Abstract
Activation of Notch signaling has been implicated in pathogenesis of various hematologic tumors including leukemias, lymphomas, and multiple myeloma. Pre-clinical studies have suggested that inhibition of Notch could be an attractive new approach to treatment of hematologic malignancies. This review discusses most recent findings in the field and potential role of Notch signaling as a therapeutic target focusing on the effects of γ-secretase inhibitors.
Keywords: Notch, Gamma-secretase inhibitor (GSI), Hematological malignancies, T-ALL, Multiple myeloma, B-cell malignancies
1. Introduction
Notch is a family of transmembrane proteins that function both as cell surface receptors and transcription regulators. The first gene of this family was cloned in the mid-1980s (Wharton et al., 1985) and identified as a gene responsible for a specific phenotype observed in one strain of Drosophila, precisely an unusual shape of Drosophila wing blades that had notches at the end. Since then Notch proteins have been identified in various species, including mammals. The first human gene of this family, Notch-1 was discovered in 1991 through the analysis of the chromosomal translocation t(7;9)(q34;q34.3) observed in patients with T-cell acute lymphoblastic leukemia (T-ALL). It was then found that the locus on chromosome 9 contains a gene highly homologous to the Drosophila gene Notch (Ellisen et al., 1991). Since then Notch signaling has been implicated in multiple processes that govern normal morphogenesis, differentiation, cell proliferation and apoptosis. At the same time evidence has accumulated that aberrant Notch activation contributes to tumorigenesis in various solid and hematological tumors. Targeting of the Notch pathway therefore may represent a promising approach in anti-cancer therapy. This review is focused on the role of Notch in hematological malignancies and discusses the clinical prospects of Notch inhibitors.
2. The Notch pathway
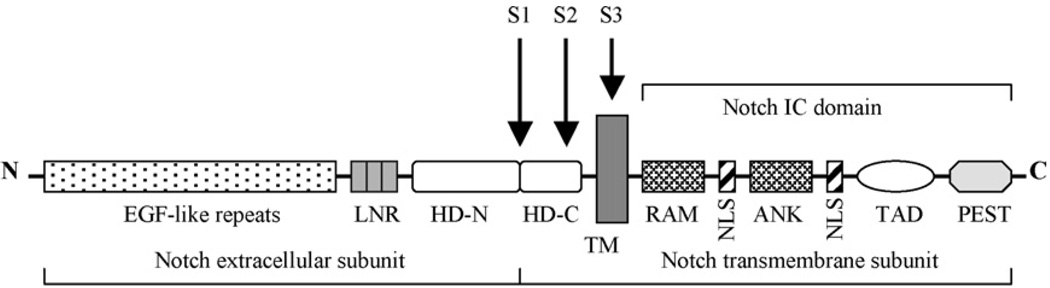
In mammals, the Notch family of receptors includes four members: Notch-1, -2, -3, and -4. Each member is a large heterodimeric protein comprised of extracellular, transmembrane, and intracellular domains in which distinct structural regions are associated with different function (Fig. 1). The extracellular domain contains a variable number of epidermal growth factor-like (EGF) repeats (36 in Notch-1 and Notch-2; 34 in Notch-3, and 29 in Notch-4) involved in ligand binding. The N-terminal localized EGF repeats are followed by a negative regulatory region that is responsible for maintaining the receptor in a resting state prior to ligand binding. This region includes three Notch family specific Lin12/Notch repeats (LNRs) near the C-terminus of the extracellular domain and a heterodimerization domain (HD) (Vardar et al., 2003). The intracellular domain contains two protein–protein interaction domains (RAM domain and ankyrin repeats), two nuclear localization signals, a transactivation domain, and a C-terminal PEST (polypeptide enriched in proline, glutamate, serine, and threonine residues) sequence involved in Notch protein degradation.
Fig. 1.
Schematic organization of Notch receptor. IC-intracellular domain; EGF, epidermal growth factor; LNR, Lin12/Notch repeats; HD-N, N-terminal region of heterodimerization domain; HD-C, C-terminal region of heterodimerization domain; TM-transmembrane domain of Notch; NLS, nuclear localizing signals; RAM, RAM domain; ANK, Ankyrin repeat domain; TAD, transactivation domain; PEST, a region rich in proline (P), glutamine (E), serine (S), and threonine (T) residues. The cleavage sites for furin-like proteases (S1), ADAM-type metalloproteases (S2), and γ-secretase (S3) are shown.
Notch proteins are initially synthesized as ~300–350 kDa full-length unprocessed precursors which then undergo proteolytic cleavage in the trans-Golgi network before reaching the cell surface. This cleavage is mediatedby a furin-like convertase and occurs within HD at a site referred to as the S1 cleavage site (Blaumueller et al., 1997; Logeat et al., 1998). This creates a Notch heterodimer consisting of extracellular and transmembrane subunits where N- and C-terminal halves of the HD are non-covalently associated (Rand et al., 2000). The extracellular subunit includes the majority of the extracellular domain. The transmembrane subunit consists of the part of the HD and the complete transmembrane and intracellular domains. Notch heterodimer is subsequently transported to the cell membrane and expressed as a cell surface receptor.
