Abstract
Reverse engineering of high-throughput omics data to infer underlying biological networks is one of the challenges in systems biology. However, applications in the field of metabolomics are rather limited. We have focused on a systematic analysis of metabolic network inference from in silico metabolome data based on statistical similarity measures. Three different data types based on biological/environmental variability around steady state were analyzed to compare the relative information content of the data types for inferring the network. Comparing the inference power of different similarity scores indicated the clear superiority of conditioning or pruning based scores as they have the ability to eliminate indirect interactions. We also show that a mathematical measure based on the Fisher information matrix gives clues on the information quality of different data types to better represent the underlying metabolic network topology. Results on several datasets of increasing complexity consistently show that metabolic variations observed at steady state, the simplest experimental analysis, are already informative to reveal the connectivity of the underlying metabolic network with a low false-positive rate when proper similarity-score approaches are employed. For experimental situations this implies that a single organism under slightly varying conditions may already generate more than enough information to rightly infer networks. Detailed examination of the strengths of interactions of the underlying metabolic networks demonstrates that the edges that cannot be captured by similarity scores mainly belong to metabolites connected with weak interaction strength.
Electronic supplementary material
The online version of this article (doi:10.1007/s11306-009-0156-4) contains supplementary material, which is available to authorized users.
Keywords: Network inference, Interaction strength, Metabolome modeling, Indirect interactions, Biological/environmental variability, Similarity scores
Introduction
The cell’s phenotype emerges from the coordinated behaviour of a web of interactions among its genes, proteins and metabolites. This implies a close relationship between the structure of interaction networks and functionality (Futschik et al.2007; Stelling et al.2002; Vazquez et al.2003). Therefore, one of the challenges in systems biology is to infer cellular networks from data collected through high-throughput techniques. The so-called ‘omics’ data hold information on the network from which they are derived. A proper analysis of such data, therefore, can reveal the structural properties of the network in question, enabling discovery of direct interactions among the measured transcripts, proteins or metabolites. In this regard, network inference is a step towards elucidating functional properties in cellular systems since the network structure is the backbone behind normal as well as abnormal phenotypic states such as disease, malfunctioning, and overproduction.
Network inference approaches are highly popular in transcriptomics to infer genetic regulatory networks (Bansal et al.2007; Soranzo et al.2007). In this study, we focus on a relatively untouched area with the overall goal of inferring metabolic networks from metabolome data. The reverse engineering approach employed is a top–down approach to network reconstruction. In the widely used bottom–up approach the metabolic network topology is compiled from the literature and is later used as a scaffold in analyzing omics data (Gonzalez et al.2008; Notebaart et al.2006; Price et al.2004; Rahnenführer et al.2004), leading to the construction of genome-scale metabolic models. Such bottom–up models are mainly limited to stoichiometric interactions between metabolites, ignoring the interactions due to regulatory mechanisms such as inhibition or activation. Additionally, stoichiometric interactions in such models are not complete as revealed by the presence of a considerable percentage of totally inactive ‘dead-end’ metabolites (Förster et al.2003). These facts are the main reason why the reconstructed bottom–up models lead to some erroneous predictions of phenotypic states (Forster et al.2003). The top–down approach, on the other hand, does not have these limitations provided that the collected data capture the variation in all pathways.
Two major issues in the top–down approach are the type of experiment or perturbation to be performed and the type of network inference method to be used. Biological data collected in different ways (steady-state or dynamic experiments, under genetic or environmental perturbations) differ in the information content they carry about the underlying network (Soranzo et al.2007). Some researchers focus on methods that require complicated experimental design such as perturbation of each node in the system separately (Sontag et al.2004; de la Fuente et al.2002). On the contrary, we concentrate on analyzing the information content of observational metabolome data based on emerging biological or environmental variations around steady state without any sophisticated targeted design. Thereby, we aim to answer the question whether natural variation observed around steady state, which is the simplest experimental analysis, is informative enough to reveal the connectivity of the underlying metabolic network. Various reverse engineering methods of omics data exist in the literature (Bansal et al.2007; Markowetz and Spang 2007). We choose statistical similarity measures as a network inference tool since they are widely employed (Margolin et al.2006; Nemenman et al.2007; Soranzo et al.2007; Werhli et al.2006; de la Fuente et al.2004), and they best suit analysis of steady-state data. Moreover, a detailed application of similarity measures on metabolome data is missing in the literature unlike the popular usage in transcriptome data-based genetic network inference attempts (Markowetz and Spang 2007; Soranzo et al.2007). The amount of applications for metabolomics so far has been limited (Nemenman et al.2007), with no detailed comparative investigation of non-linear measures or conditioning and pruning approaches which eliminate indirect interactions.
A good starting point for metabolic network inference is the use of in silico generated metabolome data from kinetic metabolic models available in literature (Mendes et al. 2003). This approach facilitates to draw conclusions on the quality of data and perturbation needed for metabolic network inference of real systems as well as enabling quick testing of inference quality. Kinetic models of three example systems (threonine synthesis pathway of E. coli consisting of 4 metabolites, S. cerevisiae glycolysis pathway with 13 metabolites, and E. coli central metabolism pathway with 18 metabolites) were used in this study for in silico data generation. We test the effect of the following parameters on network inference: (a) different types of (natural) variability, (b) different types of similarity measures and (c) elimination of indirect interactions through conditioning and/or pruning.
Materials and methods
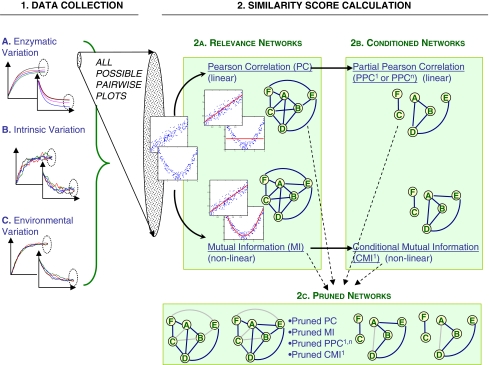
The general computational approach followed is depicted in Fig. 1.
