shown that the efficiency of the polymerase is an important determinant for the dissemination of influenza virus within the organism. High polymerase efficiency leads to high virus loads in the respiratory tract that enable the virus to enter the vascular system and to spread via infected lymphocytes and mononuclear cells to distant organs. As a result of infection, there is a depletion of lymphocytes and an impaired immune response. Reduction of high virus loads by targeting the viral polymerase may play an important role in the treatment of human influenza with systemic virus spread.
Acknowledgments
We thank Siegfried Schulz (Veterinarian service and central animal protection, Philipps University Marburg) for kindly providing support and access to the ABC blood cell counter.
Footnotes
Address reprint requests to Gülsah Gabriel, Heinrich-Pette-Institut für Experimentelle Virologie und Immunologie, an der Universität Hamburg, Martinistr. 52, 20251 Hamburg, Germany. E-mail: guelsah.gabriel@hpi.uni-hamburg.de.
Supported by grants from the Deutsche Forschungsgemeinschaft (SFB 593-TPB1 and SFB-TR19), the European Commission (CP5B-CT-2006-044263, FLUPOL, and FLUINNATE) and the BBSRC (studentship to K.B.).
References
- Claas EC, Osterhaus AD, van Beek R, De Jong JC, Rimmelzwaan GF, Senne DA, Krauss S, Shortridge KF, Webster RG. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- Peiris JS, Yu WC, Leung CW, Cheung CY, Ng WF, Nicholls JM, Ng TK, Chan KH, Lai ST, Lim WL, Yuen KY, Guan Y. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet. 2004;363:617–619. doi: 10.1016/S0140-6736(04)15595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KF, Chan PK, Chan KF, Lee WK, Lam WY, Wong KF, Tang NL, Tsang DN, Sung RY, Buckley TA, Tam JS, Cheng AF. Pathology of fatal human infection associated with avian influenza A H5N1 virus. J Med Virol. 2001;63:242–246. doi: 10.1002/1096-9071(200103)63:3<242::aid-jmv1007>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha do Q, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Xie Z, Gao Z, Liu J, Korteweg C, Ye J, Lau LT, Lu J, Gao Z, Zhang B, McNutt MA, Lu M, Anderson VM, Gong E, Yu AC, Lipkin WI. H5N1 infection of the respiratory tract and beyond: a molecular pathology study. Lancet. 2007;370:1137–1145. doi: 10.1016/S0140-6736(07)61515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uiprasertkul M, Puthavathana P, Sangsiriwut K, Pooruk P, Srisook K, Peiris M, Nicholls JM, Chokephaibulkit K, Vanprapar N, Auewarakul P. Influenza A H5N1 replication sites in humans. Emerg Infect Dis. 2005;11:1036–1041. doi: 10.3201/eid1107.041313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, Koch G, Bosman A, Koopmans M, Osterhaus AD. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome, Proc Natl Acad Sci USA. 2004;101:1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Human and avian influenza viruses target different cell types in cultures of human airway epithelium, Proc Natl Acad Sci USA. 2004;101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A. Antiviral response in pandemic influenza viruses. Emerg Infect Dis. 2006;12:44–47. doi: 10.3201/eid1201.051186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss GK, Salvatore M, Tumpey TM, Carter VS, Wang X, Basler CF, Taubenberger JK, Bumgarner RE, Palese P, Katze MG, Garcia-Sastre A. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza, Proc Natl Acad Sci USA. 2002;99:10736–10741. doi: 10.1073/pnas.112338099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao P, Tian G, Li Y, Deng G, Jiang Y, Liu C, Liu W, Bu Z, Kawaoka Y, Chen H. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J Virol. 2008;82:1146–1154. doi: 10.1128/JVI.01698-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug RM, Yuan W, Noah DL, Latham AG. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology. 2003;309:181–189. doi: 10.1016/s0042-6822(03)00119-3. [DOI] [PubMed] [Google Scholar]
- Seo SH, Hoffmann E, Webster RG. Lethal H5N1 influenza viruses escape host antiviral cytokine responses. Nat Med. 2002;8:950–954. doi: 10.1038/nm757. [DOI] [PubMed] [Google Scholar]
- Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host, Proc Natl Acad Sci USA. 2005;102:18590–18595. doi: 10.1073/pnas.0507415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon R, Franks J, Govorkova EA, Ilyushina NA, Yen HL, Hulse-Post DJ, Humberd J, Trichet M, Rehg JE, Webby RJ, Webster RG, Hoffmann E. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J Exp Med. 2006;203:689–697. doi: 10.1084/jem.20051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Herwig A, Klenk HD. Interaction of polymerase subunit PB2 and NP with importin α1 is a determinant of host range of influenza A virus. PLoS Pathog. 2008;4:e11. doi: 10.1371/journal.ppat.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Abram M, Keiner B, Wagner R, Klenk HD, Stech J. Differential polymerase activity in avian and mammalian cells determines host range of influenza virus. J Virol. 2007;81:9601–9604. doi: 10.1128/JVI.00666-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Nordmann A, Stein DA, Iversen PL, Klenk HD. Morpholino oligomers targeting the PB1 and NP genes enhance the survival of mice infected with highly pathogenic influenza A H7N7 virus. J Gen Virol. 2008;89:939–948. doi: 10.1099/vir.0.83449-0. [DOI] [PubMed] [Google Scholar]