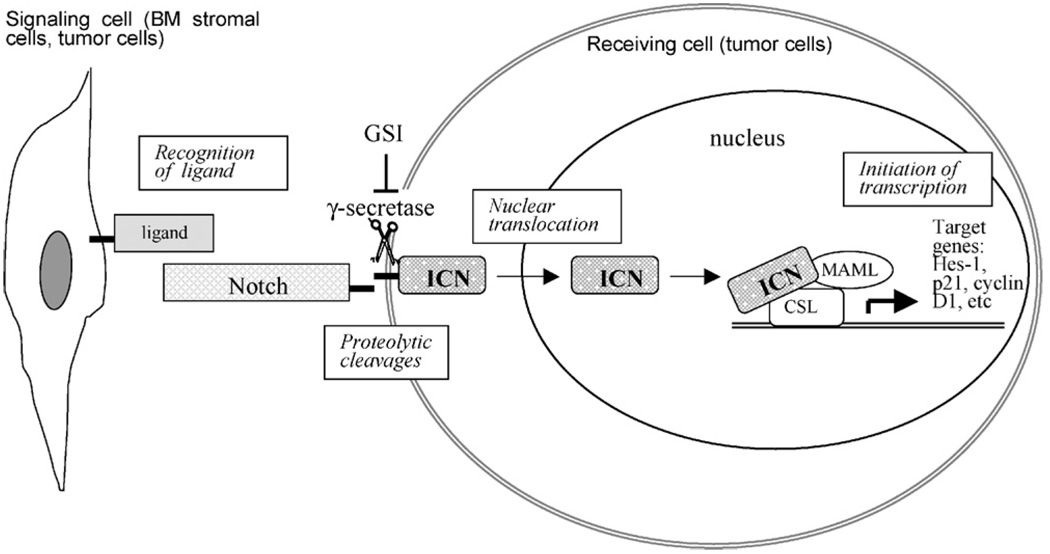
Activation of Notch signaling is initiated by binding of EGF-like repeats of Notch receptors to its ligands (Fig. 2). At present two Notch ligand families, Jagged and Delta-like have been described in mammals with a total of five Notch ligands identified (Jagged-1, -2, Delta-1, -3, and -4). Multiple lines of evidence have converged on a generally accepted mechanism of Notch activation that occurs in ligand-dependent fashion and involves two successive proteolytic cleavage steps (Brown et al., 2000; Mumm and Kopan, 2000). The first cleavage step is mediated by ADAM/TACE (A disintegrin and metalloprotease/tumor-necrosis-factor α converting enzyme) family and occurs within the extracellular domain just external to the transmembrane domain at a site referred to as the S2 cleavage site (Brou et al., 2000; Mumm et al., 2000). Subsequently, the second cleavage occurs at the site S3 located within the transmembrane domain. This cleavage step is mediated by a multisubunit protease complex possessing γ-secretase activity and comprising of presenilin 1 and 2, nicastrin, Pen-2, and Aph-1 (De Strooper et al., 1999; Schroeter et al., 1998). As a result of this cleavage the intracellular active domain of Notch (ICN) is liberated and rapidly translocates to the nucleus where it binds a transcriptional repressor RBP-Jk (recombination-binding protein Jk, also known as C promoter-binding factor (CBF-1), or CSL (CBF-1/Supressor of Hairless/Lag1)) through RAM and ankyrin repeat domains. In the absence of ICN, CBF-1 acts as a transcriptional repressor due to its ability to bind several transcriptional co-repressor complexes including histone deacetylase-1 and -2, CIR, SKIP, SMRT (Mumm and Kopan, 2000). Binding of ICN to CBF-1 displaces co-repressor complexes, thereby de-repressing transcription from promoters with CBF-1 binding elements (Mumm and Kopan, 2000). In addition, ICN/CBF-1 complexes recruits Mastermind-like (MAML) family of transcriptional activator proteins (Wu and Griffin, 2004). In turn, MAML bind several different transcriptional co-activators including histone acetyltransferase p300 and PCAF providing an additional stimulus for transcription of genes harboring CBF-1-binding sites (Artavanis-Tsakonas et al., 1999). The targets of ICN/CBF-1 signals in humans and mice include genes of the two families of basic helix-loop-helix transcriptional repressors HES (hairy/enhancer-of-split) and HERP (HES-related repressor protein, also known as HEY), as well as cyclins D1, p21, and NF-κB (Cheng et al., 2001; Iso et al., 2003; Rangarajan et al., 2001; Ronchini and Capobianco, 2001).
Fig. 2.
Canonical Notch signaling pathway. Upon binding to its ligand Jagged or Delta, Notch receptor undergoes two proteolytic cleavages, the last one is mediated by γ-secretase. This leads to the release of the intracellular ICN domain which then translocates to the nucleus and binds DNA-binding protein CSL(CBF-1) displacing co-repressor complex and recruiting co-activators including MAML, thereby converting CSL from a repressor to an transcriptional activator. ICN, Notch intracellular domain; GSI, γ-secretase inhibitor; MAML, mastermind-like coactivator; CSL(CBF-1), suppressor of hairless.
In addition to the described above canonical activation of Notch there is evidence that alternative CBF-1-independent pathways of Notch activation also exists (Martinez Arias et al., 2002). Deltex has been identified as one of the key mediators of CBF-1 independent Notch signaling (Deftos et al., 2000).
3. Notch signaling in hematological malignancies
3.1. T-cell acute lymphoblastic leukemia
The prominent role of Notch signaling in the pathogenesis of T-cell malignancies was underscored in several recently published reviews (Aster et al., 2008; Staal and Langerak, 2008). Here, we provide a brief overview of the data presently available.
