Abstract
Lung cancer is the leading cause of cancer death in the US and the world. The high mortality rate results, in part, from the lack of effective tools for early detection and the inability to identify subsets of patients who would benefit from adjuvant chemotherapy or targeted therapies. The development of high-throughput genome-wide technologies for measuring gene expression, such as microarrays, have the potential to impact the mortality rate of lung cancer patients by improving diagnosis, prognosis, and treatment. This review will highlight recent studies using high-throughput gene expression technologies that have led to clinically relevant insights into lung cancer. The hope is that diagnostic and prognostic biomarkers that have been developed as part of this work will soon be ready for wide-spread clinical application and will have a dramatic impact on the evaluation of patients with suspect lung cancer, leading to effective personalized treatment regimens.
Introduction
Lung cancer is the leading cause of cancer death in the US and the world. The high mortality rate (80–85% within 5 years) results from the lack of effective screening tools and tools for early-stage diagnosis (1–3), the inability to identify subsets of patients who would benefit from adjuvant chemotherapy or adjuvant targeted therapies, and the slow development new drug therapies. The development of high-throughput genome-wide technologies for measuring gene expression, such as microarrays, have the potential to impact the mortality rate of lung cancer patients by improving diagnosis, prognosis, and treatment.
The use of high-throughput technologies in breast cancer illustrates the potential impact that similar approaches may have in thoracic oncology. DNA microarrays have been used to identify gene expression signatures comprised of multiple genes that indicate which estrogen receptor-positive and auxiliary node-negative patients may benefit from additional chemotherapy. The are currently three commercially available gene expression based prognostic tests for breast cancer – Oncotype DX, a 21-gene assay (4) (Genomic Health, Redwood City, CA), MammaPrint, a 70-gene assay (5) (Agendia BV, Amsterdam, the Netherlands), and H/I, a 2-gene ratio assay (6) (AvariaDx, Carlsbad, California). Currently, there are two ongoing prospective randomized trials, Trial Assigning Individual Options for Treatment (TAILORx) to evaluate OncotypeDx, and Microarray in Node-negative Disease may Avoid ChemoTherapy (MINDACT) to evaluate MammaPrint versus a prognostic clinical algorithm (7). If these tests prove efficacious, they will be the first of many prognostic and diagnostic tests based on high-throughput gene expression measurements. The advantage of multi-gene biomarkers is that they are able to achieve higher accuracy than would be possible from a single gene measure (Figure 1).
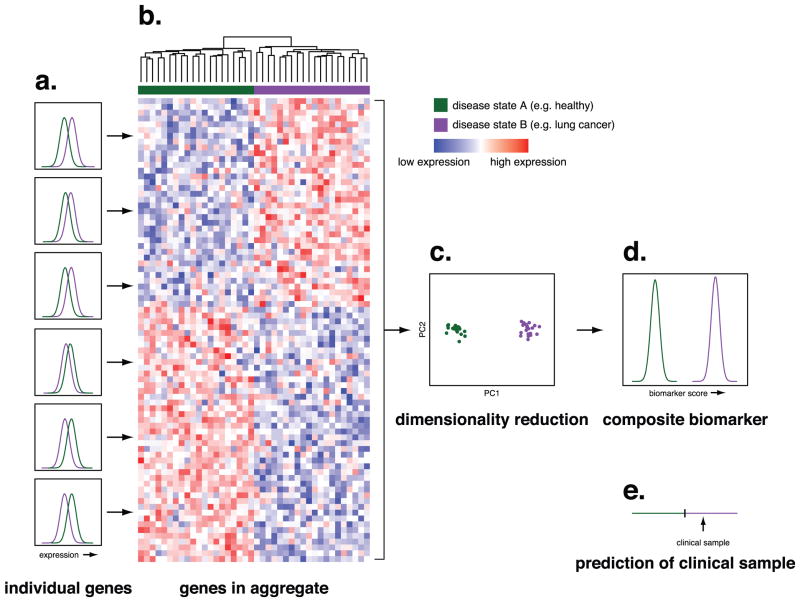
Figure 1.