Fig. 1.
The approach followed for metabolic network inference. Three datasets with different variability properties are collected in silico. Each dataset is processed to calculate similarity scores with linear and nonlinear methods (relevance networks). The alternative scores which remove indirect interactions are also applied (conditioned networks). All these networks are fed into pruning algorithm which checks data processing inequality (DPI). See Sect. 2 for details
In silico data generation
Kinetic details of models describing the studied systems were taken from JWS Online (Olivier and Snoep 2004). The systems were solved either using MATLAB’s built-in ordinary differential equation (ODE) solver ode15s for the enzymatic variability data or using the Milstein method of the stochastic differential equation (SDE) Toolbox (Picchini 2007) for the intrinsic variability and the environmental variability data (see next subsection for details of the data types). A thousand steady-state data points were collected from independent runs for each variability type analyzed. Initial values of concentrations were kept the same among different independent runs since they were found to have no effect on steady-state concentrations. For stochastic simulations, a real steady state is not possible due to fluctuating profiles, and data was collected after a few seconds of simulations starting from different near-steady-state concentrations to assure that the fluctuations were stabilized.
Biological/environmental variability
Metabolomics experiments conducted under identical conditions with the same genetic background do not necessarily lead to identical results (Fiehn et al.2000; Martins et al.2004). This has been attributed to natural variability inherent to living organisms originating from systems properties, leading to consistent correlation patterns among metabolites (Steuer 2006). In this study, we focus on three common factors causing this variability. Our goal is to compare the relative information content of each of these factors in revealing true systemic interactions.
Enzymatic variability
This type of variability is induced due to slight variation of enzyme concentrations, [E], or reaction rate constants, k, between replicate experiments. Each rate expression in the kinetic models has a parameter, νmax, which comprises both of these effects (νmax = k·[E]). A random variation of approximately ±10% was introduced to the νmax parameter of each rate expression in the models to mimic biological variability and to generate replicate metabolome data (Fig. 1) (Camacho et al.2005; Martins et al.2004). This was done by multiplying each νmax value with a random number, n, from a Normal distribution with unit mean and 0.05 standard deviation. The mathematical expression of the corresponding ODE set is:
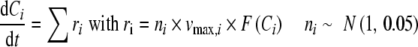 |
where Ci corresponds to the concentration of metabolite i and t is time. The variation can be much higher for mammalian systems (Margolin et al.2006) because there is considerable difference between individuals in terms of gene copy numbers and single-nucleotide-polymorphism (SNP) occurrences, affecting enzyme concentrations [E] and efficiency (k).
Intrinsic variability
Within an experiment, intracellular metabolites can exhibit true fluctuations over time (Kresnowati et al.2006; Wu et al.2005) (Fig. 1), which are not due to technical or experimental errors but because of fluctuations within cellular processes, also due to complex regulation patterns (Steuer et al.2003). Such fluctuating profiles can be natural, or can also be induced on purpose by introducing small fluctuations to temperature, pH or dissolved oxygen concentration of, e.g., a biotechnological system. A mathematical way of mimicking such ‘noisy’ profiles using kinetic models is to add a stochastic term to each ODE of the system. Thereby, the system is converted into an SDE set. Stochastic modeling is also a common way of introducing variability for model-based genetic network inference studies (Wang et al.2006; Yeung et al.2002). Mathematically expressed;
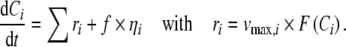 |
The stochastic term, ηi, is a random number from unit Normal distribution, and f is a system-specific weight. The weight, f, was chosen as 0.001, 0.1, and 0.01 for the 4-metabolite, 13-metabolite and 18-metabolite systems, respectively, to induce variation in metabolite levels similar in range to the metabolite levels of enzymatic variability case.
Environmental variability
Fluctuations can also be due to small changes in the extracellular substrate/nutrient concentrations, and these changes are transmitted and propagated within the cell, causing variations in the levels of internal metabolites (Steuer et al.2003). This approach results in fluctuations with noticeably smaller amplitude relative to the intrinsic variability for intermediate metabolites (Fig. 1). We have generated the third in silico metabolome dataset by adding a stochastic term only to the ODE of the initial metabolite in the considered system. In other words, this SDE represents the effect of the propagated substrate variations within the cell that is transmitted to the considered system:
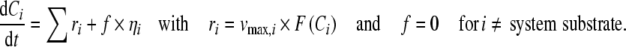 |
Similarity measures
Relevance networks
Pearson correlation (PC) was used as a linear statistical similarity measure. Spearman correlation practically gave the same results (results not shown). Entropy-based mutual information (MI) was used as a nonlinear similarity measure:
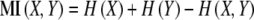 |
1 |
with H being the entropy calculated based on the b-spline interpolation algorithm of (Daub et al.2004), implemented in MATLAB. Spline parameters that were used are 3 for spline order, and 10 for the number of bins; see (Daub et al.2004) for explanations.
Conditioned networks
Conditioning is an approach enabling identification of indirect interactions in similarity networks. Elimination of such interactions can lead to a more refined network (see also de la Fuente et al. 2004). As a linear conditional similarity measure, two different scores were used. First order partial correlation is based on an exact formulation:
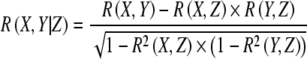 |
2 |
with R denoting zero-th order Pearson correlation. This score is calculated for every single Z outside the (X, Y) pair. The minimum of such scores is the first order partial correlation between (X, Y), and denoted as PPC1. The graphical Gaussian modeling (GGM) framework has also been employed as a linear conditioning approach, which applies conditioning on all remaining metabolites simultaneously (PPCn). It is straightforward to calculate partial Pearson correlation scores through the GGM approach by simple inversion and normalization: the inverse of the zero-th order Pearson correlation matrix is taken, and the resulting matrix is normalized to have diagonals-1 (Schäfer and Strimmer 2005).