In human leukemias, Notch-1 activation was initially demonstrated in T-ALL harboring the t(7;9)(q34;q34.3), a rare chromosomal translocation identified in less than 1% of T-ALL cases. As a result of this rearrangement a truncated Notch-1 gene is juxtaposed next to the T-cell receptor β locus, leading to the ligand-independent aberrant expression of a constitutively active form of Notch-1 (Ellisen et al., 1991). Following this initial discovery, activating mutations in the Notch-1 gene were found in more than 50% of human T-ALL (Breit et al., 2006; Weng et al., 2004). Activating mutations in Notch-1 are concentrated in exons 26 and 27, encoding the HD domain, and in exon 34, encoding the PEST domain (Weng et al., 2004). HD mutations are typically single amino acid substitutions and small in-frame deletions and insertions that induce ligand-independent activation of Notch, either due to poor S1 processing or by destabilizing the HD-LNR interaction, resulting in increased dissociation of the heterodimer or by displacing the S2 site away from the protective effect of the HD-LNR complex and exposing it to proteolytic cleavage (Malecki et al., 2006; Weng et al., 2004). In contrast to HD mutations, PEST mutations encode premature stop codons and lead to generation of truncated forms of Notch lacking the PEST domain, resulting in an increased level of active Notch due to its impaired proteosomal degradation (Weng et al., 2004). Recently, another class of activating mutations within the extracellular juxtamembrane region of Notch-1 was identified in T-ALL (Sulis et al., 2008). These juxtamembrane expansion mutations (JEMs) are generated by internal tandem duplications in the 3′-end of intron 27 and/or in the proximal region of exon 28 and resulted in the insertion of relatively long peptides around position 1740 of Notch-1. Thus, JEMs distance the entire HD-LNR complex from the membrane allowing ligand-independent proteolytic processing of S2 (Sulis et al., 2008). It is commonly accepted that deregulated Notch-1 signaling is major contributor in the pathogenesis of T-ALL.
3.2. Acute myeloid leukemia (AML)
The role of Notch signaling in AML is not that well established as in T-ALL. Although activating mutations of Notch have been reported for AML they are a rare event. Sequencing of HD and PEST domains of Notch-1 in 23 AML cell lines reveals truncating mutations in the PEST domain in 3 of them. Out of these three cell lines, two were derived from an AML relapse in a patient initially diagnosed with T-ALL (Palomero et al., 2006b). Mutation analysis of the Notch-1 gene in 121 AML and myelodysplastic syndrome (MDS) patients demonstrated the presence of a HD domain activating mutation in only one case of AML (undifferentiated M0 AML) (Palomero et al., 2006b). A missense mutation was found in the PEST domain in one out of 12 primary AML samples, but the precise role of the mutation has not been determined (Fu et al., 2006). No mutations were identified in the ankyrin repeats and PEST domain in 2 primary samples (Chen et al., 2008).
Although the majority of AML cell lines and primary samples express Notch-1 as well as its ligand Jagged-1 (Chiaramonte et al., 2005; Tohda and Nara, 2001), the role of Notch signaling in AML has to be determined. Despite the well-accepted notion that activated Notch supports cell self-renewal and tumorigenesis, Tohda et al. failed to demonstrate the ability of Notch ligands to promote self-renewal of primary AML cells. Instead, activation of Notch tended to induce differentiation of leukemic cells (Tohda et al., 2005).
3.3. B-cell malignancies
The majority of B-cell malignancies were reported to over-express Notch proteins. Thus, high level of active Notch was demonstrated in Hodgkin lymphoma, multiple myeloma (MM), B-cell chronic lymphocytic leukemia (Hubmann et al., 2002; Jundt et al., 2002, 2004; Nefedova et al., 2004) and weak but still detectable expression of Notch was observed in non-Hodgkin lymphoma cells (Jundt et al., 2002). However, in contrast to T-cell malignancies the role of Notch in B-cell tumors remains controversial. Several studies suggested that constitutively active Notch signaling leads to growth inhibition and apoptosis in malignant B-cells (Morimura et al., 2000; Nefedova et al., 2004; Romer et al., 2003; Zweidler-McKay et al., 2005). Zweidler-McKay demonstrated growth arrest and/or apoptosis as functional consequences of Notch activation in 13 cell lines representing multiple subclasses of B-cell neoplasia (murine and human preB-ALL, human Hodgkin, biphenotypic mixed-lineage leukemia and MM cells lines). This effect was observed by both expression of a constitutively active form of Notch (ICN) as well as ligand-induced activation of Notch signaling. Furthermore, all four Notch members were able to induce growth inhibition and apoptosis (Zweidler-McKay et al., 2005). In contrast, several studies have suggested that Notch signaling promotes proliferation in malignant B cells, such as a role of Notch-2 in CD23a-mediated proliferation in B-cell chronic lymphocytic leukemia (Hubmann et al., 2002). A proliferative effect of exposure to the Notch ligand Jagged-1 was demonstrated in several Hodgkin and MM cell lines (Jundt et al., 2002, 2004).
3.4. Notch signaling and microenvironment
The tumor microenvironment is recognized as an important factor supporting tumor development and progression. In contrast to T-ALL where Notch is activated primarily due to mutations, in B-cell malignancies Notch signaling might be activated by tumor cell–cell and tumor cell–microenvironment interactions. Several studies demonstrated the importance of Notch signaling in cross-talk between MM cells and their microenvironment. MM cells reside preferentially in the BM and surrounding stromal fibroblast-like cells, osteoblasts and osteoclasts (OCL), as well as soluble factors all affect MM growth and response to the treatment. BM stromal cells (fibroblasts) express both Notch ligands – Jagged and Delta – and are able to activate Notch signaling in MM cells in a co-culture system (Nefedova et al., 2004). Besides, Notch could be activated by interaction between MM cells themselves since they express both Notch ligands and the receptors. Activation of Notch signaling was suggested to be one of the mechanisms of de novo BM stroma-mediated resistance of MM cells to apoptosis induced by chemotherapeutic drugs (Nefedova et al., 2004). However, not only BM stroma induces Notch signaling in MM cells but also tumor cells could affect Notch signaling in the microenvironment. Hypomethylation of the Jagged-2 promoter in malignant primary plasma cells and MM cell lines resulted in overexpression of the Notch ligand Jagged-2 and in a co-culture assay Jagged-2 expressed by the MM cells, activated Notch signaling in BM stromal cells and thereby induced secretion of interleukin-6 (IL-6), vascular endothelial growth factor, and insulin-like growth factor-1 by the stromal cells (Houde et al., 2004). OPM-2 MM cells were able to activate Notch signaling in OCL in an in vitro co-culture system resulting in increased tartrate-resistant acid phosphatase-5 mRNA expression, an indicator of OCL activity (Schwarzer et al., 2008).