Overview of multi-gene biomarkers. A. While individual genes may show significantly different expression levels between patients in two disease states (e.g. healthy patients and patients with lung cancer), the distribution of expression levels for any single gene may overlap sufficiently that no single gene can function as an accurate marker of disease state. The histograms plot the number of individuals (y-axis) with a given expression level of each gene (x-axis) as a function of disease state. For simplicity, this example uses two disease states, but the principle is the same for multiple disease states or a continuous outcome such as survival. B. Hierarchical clustering of gene expression levels across genes that vary between disease states shows that in aggregate these genes are able to distinguish between the two disease states. The dendrogram at the top of the panel shows the degree of relatedness between samples. C. A critical step in creating a biomarker from the expression levels of multiple genes is combining the expression levels into a single metric that associates with disease status. In this example, we show how following Principal Component Analysis, the values of the first component (x-axis) distinguish between the two disease states. While here we illustrate the use of Principal Component Analysis, there are many methods for performing this sort of dimensionality reduction. D. The distribution of multi-gene biomarker scores as a function of disease state shows that compared with the distribution of expression levels in individual genes, the multi-gene biomarker can distinguish between the two disease states with a high degree of accuracy. E. When the expression levels of the genes in the biomarker are measured in a clinical sample, a prediction can be made as to the disease state of the patient from which the sample was obtained based on the biomarker score.
This review will highlight recent studies using high-throughput gene expression technologies that have led to clinically relevant insights into lung cancer. The studies present molecular markers for the diagnosis of lung cancer, the prognosis of early-stage lung cancer, and sensitivity and response to chemotherapeutic agents (Table 1). Due to the clinical focus of this review, mechanistic insights into lung cancer biology and pathogenesis using high-throughput gene expression technologies (examples include (8–10)) as well as the technical, computational and analytic challenges inherent in processing and analyzing high-throughput data will not be discussed. Also, the application of the high-throughput technologies to study SNPs, DNA methylation, alternative splicing, and protein expression in lung cancer is discussed elsewhere (11–14).
Table 1.
Summary of the high-throughput gene expression lung cancer studies highlighted in this review
| Clinical Application | Publication | Publication Date | Tissue | Technology | Samples | Results |
|---|---|---|---|---|---|---|
| Diagnosis | Spira et al.(34) | 2007 | Large Airway Epithelium | Affymetrix Hu133A | 87 Smokers with lung cancer, 77 Smokers without lung cancer | Gene expression diagnostic for the evaluation of smokers with suspect lung cancer |
| Beane et al.(35) | 2008 | Large Airway Epithelium | Affymetrix Hu133A | Spira et al.(34) samples | A clinicogenomic model for | |
| Molecular Classification and Characterization | Garber et al.(37) | 2001 | Lung Tissue | 24,000-element cDNA microarray | 41 AD, 16 SCC, 5 LCC, 5 SCLC, 5 normal lung, 1 fetal lung, replicates | Discovery of molecular subtypes of lung tumors |
| Bhattacharjee et al.(38) | 2001 | Lung Tissue | Affymetrix Hu95A | 127 AD, 21 SCC, 20 Carcinoids, 6 SCLC, 12 AD of non-lung origin, 17 normal lung, replicates | Discovery of molecular subtypes of lung tumors | |
| Ramaswamy et al.(49) | 2002 | Tumor Tissue | Affymetrix Hu6800, Hu35KsubA, Hu95A, Rosetta oligonucleotide microarray | Lung, prostate, breast, CNS, and blood cancers | 17-gene signature associated with metastasis | |
| Hoang et al.(52) | 2004 | Lung Tissue | 2,400-element cDNA microarray | 12 AD, 3 SCC | Gene signature for metastasis, micrometastasis, and over metastasis | |
| Xi et al.(53) | 2005 | Lung Tissue | Affymetrix Hu6800, Hu95A | Beer et al.(54) and Bhattacharjee et al.(38) samples | Lymph node positive gene signature | |