First order conditional mutual information (CMI1) was used as a nonlinear conditional similarity measure. For a metabolite pair (X, Y) a set of CMI scores is calculated by conditioning with respect to each of the remaining metabolites (Z) one by one using the following expression:
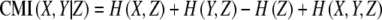 |
3 |
The minimum of those scores is considered as the CMI1 score of that pair. Higher order nonlinear conditioning was not used due to high computational time requirements.
Pruned networks
Pruning is an alternative approach to remove indirect interactions. The algorithm accepts a network graph as an input where all interactions with significant similarity scores are represented as an edge. Data processing inequality (DPI) is the method employed to prune this network, and is applied after all edges with insignificant scores (see below) are removed from the network. It is based on the comparison of pairwise similarity scores among every fully connected three metabolites (Margolin et al.2006). The edge with the lowest score in the checked triplets is assumed to be indirect and removed since there is a higher-score two-edge path connecting the two metabolites. A tolerance parameter of 0.10 was used in DPI-pruning calculations to prevent too stringent pruning (Margolin et al.2006). Mathematically expressing; for a triplet of metabolites X, Y, Z, if the edge between X and Z obeys the following inequality, then it is removed from the network:
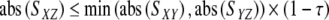 |
4 |
with S indicating similarity score, and τ the tolerance parameter. DPI approaches with higher order alternative path checks are also available (Chen 1998; Patil and Kulkarni 2007) under the name of pathfinder network scaling approach. Pathfinder network scaling is used widely in information visualization, citation pattern analysis and knowledge acquisition (White 2003; de Moya-Anegon et al.2007). In addition to triplet checks, we have also applied rectangular inequality checks for comparison. We have applied the pruning approach to all networks constructed based on unconditioned and conditioned scores (PC, MI, PPC1, PPCn and CMI1). The ARACNE approach (Margolin et al.2006) corresponds to the case where Eq. 4 is based on the MI score.
Significance measure of similarity scores
A distribution-free test, the permutation test, was applied to the collected in silico data to assign a P-value to each possible edge by shuffling the data 5,000 times. A P-value cut-off of 0.01 was used to select edges with significant similarity scores. These selected edges are combined to give the connectivity pattern of the inferred network, which then can be compared with the actual metabolic network derived from the in silico model. The formation of actual metabolic interaction network is based on the ODE balances around metabolites, which shows if the level of one metabolite is influenced by the level of others (calculation of Jacobean matrix of the system gives the same information quantitatively, see Sect. 2.5). In this way, not only the intuitive substrate-product interactions are counted, but also the influences between substrates of the reactions are covered. This corresponds to substrate-graph representation of metabolic networks in graph theoretical analyses (Wagner and Fell 2001).
Receiving-Operator Characteristic (ROC) curves were used as a global measure of network inference quality for larger systems, by plotting true-positive rates (TPR) and false-positive rates (FPR) against each other. The geometric mean of specificity and sensitivity is another measure which can be used to evaluate the quality of classifications at a given P-value threshold (e.g. P = 0.01) (Kubat et al.1998). ROC curves enable a global comparison whereas the geometric mean score allows the comparison of method performance based on a single score. It is calculated as:
 |
5 |
The score changes between 0 and 1, with 1 corresponding to perfect inference, and 0 to worst inference.
Effect of strength of interactions on network inference
As an independent tool to validate the results of the metabolic network inference, we use the interaction strength. The strength of interactions in a cellular network is not the same for all edges in the network. A practical way of quantifying the interaction strength between metabolite pairs in in silico kinetic systems is via the calculation of Jacobian matrix corresponding to the right-hand-side of the ODE system. The (i, j)th entry of the Jacobian matrix corresponds to the magnitude of change in the time behavior of metabolite i in response to an infinitesimal change in the level of metabolite j. Mathematically speaking;
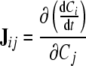 |
6 |
with C representing concentration.
To calculate the Jacobian strength of an interaction, (a) we have calculated the Jacobian matrix of the system at steady state based on Eq. 6 and (b) we have assigned the absolute maximum of upper- and lower-diagonal entries as the Jacobian strength of each metabolite pair since the two entries may differ depending on the reversibility of interactions.
Results and discussion
Threonine synthesis pathway in E. coli
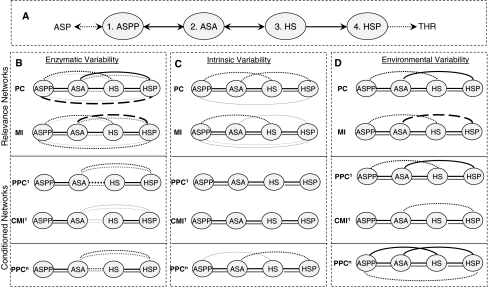
For illustrative purposes we start with a small example of four metabolites: the threonine synthesis pathway in E. coli (Chassagnole et al. 2001a, b). The system is a linear pathway with four metabolites and five reactions (Fig. 2a). The pathway has three true edges (E12, E23, E34) whereas the number of all possible edges is six (including false edges of E13, E14, E24). The three approaches to induce variability at steady state were applied to this system, and all related similarity measures were calculated as mentioned in the Sect. 2, enabling a thorough investigation of similarity network approaches. Resulting network configurations corresponding to each similarity analysis are given in Fig. 2 for each variability approach.
Fig. 2.