4. Notch signaling inhibition as therapy
Because Notch signaling is of critical importance in the pathogenesis of hematological as well as solid malignancies, inhibition of this pathway was proposed as an emerging strategy for cancer treatment. As described above, Notch activation includes two consecutive proteolytic cleavage steps followed by translocation of the active ICN to the nucleus, binding with CBF-1 transcriptional factor and co-activators of MAML family, and transcription of target genes. Any pharmacological intervention that interferes with these steps could result in inhibition of Notch signaling. So far, blocking the final intra-membranous cleavage of Notch mediated by the γ-secretase complex has been the most successful strategy for development of targeted therapeutics.
γ-Secretase is a large intramembranous aspartyl protease that processes a wide range of type I membrane proteins, including β-amyloid precursor protein (APP), Notch family members, as well as low density lipoprotein family members, cadherins, CD44, ErbB-4, CSF-1, and syndecans among 25 substrates identified so far (Lleó, 2008). The γ-secretase complex is composed of presenelin (PS)-1, PS-2, nicastrin, Aph-1, and Pen-2. Recent studies have indicated the critical role of PS1 for the activity of this complex (Lleó, 2008). Initially, interest to γ-secretase came from the fact that this complex is responsible for the cleavage of APP that generates amyloid-β peptide (Aβ), a primary component of the amyloid plaques. Abnormal accumulation of Aβ peptides and formation of amyloid plaques is considered to play a leading role in pathogenesis of Alzheimer’s disease (Selkoe and Kopan, 2003). Due to its role in Aβ peptides generation, γ-secretase complex became an attractive target for developing therapeutics for Alzheimer’s disease (reviewed in (Lleó, 2008). Most γ-secretase inhibitors (GSI) tested have also affected the cleavage of Notch receptors, thus providing a rationale for testing the anti-tumor activity of these compounds.
The first data revealing the anti-tumor effect of GSI were obtained in solid tumors. The tripeptide GSI-I (Z-Leu-Leu-Nle-CHO, Calbiochem) as well as LY-411,575 blocked Notch activation and resulted in apoptosis in primary and immortalized Kaposi’s sarcoma cells in vitro and tumor growth inhibition in xenograft mouse model in vivo (Curry et al., 2005). Inhibition of the Notch pathway with the dipeptide GSI DAPT (N-[N-(3,5-difluorophenacetyl-l-alanyl)]-S-phenylglycine t-butyl ester, GSI-IX, Calbiochem) in medulloblastoma cell lines and primary tumor cultures resulted in a significant reduction of viable cell numbers. In vivo, short-term treatment of mice carrying D283 medulloblastoma xenografts using DAPT resulted in markedly decreased tumor cell proliferation and increased apoptosis (Hallahan et al., 2004). However, a longer 4-week study of the activity of this GSI in vivo was inconclusive, because the drug was not effective in inhibiting the Notch pathway activity after 2 weeks as was evidenced by failure to suppress Hes-1 expression in the marrow. The ability of different GSI to inhibit tumor growth was demonstrated in vitro and in vivo for other solid tumors including lung, ovarian cancer, and melanoma (Konishi et al., 2007; Park et al., 2006; Qin et al., 2004).
4.1. T-ALL
The high occurrence of Notch-activating mutations in TALL suggested targeting of this pathway as a promising therapeutic strategy in T-ALL. The first evidence of the effect of GSI was provided by the observation that five human TALL cell lines lacking t(7;9) underwent G0/G1 cell cycle arrest after 4–8 days of treatment with GSI Compound E (Weng et al., 2004) (Table 1). Several other studies confirmed this initial observation and described induction of cell cycle arrest followed by apoptosis after prolonged (4 days for CUTLL1 cell line and 7 days or more for other cell lines) treatment of “sensitive” Notch-1 mutation-positive T-ALL cell lines with GSI (De Keersmaecker et al., 2008; Kogoshi et al., 2007; O’Neil et al., 2006; Palomero et al., 2006a). In addition, GSI (Compound E (GSI-XXI); Calbiochem) was able to sensitize T-ALL cells to the effect of dexamethasone and increase the anti-proliferative effect of imatinib in T-ALL cells with ABL1 fusions (De Keersmaecker et al., 2008). Recently MRK-003, a new cyclic sulfamide GSI was developed by Merck (Lewis et al., 2007). Treatment with MRK-003 of three T-ALL cell lines (DND-41, HPB-ALL, and TALL-1) resulted in prolonged G0/G1 cell cycle arrest followed by induction of apoptosis (Lewis et al., 2007).
Table 1.
Gamma-secretase inhibitors used for studies in hematological malignancies.