- Feldmann A, Schafer MK, Garten W, Klenk HD. Targeted infection of endothelial cells by avian influenza virus A/FPV/Rostock/34 (H7N1) in chicken embryos. J Virol. 2000;74:8018–8027. doi: 10.1128/jvi.74.17.8018-8027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingel K, Hohenadl C, Canu A, Albrecht M, Seemann M, Mall G, Kandolf R. Ongoing enterovirus-induced myocarditis is associated with persistent heart muscle infection: quantitative analysis of virus replication, tissue damage, and inflammation, Proc Natl Acad Sci USA. 1992;89:314–318. doi: 10.1073/pnas.89.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiblauer H, Kendal AP, Rott R. Pathogenicity of influenza A/Seal/Mass/1/80 virus mutants for mammalian species. Arch Virol. 1995;140:341–348. doi: 10.1007/BF01309867. [DOI] [PubMed] [Google Scholar]
- Korteweg C, Gu J. Pathology, molecular biology, and pathogenesis of avian influenza A (H5N1) infection in humans. Am J Pathol. 2008;172:1155–1170. doi: 10.2353/ajpath.2008.070791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby AW. America’s Forgotten Pandemic. Cambridge, UK,: Cambridge University Press,; 1989:pp 22–58. [Google Scholar]
- LeCount E. The pathologic anatomy of influenzal bronchopneumonia. J Am Med Assoc. 1919;72:650–652. [Google Scholar]
- Winternitz M, Wason I, McNamara F. The Pathology of Influenza. New Haven, CT,: Yale University Press,; 1920:pp 34–39. [Google Scholar]
- Wolbach S. Comments on the pathology and bacteriology of fatal influenza cases, as observed at Camp Devens, Mass. Johns Hopkins Hospital Bull. 1919;30:104–109. [Google Scholar]
- Pappas C, Aguilar PV, Basler CF, Solorzano A, Zeng H, Perrone LA, Palese P, Garcia-Sastre A, Katz JM, Tumpey TM. Single gene reassortants identify a critical role for PB1, HA, and NA in the high virulence of the 1918 pandemic influenza virus. Proc Natl Acad Sci USA. 2008;105:3064–3069. doi: 10.1073/pnas.0711815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenk HD. Infection of the endothelium by influenza viruses. Thromb Haemost. 2005;94:262–265. doi: 10.1160/TH05-04-0264. [DOI] [PubMed] [Google Scholar]
- Perrone LA, Plowden JK, Garcia-Sastre A, Katz JM, Tumpey TM. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 2008;4:e1000115. doi: 10.1371/journal.ppat.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am J Pathol. 2007;171:1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumpey TM, Lu X, Morken T, Zaki SR, Katz JM. Depletion of lymphocytes and diminished cytokine production in mice infected with a highly virulent influenza A (H5N1) virus isolated from humans. J Virol. 2000;74:6105–6116. doi: 10.1128/jvi.74.13.6105-6116.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobasa D, Jones SM, Shinya K, Kash JC, Copps J, Ebihara H, Hatta Y, Kim JH, Halfmann P, Hatta M, Feldmann F, Alimonti JB, Fernando L, Li Y, Katze MG, Feldmann H, Kawaoka Y. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- Hofmann SR, Ettinger R, Zhou YJ, Gadina M, Lipsky P, Siegel R, Candotti F, O'Shea JJ. Cytokines and their role in lymphoid development, differentiation and homeostasis. Curr Opin Allergy Clin Immunol. 2002;2:495–506. doi: 10.1097/00130832-200212000-00004. [DOI] [PubMed] [Google Scholar]
- Monteiro JM, Harvey C, Trinchieri G. Role of interleukin-12 in primary influenza virus infection. J Virol. 1998;72:4825–4831. doi: 10.1128/jvi.72.6.4825-4831.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MA, Hural J. Functions of IL-4 and control of its expression. Crit Rev Immunol. 1997;17:1–32. doi: 10.1615/critrevimmunol.v17.i1.10. [DOI] [PubMed] [Google Scholar]
- Winsor GL, Waterhouse CC, MacLellan RL, Stadnyk AW. Interleukin-4 and IFN-γ differentially stimulate macrophage chemoattractant protein-1 (MCP-1) and eotaxin production by intestinal epithelial cells. J Interferon Cytokine Res. 2000;20:299–308. doi: 10.1089/107999000312432. [DOI] [PubMed] [Google Scholar]
- Leeuwenberg JF, Dentener MA, Buurman WA. Lipopolysaccharide LPS-mediated soluble TNF receptor release and TNF receptor expression by monocytes: role of CD14, LPS binding protein, and bactericidal/permeability-increasing protein. J Immunol. 1994;152:5070–5076. [PubMed] [Google Scholar]
- Pinckard JK, Sheehan KC, Arthur CD, Schreiber RD. Constitutive shedding of both p55 and p75 murine TNF receptors in vivo. J Immunol. 1997;158:3869–3873. [PubMed] [Google Scholar]
- Huaux F, Arras M, Vink A, Renauld JC, Lison D. Soluble tumor necrosis factor (TNF) receptors p55 and p75 and interleukin-10 down-regulate TNF-α activity during the lung response to silica particles in NMRI mice. Am J Respir Cell Mol Biol. 1999;21:137–145. doi: 10.1165/ajrcmb.21.1.3570. [DOI] [PubMed] [Google Scholar]