Inference of threonine synthesis pathway in E. coli by different similarity approaches. (a) The real pathway, (b) network inference by enzymatic variability, (c) network inference by intrinsic variability (d) network inference by environmental variability. Black lines are edges for non-pruned networks whereas gray lines show edges for pruned networks. Dashed and dotted lines imply ambiguity regarding the corresponding edges (i.e. edge is absent or present depending on different realizations). Dashed lines mean presence of the edge in at least 90% of 100 realizations, and thin dotted lines mean presence of the edge in only at most 10% of the cases. Normal dotted lines correspond to cases in between. ASP aspartate, ASPP aspartyl-P, ASA aspartic semialdehyde, HS homoserine, HSP O-phospho-homoserine, THR threonine
Similarity score calculations are based on a dataset of 1,000 generated data points. A hundred such datasets were generated to test the reproducibility of the resulting networks. The solid lines in Fig. 2 correspond to perfectly reproducible edges whereas the presence of dotted or dashed edges indicates variability in the inference results among the 100 independent datasets (see legend to Fig. 2 for details). Figure 2 reveals that conditioning reduces the number of false positives: PPC1 and CMI1 perform noticeably better compared to the non-conditioned counterparts that give networks with more connectivity. The performance of GGM based PPCn is also comparable. Additionally, DPI pruning is very effective, especially with the intrinsic and environmental variability approaches (Fig. 2c, d), leading to the full inference of the original network by all used similarity measures without leaving any ambiguous edges behind. This shows the refining power of pruning on the inferred network. It is more obvious for environmental variations where the non-pruned results are relatively less promising, especially for the linear measure tests. Even conditioned approaches lead to a set of false-positive edges for this type of data. Application of DPI pruning (gray lines), on the other hand, successfully infers the original network for all types of similarity measures (Fig. 2d).
In terms of linear vs. nonlinear measures, no clear difference was observed for the two systems between PC and MI, or PPC1 and CMI1, regardless of the variability approach. This implies that relationships between metabolites around steady state are mainly linear for the analyzed conditions, in parallel with previous findings for transcriptome data (Steuer et al.2002).
The three data types used in this study can be grouped in two classes. Enzymatic variability data is based on variations of enzymatic properties across different experiments leading to slight differences in individual reaction rates, and that, in turn, causes variability in metabolite levels. Intrinsic and environmental variability, on the other hand, cover net effect of several types of dynamic fluctuations on metabolite levels. It was shown for Vmax-dependent enzymatic variability (Camacho et al.2005) that two neighbouring metabolites in the network may have little or no similarity when the enzymes that regulate them vary in different directions causing a low correlation. That said, it may not be possible to have a perfect network inference based on enzymatic variability as it is dependent on enzyme mechanisms behind metabolic conversions. In other words, enzyme mechanisms play an important role in metabolic network inference. This was partly observed when conditioning or pruning is applied to the edge E23 in Fig. 2b, leading to ambiguous edges for neighbouring metabolites. The other two data types (Fig. 2c, d), on the other hand, did not have this limitation. This suggests that a different data type makes it possible to break the barriers due to enzyme mechanisms and to infer the edges connecting metabolites whose co-response are controlled by multiple enzymes in different directions.
Comparison of the three variability approaches indicate that, for this small example, intrinsic variability leads to the best results, with identification of the original network not only by conditioning but also by pruning regardless of the similarity method employed.
Application to larger networks: glycolytic pathway in S. cerevisiae and central metabolism in E. coli
The next examples are of larger networks. The first one is the glycolysis pathway of S. cerevisiae which consists of 13 metabolites and 18 reactions (Teusink et al.2000). The 13 metabolites correspond to 78 possible interactions, whereas the number of real edges in the network is 21. Additionally, the 18-metabolite and 30-reaction network of E. coli central carbon metabolism was considered (Chassagnole et al.2002), which has 153 possible pairwise interactions, of which 39 are genuine.
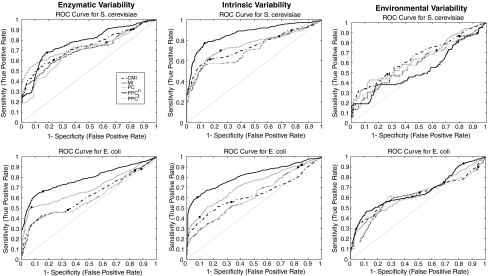
Receiving-Operator Characteristic curves were created for both microorganisms for an overall comparison of different employed approaches (Fig. 3). The curves are based on average true-positive and false-positive counts of 10 independent datasets. The diagonal line in ROC curves corresponds to cases where true-positive rate (TPR) and false-positive rate (FPR) are equal to each other, and known as random scenario. The more distant an ROC curve to the random scenario line in the upper diagonal area, the better the performance of the corresponding similarity score. In summary, the ROC curves reveal that (a) environmental variability has the worst performance, (b) PPCn is clearly superior to other approaches in either of the remaining two variability approaches since its ROC curve is most distant to the random scenario line, (c) unconditioned scores, PC and MI, have ROC curves relatively closest to random scenario line, in accordance with their low performance.
Fig. 3.
ROC curves for employed variability approaches for S. cerevisiae (1st row) and E. coli (2nd row) systems. The Black dots on the curves correspond to true positive rate and false positive rate for significance threshold of P ≤ 0.01. The dotted diagonal line corresponds to random scenario
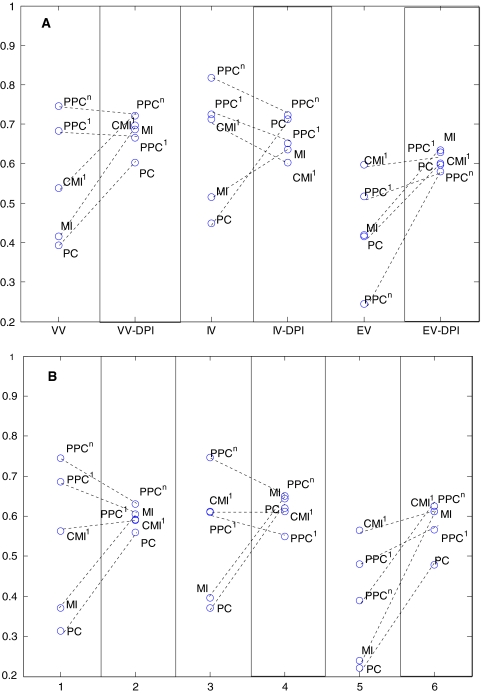
Figure 4a and b give a more focused view of the different variability methods based on the g-score (Eq. 5) at a P-value cut-off of 0.01. The detailed corresponding tables are given in Supplementary File. A superiority of intrinsic variability over others was observed (Supplementary File, Fig. 4), consistent with the results of the previous section. Pruning of the conditioned scores generally worsened the prediction or does not have any noticeable effect. The real power of pruning was observed when applied to non-conditioned scores, PC and MI. Additionally, the PPCn score without pruning was always better than any of the pruned networks (including the ARACNE approach; the pruning-applied MI) for enzymatic and intrinsic variability data. The ARACNE approach usually had a lower performance over the other DPI-pruned similarity scores, with pruned PPCn having a better inference, implying that mutual information is not always the best similarity measure to use for metabolic network inference. The use of rectangular inequality for pruning (see Sect. 2) did not lead to any significant change compared to triangular inequality (results not shown). A general observation valid for both metabolic systems and for all three variability approaches is that PC and MI measures result in highly connected networks, associated with a very high number of false positives although they are slightly better to predict positive edges.