| Compound | Type of cancer cells tested | Type of studies | Biological effect reported | Reference |
|---|---|---|---|---|
| GSI-I (Z-LLNle-CHO, Calbiochem) | Burkitt lymphoma and diffuse large | In vitro | Growth suppression | Kogoshi et al. (2007) |
| B-cell lymphoma cell lines | Tohda et al. (2006) | |||
| AML cell lines | In vitro | Growth suppression | Kogoshi et al. (2007) | |
| T-ALL cell lines | In vitro | Growth suppression | Kogoshi et al. (2007) | |
| GSI-IX (DAPT, Calbiochem) | Burkitt lymphoma and diffuse large | In vitro | Growth suppression | Kogoshi et al. (2007) |
| B-cell lymphoma cell lines | Tohda et al. (2006) | |||
| AML cell lines | In vitro | Growth suppression | Kogoshi et al. (2007) | |
| T-ALL cell lines | In vitro | Growth suppression | Kogoshi et al. (2007) | |
| Mouse T-ALL cell lines | In vitro | G0/G1 cell cycle arrest and apoptosis | O’Neil et al. (2006) | |
| GSI-X (L-685,458 Calbiochem) | T-ALL cell line ALL-SIL (express ABL1 fusion protein) |
In vitro | Antagonized the inhibitory effect of imatinib on ALL-SIL cell proliferation |
De Keersmaecker et al. (2008) |
| GSI-XII (Z-IL-CHO, Calbiochem) | Burkitt lymphoma and diffuse large | In vitro | Growth suppression and apoptosis | Kogoshi et al. (2007) |
| B-cell lymphoma cell lines | Tohda et al. (2006) | |||
| MM cell lines and primary cells | In vitro | Apoptosis; increased cytotoxicity of doxorubicin | Nefedova et al. (2008) | |
| In vivo | Inhibition of RPMI-8226 and H929 tumor growth; increased anti-MM effect of doxorubicin and melphalan |
|||
| AML cell lines | In vitro | Induction of apoptosis | Kogoshi et al. (2007) | |
| T-ALL cell lines | In vitro | Induction of apoptosis | Kogoshi et al. (2007) | |
| GSI-XXI (Compound E, | T-ALL cell lines | In vitro | Cell cycle arrest in 5 out of 30 cell lines tested | Weng et al. (2004) |
| Calbiochem) | T-ALL cell line CUTLL1 | In vitro | G0/G1 cell cycle arrest and apoptosis | Palomero et al. (2006a,b) |
| T-ALL cell lines | In vitro | Reversible G0/G1 cell cycle arrest followed by apoptosis; Stimulation of T-ALL cell lines differentiation from CD4−/CD8+− to CD4+/CD8+ (MOLT-4) or changing expression of CD3; Antagonized the inhibitory effect of imatinib on ALL-SIL cell proliferation; Sensitized cells to dexamethasone |
De Keersmaecker et al. (2008) | |
| DAPT | MM cell lines, Hodgkin lymphoma cell line KM-H2 |
In vitro | Decrease in proliferation | Jundt et al. (2004) |
| GSI-15 (Acros Organics) | MM OPM-2 cell line | In vitro | Decrease in proliferation and induction of apoptosis; Inhibition of OCL activity |
Schwarzer et al. (2008) |
| MRK-003 (Merck) | T-ALL cell lines | In vitro | G0/G1 cell cycle arrest followed by apoptosis in 5 out of 20 cell lines tested |
O’Neil et al. (2007) |
| In vitro | G0/G1 cell cycle arrest followed by apoptosis in 3 cell lines tested |
Lewis et al. (2007) | ||
| MK-0752 (Merck) | T-ALL | Clinical trial | Results are not published yet | Deangelo et al. (2006) (abstract) |
GSI, γ-secretase inhibitor; T-ALL, T-cell lymphoblastic leukaemia; AML, acute myeloid leukaemia; MM, multiple myeloma; OCL, octeoclasts.
It was anticipated that GSI therapy would be successful especially for those tumors driven by aberrant Notch signaling. However, the overall efficacy of GSI treatment was poor since only a small number of human T-ALL cell lines responded. The GSI Compound E had effect in only 5 out of 30 human T-ALL cell lines and 5 out of 20 T-ALL cell lines were sensitive to MRK-003 (O’Neil et al., 2007; Weng et al., 2004). To investigate the molecular basis of resistance to GSI, a subset of seven T-ALL cell lines expressing high level of ICN (Notch active domain) and lacking truncating mutations in the PEST C-terminal domain was analyzed. All these cell lines as well as 7 out of 81 (8.6%) primary T-ALL samples carried either a mutation or homozygous deletion of the gene FBW7, a ubiquitin ligase that plays a critical role in ICN degradation. FBW7 interacts with its substrates via WD40 propeller domain after the substrate became phosphorylated. All identified FBW7 gene mutations were localized within critical WD40 arginine residues thus preventing its binding to ICN (O’Neil et al., 2007). Using oligonucleotide microarrays in search for the differentially expressed genes associated with T-ALL sensitivity or resistance to GSI, Palomero et al. demonstrated that the loss of tumor suppressor gene PTEN due to mutations was associated with resistance to pharmacological inhibition of Notch (Palomero et al., 2007). PTEN controls multiple pathways regulating cellular growth and survival and one of its functions is inhibition of PI3K/Akt signaling. GSI-sensitive cells lines expressed normal PTEN protein, while GSI-resistant cells showed either absent or decreased PTEN protein due to mutations that encode premature stop codons. In addition to T-ALL cell lines, loss of PTEN protein was observed in 6 out of 35 (17%) T-ALL primary samples as determined by immunohistochemistry and mutationsof the PTEN gene were detected in 9 out of 111 (8%) T-ALL cases. The expression of PTEN was negatively regulated by Notch. Mutational loss of PTEN resulted in consequent hyperactivation of PI3K/Akt and resistance of T-ALL cells to GSI (Palomero et al., 2007).
A clinical trial of MK-0752, a compound similar to MRK-003, for treatment of T-ALL was initiated (Deangelo et al., 2006). The study is ongoing and the results are not published yet.
4.2. B-cell malignancies
Several studies evaluated the effect of GSI on B-cell lymphoma and MM tumor growth. Using three different GSI compounds (GSI-I, IX, and XII; Calbiochem) it was reported that in vitro growth of three Burkitt lymphoma cell lines Ramos, Daudi, and Raji, as well as the diffuse large B-cell lymphoma cell line MD901 was significantly suppressed due to induction of apoptosis (Kogoshi et al., 2007). Similarly, the growth of the newly established B-cell lymphoma TMD8 cell line was suppressed after treatment with either GSI-I, -IX, or -XII (Tohda et al., 2006).