Fig. 4.
Geometric mean score of sensitivity and specificity for aS. cerevisiae and bE. coli systems. The g-scores are for P = 0.01 threshold for similarity measures. The x-axis of figures correspond to the pruned and non-pruned versions of three variability approaches with VV: enzymatic variability, IV: intrinsic variation, EV: environmental variation; and block names with DPI corresponding to pruned versions. Pruned and non-pruned versions of the same similarity score for the same type of variability was connected to each other by dotted lines to show the effect of pruning. Higher g-scores correspond to better inference
In practice, the ROC curves are not available because the true network is unknown. Hence, one has to select a P-value and usually a value of 0.01 is chosen. The consequence of this selection is shown with the black dots on the ROC’s of Fig. 3. The choice of the cut-off point for the P-value can lead to unfavorable results, e.g., in the case of the CMI of E. coli of the enzymatic variability: a better compromise between false-positive rate and true-positive rate would have been obtained at another P-value (i.e. at another point on the ROC curve). Unfortunately, the position of the ‘P-value point’ on the ROC curve is not known for practical cases. This serves as a remark of warning for practitioners: the P-value is just an arbitrary choice and a different choice of P-value leads to different results, a more- or less-connected graph with more or less false positives and false negatives, and thereby the choice of P-value can lead to a suboptimal recovery of the underlying network.
The importance of quantitative measures for the information quality of experimental data to be used in network inference was pointed out (Camacho et al.2007). The Fisher Information Matrix has been in use for this purpose to judge the quality of experiments (Kresnowati et al.2005). The multiplication of a data matrix with its transpose is called the Fisher Information Matrix and the condition number (called modified E-optimality) of this matrix is one of the most widely used criteria for information content of data (Balsa-Canto et al.2007). In this measure, lower scores correspond to better data types. We calculated the condition number of the Fisher Information Matrices corresponding to each of the three data types for both systems. Data were standardized before the calculation of modified E-optimality score. The condition numbers of data from environmental variability are on the order of 109 and 1012 for S. cerevisiae and E. coli systems, respectively, while that of enzymatic and intrinsic variability data are at least 106 fold lower. This fact points to the low quality of the environmental variability data, in parallel with the observations in Fig. 4a and b. To further strengthen these results, environmental variability data with 50 times higher weight for the stochastic term was generated for E. coli; resulting in a dataset with much higher variation. The corresponding condition number of the Fisher Information Matrix was, albeit lower than original, still 104 fold higher than the other data matrices, suggesting that environmental variability does not result in informative data for the inference of intracellular networks.
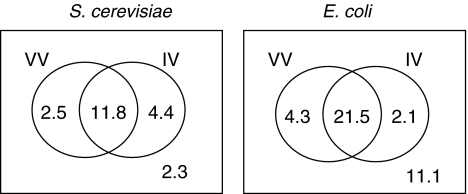
Figure 5 compares the complementary power of the two best variability approaches for the best performing similarity score, PPCn, in terms of true-positive counts. The figure indicates that some edges were inferred only by one of the two methods. The union of edges correctly inferred by both variability methods corresponded to a true-positive rate of 0.89 (0.68 and 0.77 for individual approaches) and 0.72 (0.66 and 0.61 for individual approaches) for S. cerevisiae and E. coli models respectively for PPCn score, leading to a more complete picture of underlying metabolic network. The corresponding false-positive rates were 0.28 and 0.21 for both microbial systems.
Fig. 5.
True-positive counts of enzymatic variability (VV) and intrinsic variability (IV) approaches for PPCn score. The results are given in complementary way for both microbial systems. VV and IV can capture the same 11.8 and 21.5 edges in S. cerevisiae and E. coli systems out of 21 and 39, respectively. There is a small number of edges which can only be inferred by one of the variability approaches
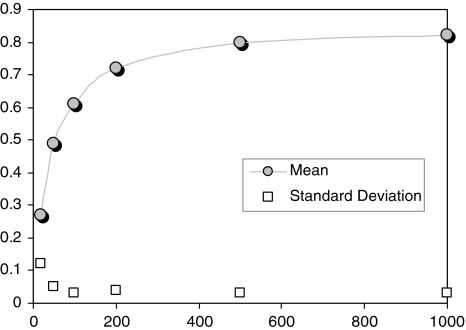
Figure 6 demonstrates the effect of the number of datapoints on the quality of network inference for the best-performing score, PPCn for intrinsic variability data of S. cerevisiae. The plot shows that 500 datapoints are sufficient to obtain the same inference quality, and there is a sharp decrease in the quality if the dataset includes less than 200 points. An important remaining question is at what sample sizes this type of network inference breaks down, but this is also largely related to the amount of natural variation included in the dataset. This should be part of further study on metabolic network inference using similarity measures. However, the requirement of a high number of replicate measurements is already a known disadvantage of similarity-based network inference approaches (Camacho et al. 2007; Soranzo et al. 2007).
Fig. 6.
The effect of number of datapoints (x-axis) on the inference quality of PPCn. The figure is based on intrinsic variability-based data of S. cerevisiae system. Y-axis shows the geometric-mean of sensitivity and specificity as introduced in Eq. 5. The scores are averages of 10 different datasets. The corresponding standard deviation is also plotted
Validation of the results
Figure 5 reveals that, especially for E. coli system, there is a number of interactions which cannot be captured by neither the enzymatic nor the intrinsic variability-based data types (false negatives). Therefore, to investigate the role of weak-strength interactions on the false negatives encountered in similarity-based inference methods, we first focus on the E. coli system. We have classified weak interactions as the ones with interaction strengths lower than 1. From the 39 interactions in E. coli system, 12 fall into this category. Further inspection of these weakest 12 interaction strengths (with a range of 8.10−6–0.17) reveals that 9 and 11 of them have insignificant PPCnP-values, respectively, for data based on enzymatic and intrinsic variability. This explains why these interactions cannot be captured by the PPCn score. Ignoring these interactions can lead to a true-positive rate of as high as 0.84, compared to current values of around 0.60 (Fig. 3, Supplementary Table 1). Further calculations of Spearman rank correlations between strengths of 39 interactions and corresponding PPCn scores gives 0.64 (P-value: 1.10−5) and 0.72 (P-value: 2.10−7), respectively, for enzymatic and intrinsic variability datasets. That is, there is a significant relationship between these two entities for both data types. For S. cerevisiae, a very low number of false negatives was observed, which is in accordance with the fact that no weak interactions were present in this system. Summarizing, false negatives in metabolic network discovery are present because of low interaction strength and not primarily because of the failure of the network inference methods.
Concluding remarks
A systematic analysis of metabolic network inference was performed based on different types of in silico steady-state metabolome data. A comprehensive investigation of similarity measures for network inference on metabolomics data enabled the testing of nonlinear measures as well as measures eliminating indirect interactions. Linear versus nonlinear similarity measures were shown not to differ noticeably implying the lack of non-linear relationships among metabolites around steady-state conditions, which is especially true for datasets with relatively small perturbations around steady state. Conditioning and pruning approaches were found to improve results considerably by eliminating a high percentage of indirect links. The false negatives encountered were shown to be related to intrinsic properties of the network, i.e. weak interactions. Along the way, we extended the ARACNE approach, which is specific to the MI scores, to other similarity scores including conditioned ones and concluded that PPCn has a better inference capacity than any of the pruned scores.
Comparison of different variability methods reveals that intrinsic variability is generally more informative. Translating this result to experimental situations, this implies that a single organism under slightly varying conditions may already generate more than enough information to rightly infer networks, without having to turn to more genetic diversity or to more complicated experimental design. However, solely perturbing substrate conditions will not reveal the underlying network.
Use of Fisher Information Matrix-based testing gave hints on the quality of different datasets, suggesting a diagnostic for the quantitative pre-inspection of data. Use of environmental variability was not promising even when conditioning was applied. Pruning, however, improved the results of this variability type considerably, albeit still being inferior to the two other variability approaches.
A disadvantage with similarity-based approaches presented here is the requirement of a high number of replicate measurements. However, no complicated experimental design is needed, making it more practical to employ this approach. Additionally, we have shown that pruning and conditioning approaches have the power to eliminate some ambiguous edges arising due to non-reproducible datasets. We have focused on data from steady-state variations without any designed perturbation since designed perturbations (e.g. knock-out or overexpression of selected enzymes) correspond to different cellular states with different similarity patterns. Therefore, one should be cautious to analyze such data as it can lead to misleading correlations (Camacho et al.2005).
It is not yet possible to have a perfect inference for metabolic networks with the presented approach. However, the finding that different data types hold different information over a network points to the importance of integrated analysis of different data types. It can be argued that all three different types of variation analyzed can be present under normal conditions. Integration of results from different data types were shown to result in much higher true-positive rates, pointing to higher information content of a dataset including the effect of all three variations. The focus on proper experimental setup for reverse engineering approaches together with the measures quantifying the information content of omics datasets will be the future trend in this top–down systems biology approach.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work is supported by Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research (NWO). Joke Blom (CWI, Amsterdam) is gratefully acknowledged for her invaluable help on the solution of stochastic differential equations. Prof. Ruud Berger (UMC Utrecht) and Prof. Joost Teixeira de Mattos (SILS, University of Amsterdam) are acknowledged for discussions on variability in mammalian and microbial systems, respectively. We thank Daniel J. Vis (SILS, University of Amsterdam) for his comments on the manuscript; Kaustubh Patil (Max-Planck Institute, Saarbruecken) for discussions on pathfinder networks and reading the manuscript; and Emrah Nikerel (Technical University of Delft) for discussions on intrinsic fluctuations.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s11306-009-0156-4) contains supplementary material, which is available to authorized users.