The anti-myeloma effect of the GSI DAPT was investigated by Jundt et al. (2004). According to this study the Notch ligand Jagged-1 stimulated proliferation of MM cell lines RPMI-8226, H929, OPM-2, LP-1 and Hodgkin cell line KM-H2 and treatment with DAPT resulted in a 40–75% decrease in proliferation rates of these cell lines.
The tumor BM microenvironment has been shown to play a critical role not only in controlling MM cell growth and survival, but also in the sensitivity to therapeutic agents (Meads et al., 2008). BM fibroblast-like stromal cells express Notch ligands and therefore can activate Notch signaling in MM cells and following Notch activation, MM cell became resistant to the chemotherapy-induced apoptosis (Nefedova et al., 2004). In MM cell lines or primary MM cells cultured with or without BM stromal cells, GSI-XII (Calbiochem) induced significant apoptosis due to upregulation of the pro-apoptotic protein Noxa. Addition of GSI increased the MM cytotoxicity of doxorubicin and overcame BM-mediated drug resistance in vitro. In in vivo mouse MM models GSI significantly increased the anti-tumor effect of doxorubicin or melphalan (Nefedova et al., 2008). Thus, addition of GSI potentiates the effect of conventional chemotherapeutics and would allow for the decrease of the effective dose of drugs which in turn would reduce the toxicity of the treatment.
Other important elements of the BM stromal microenvironment are osteoblasts and OCL. The altered balance between the activities of these two is the cause of bone disease, a typical feature of MM. Recently, the effect of GSI on MM-OCL cell interaction was evaluated. GSI-15 (RH02015SC, Maybridge, Acros Organics, Belgium) used in this study reduced the proliferation and induced apoptosis in OPM-2 MM cells cultured alone or co-cultured with OCL. Furthermore, the Notch inhibitor completely abolished increased OCL activity (Schwarzer et al., 2008).
5. Conclusions
There is now clear evidence pointing out a role of Notch in the pathogenesis of hematological malignancies, making Notch signaling an attractive target for the development of novel therapeutics. Several key steps in the activation of this pathway could potentially be pharmacologically targeted. A number of agents including ADAM inhibitors, Notch antisense, anti-Notch monoclonal antibodies and RNA interference have been suggested for targeting Notch; however, they did not reach preclinical studies so far. Relative specificity and high anti-Notch activity make GSIs that inhibit the final cleavage step of the Notch receptor, the only agents that were used in preclinical studies (summarized in Table 1). The clinical use of GSIs, however, might still be limited due to their side-effects. Thus, gastrointestinal toxicity was reported in a mouse model (van Es et al., 2005; Wong et al., 2004). This effect could be related to the blockade of the physiological effect of Notch signaling in normal tissues. In addition, it could be due to non-selectivity of GSIs since the γ-secretase complex can process more than 20 substrates in addition to Notch. The toxicity of compounds could be minimized by lowering the dose of GSIs when used in combination with other chemotherapeutic agents. This approach allowed for reducing the dose of chemotherapy and was demonstrated to be effective in preclinical studies (De Keersmaecker et al., 2008; Nefedova et al., 2008). More studies of the combined effect of GSIs with conventional and new chemotherapeutic drugs will be necessary to determine the clinical potential of such an approach.
Recently, a series of hydrocarbon stapled alpha-helical peptides targeting Notch (SAHNs) complex were synthesized (Moellering et al., 2007). SAHN1 was able to bind nuclear ICN and CSL and inhibit downstream targets of Notch signaling. Furthermore, inhibition of Notch signaling with SAHN1 conferred cytotoxicity in a panel of T-ALL cell lines including those resistant to GSIs (Moellering et al., 2007). Further development of this approach and testing it in the context of various cells might be beneficial for treatment of hematological malignancies.
References
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Aster J, Pear W, Blacklow S. Notch signaling in leukemia. Annu. Rev. Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaumueller C, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- Breit S, Stanulla M, Flohr T, Schrappe M, Ludwig W, Tolle G, Happich M, Muckenthaler M, Kulozik A. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood. 2006;108:1151–1157. doi: 10.1182/blood-2005-12-4956. [DOI] [PubMed] [Google Scholar]
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens J, Cumano A, Roux P, Black R, Israël A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- Brown M, Ye J, Rawson R, Goldstein J. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Chen P, Yen C, Wang W, Lin Y, Chu C, Chiou T, Liu J, Yang M. Down-regulation of Notch-1 expression decreases PU.1-mediated myeloid differentiation signaling in acute myeloid leukaemia. Int. J. Oncol. 2008;32:1335–1341. doi: 10.3892/ijo_32_6_1335. [DOI] [PubMed] [Google Scholar]
- Cheng P, Zlobin A, Volgina V, Gottipati S, Osborne B, Miele L, Gabrilovich DI. Notch-1 regulates NF-kappa B activity in hematopoietic progenitor cells. J. Immunol. 2001;167:4458–4467. doi: 10.4049/jimmunol.167.8.4458. [DOI] [PubMed] [Google Scholar]
- Chiaramonte R, Basile A, Tassi E, Calzavara E, Cecchinato V, Rossi V, Biondi A, Comi P. A wide role for NOTCH1 signaling in acute leukemia. Cancer Lett. 2005;219:113–120. doi: 10.1016/j.canlet.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Curry C, Reed L, Golde T, Miele L, Nickoloff B, Foreman K. Gamma secretase inhibitor blocks Notch activation and induces apoptosis in Kaposi’s sarcoma tumor cells. Oncogene. 2005;24:6333–6344. doi: 10.1038/sj.onc.1208783. [DOI] [PubMed] [Google Scholar]
- De Keersmaecker K, Lahortiga I, Mentens N, Folens C, Van Neste L, Bekaert S, Vandenberghe P, Odero M, Marynen P, Cools J. In vitro validation of gamma-secretase inhibitors alone or in combination with other anti-cancer drugs for the treatment of T-cell acute lymphoblastic leukemia. Haematologica. 2008;93:533–542. doi: 10.3324/haematol.11894. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Annaert W, Cupers P, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- Deangelo D, Stone R, Silverman L, Stock W, Arttar E, Fearen I, et al. A phase I clinical trial of the Notch inhibitor MK-0752 in patients with T-cell acute lymphoblastic leukemia/lymphoma (T-ALL) and other leukemias. J. Clin. Oncol. 2006;24:6585. (Abstract) [Google Scholar]
- Deftos M, Huang E, Ojala E, Forbush K, Bevan M. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 2000;13:73–84. doi: 10.1016/s1074-7613(00)00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellisen L, Bird J, West D, Soreng A, Reynolds T, Smith S, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- Fu L, Kogoshi H, Nara N, Tohda S. NOTCH1 mutations are rare in acute myeloid leukemia. Leuk. Lymphoma. 2006;47:2400–2403. doi: 10.1080/10428190600773339. [DOI] [PubMed] [Google Scholar]
- Hallahan A, Pritchard J, Hansen S, Benson M, Stoeck J, Hatton B, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- Houde C, Li Y, Song L, Barton K, Zhang Q, Godwin J, et al. Overexpression of the NOTCH ligand JAG2 in malignant plasma cells from multiple myeloma patients and cell lines. Blood. 2004;104:3697–3704. doi: 10.1182/blood-2003-12-4114. [DOI] [PubMed] [Google Scholar]
- Hubmann R, Schwarzmeier J, Shehata M, Hilgarth M, Duechler M, Dettke M, Berger R. Notch2 is involved in the overexpression of CD23 in B-cell chronic lymphocytic leukemia. Blood. 2002;99:3742–3747. doi: 10.1182/blood.v99.10.3742. [DOI] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- Jundt F, Anagnostopoulos I, Forster R, Mathas S, Stein H, Dorken B. Activated Notch1 signaling promotes tumor cell proliferation and survival in Hodgkin and anaplastic large cell lymphoma. Blood. 2002;99:3398–3403. doi: 10.1182/blood.v99.9.3398. [DOI] [PubMed] [Google Scholar]
- Jundt F, Probsting KS, Anagnostopoulos I, Muehlinghaus G, Chatterjee M, Mathas S, Bargou RC, Manz R, Stein H, Dorken B. Jagged1-induced Notch signaling drives proliferation of multiple myeloma cells. Blood. 2004;103:3511–3515. doi: 10.1182/blood-2003-07-2254. [DOI] [PubMed] [Google Scholar]
- Kogoshi H, Sato T, Koyama T, Nara N, Tohda S. Gamma-secretase inhibitors suppress the growth of leukemia and lymphoma cells. Oncol. Rep. 2007;18:77–80. [PubMed] [Google Scholar]
- Konishi J, Kawaguchi K, Vo H, Haruki N, Gonzalez A, Carbone D, Dang T. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 2007;67:8051–8057. doi: 10.1158/0008-5472.CAN-07-1022. [DOI] [PubMed] [Google Scholar]
- Lewis H, Leveridge M, Strack P, Haldon C, O’neil J, Kim H, et al. Apoptosis in T cell acute lymphoblastic leukemia cells after cell cycle arrest induced by pharmacological inhibition of notch signaling. Chem. Biol. 2007;14:209–219. doi: 10.1016/j.chembiol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Lleó A. Activity of gamma-secretase on substrates other than APP. Curr. Top. Med. Chem. 2008;8:9–16. doi: 10.2174/156802608783334060. [DOI] [PubMed] [Google Scholar]
- Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah N, Israel A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki M, Sanchez-Irizarry C, Mitchell J, Histen G, Xu M, Aster J, Blacklow S. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol. Cell. Biol. 2006;26:4642–4651. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Arias A, Zecchini V, Brennan K. CSL-independent Notch signalling: a checkpoint in cell fate decisions during development? Curr. Opin. Genet. Dev. 2002;12:524–533. doi: 10.1016/s0959-437x(02)00336-2. [DOI] [PubMed] [Google Scholar]
- Meads M, Hazlehurst L, Dalton W. The bone marrow microenvironment as a tumor sanctuary and contributor to drug resistance. Clin. Cancer Res. 2008;14:2519–2526. doi: 10.1158/1078-0432.CCR-07-2223. [DOI] [PubMed] [Google Scholar]
- Moellering R, Cornejo M, Rocknik J, Hancock M, DelBianco C, Blacklow S, et al. Direct inhibition of the Notch transactivation complex with synthetic constrained peptides in T-cell acute lymphoblastic leukemia. Blood. 2007;110:2819. (Abstract) [Google Scholar]
- Morimura T, Goitsuka R, Zhang Y, Saito I, Reth M, Kitamura D. Cell cycle arrest and apoptosis induced by Notch1 in B cells. J. Biol. Chem. 2000;275:36523–36531. doi: 10.1074/jbc.M006415200. [DOI] [PubMed] [Google Scholar]
- Mumm J, Kopan R. Notch signaling: from the outside in. Dev. Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- Mumm J, Schroeter E, Saxena M, Griesemer A, Tian X, Pan D, Ray W, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- Nefedova Y, Cheng P, Alsina M, Dalton W, Gabrilovich D. Involvement of Notch-1 signaling in bone marrow stroma-mediated de novo drug resistance of myeloma and other malignant lymphoid cell lines. Blood. 2004;103:3503–3510. doi: 10.1182/blood-2003-07-2340. [DOI] [PubMed] [Google Scholar]
- Nefedova Y, Sullivan D, Bolick S, Dalton W, Gabrilovich D. Inhibition of Notch signaling induces apoptosis of myeloma cells and enhances sensitivity to chemotherapy. Blood. 2008;111:2220–2229. doi: 10.1182/blood-2007-07-102632. [DOI] [PubMed] [Google Scholar]