References
- Balsa-Canto, E., Rodriguez-Fernandez, M., & Banga, J. R. (2007). Optimal design of dynamic experiments for improved estimation of kinetic parameters of thermal degradation. Journal of Food Engineering,82, 178–188. doi:10.1016/j.jfoodeng.2007.02.006. [DOI]
- Bansal, M., Belcastro, V., Ambesi-Impiombato, A., & di Bernardo, D. (2007). How to infer gene networks from expression profiles. Molecular Systems Biology,3, 78. [DOI] [PMC free article] [PubMed]
- Camacho, D., Fuente, A., & Mendes, P. (2005). The origin of correlations in metabolomics data. Metabolomics,1, 53–63. doi:10.1007/s11306-005-1107-3. [DOI]
- Camacho, D., Licona, P. L., Mendes, P., & Laubenbacher, R. (2007). Comparison of reverse-engineering methods using an in silico network. Annals of the New York Academy of Sciences,1115, 73–89. doi:10.1196/annals.1407.006. [DOI] [PubMed]
- Chassagnole, C., Fell, D. A., Raïs, B., Kudla, B., & Mazat, J. P. (2001a). Control of the threonine-synthesis pathway in Escherichia coli: A theoretical and experimental approach. The Biochemical Journal,356, 433–444. doi:10.1042/0264-6021:3560433. [DOI] [PMC free article] [PubMed]
- Chassagnole, C., Noisommit-Rizzi, N., Schmid, J. W., Mauch, K., & Reuss, M. (2002). Dynamic modeling of the central carbon metabolism of Escherichia coli. Biotechnology and Bioengineering,79, 53–73. doi:10.1002/bit.10288. [DOI] [PubMed]
- Chassagnole, C., Raïs, B., Quentin, E., Fell, D. A., & Mazat, J. P. (2001b). An integrated study of threonine-pathway enzyme kinetics in Escherichia coli. The Biochemical Journal,356, 415–423. doi:10.1042/0264-6021:3560415. [DOI] [PMC free article] [PubMed]
- Chen, C. (1998). Generalised similarity analysis and pathfinder network scaling. Interacting with Computers,10, 107–128. doi:10.1016/S0953-5438(98)00015-0. [DOI]
- Daub, C. O., Steuer, R., Selbig, J., & Kloska, S. (2004). Estimating mutual information using b-spline functions–an improved similarity measure for analysing gene expression data. BMC Bioinformatics,5, 118. doi:10.1186/1471-2105-5-118. [DOI] [PMC free article] [PubMed]
- de la Fuente, A., Bing, N., Hoeschele, I., & Mendes, P. (2004). Discovery of meaningful associations in genomic data using partial correlation coefficients. Bioinformatics,20, 3565–3574. [DOI] [PubMed]
- de la Fuente, A., Brazhnik, P., & Mendes, P. (2002). Linking the genes: inferring quantitative gene networks from microarray data. Trends in Genetics,18, 395–398. [DOI] [PubMed]
- de Moya-Anegon, F., Vargas-Quesada, B., Chinchilla-Rodriguez, Z., Corera-Alvarez, E., Munoz-Fernandez, F. J., & Herrero-Solena, V. (2007). Visualizing the marrow of science. Journal of the American Society for Information Science and Technology,58, 2167–2179. [DOI]
- Fiehn, O., Kopka, J., Dörmann, P., Altmann, T., Trethewey, R. N., & Willmitzer, L. (2000). Metabolite profiling for plant functional genomics. Nature Biotechnology,18, 1157–1161. doi:10.1038/81137. [DOI] [PubMed]
- Förster, J., Famili, I., Fu, P., Palsson, B. Ø., & Nielsen, J. (2003). Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Research,13, 244–253. doi:10.1101/gr.234503. [DOI] [PMC free article] [PubMed]
- Forster, J., Famili, I., Palsson, B. O., & Nielsen, J. (2003). Large-scale evaluation of in silico gene deletions in saccharomyces cerevisiae. Omics,7, 193–202. doi:10.1089/153623103322246584. [DOI] [PubMed]
- Futschik, M. E., Chaurasia, G., & Herzel, H. (2007). Comparison of human protein–protein interaction maps. Bioinformatics (Oxford, England),23, 605–611. doi:10.1093/bioinformatics/btl683. [DOI] [PubMed]
- Gonzalez, O., Gronau, S., Falb, M., Pfeiffer, F., Mendoza, E., Zimmer, R., et al. (2008). Reconstruction, modeling & analysis of halobacterium salinarum r-1 metabolism. Molecular BioSystems,4, 148–159. doi:10.1039/b715203e. [DOI] [PubMed]
- Kresnowati, M. T. A. P., van Winden, W. A., Almering, M. J. H., ten Pierick, A., Ras, C., Knijnenburg, T. A., et al. (2006). When transcriptome meets metabolome: fast cellular responses of yeast to sudden relief of glucose limitation. Molecular Systems Biology,2, 49. doi:10.1038/msb4100083. [DOI] [PMC free article] [PubMed]
- Kresnowati, M. T. A. P., van Winden, W. A., & Heijnen, J. J. (2005). Determination of elasticities, concentration and flux control coefficients from transient metabolite data using linlog kinetics. Metabolic Engineering,7, 142–153. doi:10.1016/j.ymben.2004.12.002. [DOI] [PubMed]
- Kubat, M., Holte, R., & Matwin, S. (1998). Machine learning for the detection of oil spills in satellite radar images. Machine Learning,30, 195–215. doi:10.1023/A:1007452223027. [DOI]
- Margolin, A. A., Nemenman, I., Basso, K., Wiggins, C., Stolovitzky, G., Dalla Favera, R., et al. (2006). Aracne: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics,7(Suppl 1), S7. doi:10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed]
- Markowetz, F., & Spang, R. (2007). Inferring cellular networks—A review. BMC Bioinformatics,8(Suppl 6), S5. doi:10.1186/1471-2105-8-S6-S5. [DOI] [PMC free article] [PubMed]
- Martins, A., Camacho, D., Shuman, J., Sha, P., Mendes, P., & Shulaev, V. (2004). A systems biology study of two distinct growth phases of Saccharomyces cerevisiae cultures. Current Genomics,5, 649–663. doi:10.2174/1389202043348643. [DOI]
- Mendes, P., Sha, W., & Ye, K. (2003). Artificial gene networks for objective comparison of analysis algorithms. Bioinformatics (Oxford, England),19(Suppl. 2), ii122–ii129. doi:10.1093/bioinformatics/btg1069. [DOI] [PubMed]
- Nemenman, I., Escola, G. S., Hlavacek, W. S., Unkefer, P. J., Unkefer, C. J., & Wall, M. E. (2007). Reconstruction of metabolic networks from high-throughput metabolite profiling data: In silico analysis of red blood cell metabolism. Annals of the New York Academy of Sciences,1115, 102–115. doi:10.1196/annals.1407.013. [DOI] [PubMed]
- Notebaart, R. A., van Enckevort, F. H. J., Francke, C., Siezen, R. J., & Teusink, B. (2006). Accelerating the reconstruction of genome-scale metabolic networks. BMC Bioinformatics,7, 296. doi:10.1186/1471-2105-7-296. [DOI] [PMC free article] [PubMed]
- Olivier, B. G., & Snoep, J. L. (2004). Web-based kinetic modelling using jws online. Bioinformatics (Oxford, England),20, 2143–2144. doi:10.1093/bioinformatics/bth200. [DOI] [PubMed]
- Patil, K. R., & Kulkarni, A. J. (2007). A simple visualization technique to understand the system dynamics in bioreactors. Biotechnology Progress,23, 1101–1105. [DOI] [PubMed]
- Picchini, U. (2007). Sde toolbox: Simulation and estimation of stochastic differential equations with matlab. Retrieved from http://sdetoolbox.sourceforge.net.