- O’Neil J, Calvo J, McKenna K, Krishnamoorthy V, Aster J, Bassing C, Alt F, Kelliher M, Look A. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2006;107:781–785. doi: 10.1182/blood-2005-06-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J. Exp. Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T, Barnes K, Real P, Bender J, Sulis M, Murty V, Colovai A, Balbin M, Ferrando A. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006a;20:1279–1287. doi: 10.1038/sj.leu.2404258. [DOI] [PubMed] [Google Scholar]
- Palomero T, McKenna K, O-Neil J, Galinsky I, Stone R, Suzukawa K, et al. Activating mutations in NOTCH1 in acute myeloid leukemia and lineage switch leukemias. Leukemia. 2006b;20:1963–1966. doi: 10.1038/sj.leu.2404409. [DOI] [PubMed] [Google Scholar]
- Palomero T, Sulis M, Cortina M, Real P, Barnes K, Ciofani M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat. Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Li M, Nakayama K, Mao T, Davidson B, Zhang Z, Kurman R, Eberhart C, Shih I, Wang T. Notch3 gene amplification in ovarian cancer. Cancer Res. 2006;66:6312–6318. doi: 10.1158/0008-5472.CAN-05-3610. [DOI] [PubMed] [Google Scholar]
- Qin JZ, Stennett L, Bacon P, Bodner B, Hendrix MJ, Seftor RE, et al. p53-independent NOXA induction overcomes apoptotic resistance of malignant melanomas. Mol. Cancer Ther. 2004;3:895–902. [PubMed] [Google Scholar]
- Rand M, Grimm L, Artavanis-Tsakonas S, Patriub V, Blacklow S, Sklar J, Aster J. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol. Cell. Biol. 2000;20:1825–1835. doi: 10.1128/mcb.20.5.1825-1835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20:3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer S, Saunders U, Jäck H, Jehn B. Notch1 enhances B-cell receptor-induced apoptosis in mature activated B cells without affecting cell cycle progression and surface IgM expression. Cell Death Differ. 2003;10:833–844. doi: 10.1038/sj.cdd.4401253. [DOI] [PubMed] [Google Scholar]
- Ronchini C, Capobianco AJ. Induction of cyclin d1 transcription and cdk2 activity by Notch (IC): implication for cell cycle distribution in transformation by Notch (IC) Mol. Cell. Biol. 2001;21:5925–5934. doi: 10.1128/MCB.21.17.5925-5934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter E, Kisslinger J, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- Schwarzer R, Kaiser M, Acikgoez O, Heider U, Mathas S, Preissner R, Sezer O, Doerken B, Jundt F. Notch inhibition blocks multiple myeloma cell-induced osteoclast activation. Leukemia. 2008 doi: 10.1038/leu.2008.138. [DOI] [PubMed] [Google Scholar]
- Selkoe D, Kopan R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu. Rev. Neurosci. 2003;26:565–597. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- Staal F, Langerak A. Signaling pathways involved in the development of T-cell acute lymphoblastic leukemia. Haematologica. 2008;93:493–497. doi: 10.3324/haematol.12917. [DOI] [PubMed] [Google Scholar]
- Sulis M, Williams O, Palomero T, Tosello V, Pallikuppam S, Real P, Barnes K, Zuurbier L, Meijerink J, Ferrando A. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood. 2008;112:733–740. doi: 10.1182/blood-2007-12-130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohda S, Nara N. Expression of Notch1 and Jagged1 proteins in acute myeloid leukemia cells. Leuk. Lymphoma. 2001;42:467–472. doi: 10.3109/10428190109064603. [DOI] [PubMed] [Google Scholar]
- Tohda S, Kogoshi H, Murakami N, Sakano S, Nara N. Diverse effects of the Notch ligands Jagged1 and Delta1 on the growth and differentiation of primary acute myeloblastic leukemia cells. Exp. Hematol. 2005;33:558–563. doi: 10.1016/j.exphem.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Tohda S, Sato T, Kogoshi H, Fu L, Sakano S, Nara N. Establishment of a novel B-cell lymphoma cell line with suppressed growth by gamma-secretase inhibitors. Leuk. Res. 2006;30:1385–1390. doi: 10.1016/j.leukres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- van Es J, van Gijn M, Riccio O, van den Born M, Vooijs M, Begthel H, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- Vardar D, North C, Sanchez-Irizarry C, Aster J, Blacklow S. Nuclear magnetic resonance structure of a prototype Lin12-Notch repeat module from human Notch1. Biochemistry. 2003;42:7061–7067. doi: 10.1021/bi034156y. [DOI] [PubMed] [Google Scholar]
- Weng A, Ferrando A, Lee W, Morris J, Silverman L, Sanchez-Irizarry C, Blacklow S, Look A, Aster J. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Wharton K, Johansen K, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- Wong G, Manfra D, Poulet F, Zhang Q, Josien H, Bara T, et al. Chronic treatment with the gamma-secretase inhibitor LY-411, 575 inhibits beta-amyloid peptide production and alters lymphopoiesis and intestinal cell differentiation. J. Biol. Chem. 2004;279:12876–12882. doi: 10.1074/jbc.M311652200. [DOI] [PubMed] [Google Scholar]
- Wu L, Griffin J. Modulation of Notch signaling by mastermind-like (MAML) transcriptional co-activators and their involvement in tumori-genesis. Semin. Cancer Biol. 2004;14:348–356. doi: 10.1016/j.semcancer.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Zweidler-McKay P, He Y, Xu L, Rodriguez C, Karnell F, Carpenter A, Aster J, Allman D, Pear W. Notch signaling is a potent inducer of growth arrest and apoptosis in a wide range of B-cell malignancies. Blood. 2005;106:3898–3906. doi: 10.1182/blood-2005-01-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]