- Price, N. D., Reed, J. L., & Palsson, B. O. (2004). Genome-scale models of microbial cells: evaluating the consequences of constraints. Nature Reviews. Microbiology,2, 886–897. doi:10.1038/nrmicro1023. [DOI] [PubMed]
- Rahnenführer, J., Domingues, F. S., Maydt, J., & Lengauer, T. (2004). Calculating the statistical significance of changes in pathway activity from gene expression data. Statistical Application in Genetics and Molecular Biology3, Article16. [DOI] [PubMed]
- Schäfer, J., & Strimmer, K. (2005). An empirical Bayes approach to inferring large-scale gene association networks. Bioinformatics (Oxford, England),21, 754–764. doi:10.1093/bioinformatics/bti062. [DOI] [PubMed]
- Sontag, E., Kiyatkin, A., & Kholodenko, B. N. (2004). Inferring dynamic architecture of cellular networks using time series of gene expression, protein and metabolite data. Bioinformatics (Oxford, England),20, 1877–1886. doi:10.1093/bioinformatics/bth173. [DOI] [PubMed]
- Soranzo, N., Bianconi, G., & Altafini, C. (2007). Comparing association network algorithms for reverse engineering of large-scale gene regulatory networks: Synthetic versus real data. Bioinformatics (Oxford, England),23, 1640–1647. doi:10.1093/bioinformatics/btm163. [DOI] [PubMed]
- Stelling, J., Klamt, S., Bettenbrock, K., Schuster, S., & Gilles, E. D. (2002). Metabolic network structure determines key aspects of functionality and regulation. Nature,420, 190–193. doi:10.1038/nature01166. [DOI] [PubMed]
- Steuer, R. (2006). Review: On the analysis and interpretation of correlations in metabolomic data. Briefings in Bioinformatics,7, 151–158. doi:10.1093/bib/bbl009. [DOI] [PubMed]
- Steuer, R., Kurths, J., Daub, C. O., Weise, J., & Selbig, J. (2002). The mutual information: detecting and evaluating dependencies between variables. Bioinformatics (Oxford, England),18(Suppl 2), S231–S240. [DOI] [PubMed]
- Steuer, R., Kurths, J., Fiehn, O., & Weckwerth, W. (2003). Observing and interpreting correlations in metabolomic networks. Bioinformatics Oxford, England,19, 1019–1026. doi:10.1093/bioinformatics/btg120. [DOI] [PubMed]
- Teusink, B., Passarge, J., Reijenga, C. A., Esgalhado, E., van der Weijden, C. C., Schepper, M., et al. (2000). Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. European Journal of Biochemistry,267, 5313–5329. doi:10.1046/j.1432-1327.2000.01527.x. [DOI] [PubMed]
- Vazquez, A., Flammini, A., Maritan, A., & Vespignani, A. (2003). Global protein function prediction from protein–protein interaction networks. Nature Biotechnology,21, 697–700. doi:10.1038/nbt825. [DOI] [PubMed]
- Wagner, A., & Fell, D. A. (2001). The small world inside large metabolite networks. Proceedings of the Royal Society of London Series B. Biological Sciences,268, 1803–1810. doi:10.1098/rspb.2001.1711. [DOI] [PMC free article] [PubMed]
- Wang, Y., Joshi, T., Zhang, X., Xu, D., & Chen, L. (2006). Inferring gene regulatory networks from multiple microarray datasets. Bioinformatics (Oxford, England),22, 2413–2420. doi:10.1093/bioinformatics/btl396. [DOI] [PubMed]
- Werhli, A. V., Grzegorczyk, M., & Husmeier, D. (2006). Comparative evaluation of reverse engineering gene regulatory networks with relevance networks, graphical gaussian models and bayesian networks. Bioinformatics (Oxford, England),22, 2523–2531. doi:10.1093/bioinformatics/btl391. [DOI] [PubMed]
- White, H. D. (2003). Pathfinder networks and author cocitation analysis: a remapping of paradigmatic information scientists. Journal of the American Society for Information Science and Technology,54, 423–434. doi:10.1002/asi.10228. [DOI]
- Wu, L., Mashego, M. R., van Dam, J. C., Proell, A. M., Vinke, J. L., Ras, C., et al. (2005). Quantitative analysis of the microbial metabolome by isotope dilution mass spectrometry using uniformly 13c-labeled cell extracts as internal standards. Analytical Biochemistry,336, 164–171. doi:10.1016/j.ab.2004.09.001. [DOI] [PubMed]
- Yeung, M. K. S., Tegnér, J., & Collins, J. J. (2002). Reverse engineering gene networks using singular value decomposition and robust regression. Proceedings of the National Academy of Sciences of the United States of America,99, 6163–6168. doi:10.1073/pnas.092576199. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.