| Yanaihara et al.(55) | 2006 | Lung Tissue | 352 miRNA microarray | 104 pairs of primary lung cancer and adjacent non-cancerous tissue (65 AD, 39 SCC), 32 additional AD for validation | miRNAs associated with subtypes of lung tumors | |
| Prognosis | Beer et al.(54) | 2002 | Lung Tissue | Affymetrix Hu6800 | 67 Stage I AD, 19 Stage III AD, 10 non-neoplastic lung | 50-gene lung cancer prognostic signature |
| Potti et al.(66) | 2006 | Lung Tissue | Affymetrix Hu133Aplus2 | 140 AD, 58 SCC, various stages | Prognostic lung metagene model | |
| Lu et al.(68) | 2006 | Lung Tissue | Affymetrix Hu95Av2, Hu6800, Hu133A Hu133AB, Hu133plus2 | Meta-analysis of 7 different datasets, 324 samples | 64-gene lung cancer prognostic signature | |
| Chen et al.(70) | 2007 | Lung Tissue | 9,600-element cDNA microarray | 75 AD, 70 SCC, 16 Other | 5-gene lung cancer prognostic signature | |
| Shedden et al.(75) | 2008 | Lung Tissue | Affymetrix Hu133A | 442 Stage I, II, and III AD | A multi-site, blinded study to test several lung cancer prognostic gene signatures | |
| Yanaihara et al.(55) | 2006 | Lung Tissue | 352 miRNA microarray | 104 pairs of primary lung cancer and adjacent non- cancerous tissue (65 AD, 39 SCC), 32 additional AD for validation | Identification of lung cancer prognostic miRNAs | |
| Pharmacogenomics | Staunton et al.(82) | 2001 | Lung Cancer Cell Lines | Affymetrix Hu6800 | NCI-60 (60 Lung cancer cell lines) | Gene expression prediction of chemosensitivity |
| Potti et al.(85) | 2006 | Lung Cancer Cell Lines, Lung Tissue | Affymetrix Hu95Av2, Affymetrix Hu133A | NCI-60, breast, lung, and ovarian cancer samples | Gene expression prediction of chemosensitivity to guide use of chemotherapeutics | |
| Hsu et al.(86) | 2007 | Lung Cancer Cell Lines, Lung Tissue | Affymetrix Hu95Av2, Affymetrix Hu133A | Potti et al.(85) samples | Gene expression prediction of response to cisplatin and pemetrexed | |
| Gemma et al.(88) | 2006 | Lung Cancer Cell Lines | 1300-element cDNA microarray, Affymetrix Hu133A | 29 Lung cancer cell lines | Gene expression signatures of sensitivity to several anti-cancer agents |
High-throughput Technologies
The success of the Human Genome Project coupled with a variety of technological advances such as rapid oligonucleotide synthesis and microarray chip fabrication has enabled the development of high-throughput gene expression technologies. Depending on the experimental design, RNA samples are obtained from cell cultures or surgical tissues. Prior to RNA isolation and processing, techniques such as laser capture microdissection (15) can be used to obtain a homogeneous population of cells from tissue specimens.
Microarrays are currently among the most commonly used technology for quantitatively measuring the expression of genes or miRNAs in a high throughput manner. Microarrays are orderly arrays of spots composed of oligonucleotides complementary to genes/miRNAs that are immobilized onto a solid support such as a glass slide (16;17). Microarrays take advantage of Watson-Crick base pairing, and therefore, only complementary nucleic acids will hybridize and produce a signal that can be used as a measure of expression. The production and use of microarrays requires several steps including the synthesis of probes, array fabrication, target hybridization, fluorescence scanning, and image processing to produce a numerical readout of expression. Complementary DNA (cDNA) microarrays, developed at Stanford University, use DNA clones (selected from sequence databases) between 500 and 5000 base pairs in length as probes. Oligonucleotide microarrays, as their name implies, use as probes short oligonucleotides that have been derived from gene or miRNA sequences. In addition to microarrays, other high-throughput sequence-based technologies to measure gene expression such as serial analysis of gene expression (SAGE) (18) have been used in the past, and technological advances in sequencing are leading to new massively parallel sequencing technologies (19–21) that will likely be used extensively in future research.
Lung Cancer Diagnosis
The risk for developing lung cancer increases with cumulative exposure to cigarette smoke. The incidence of lung cancer, however, even in a high-risk population of smokers is only ~15% over a lifetime (22). Currently, there are no effective diagnostic biomarkers to identify which current and former smokers are at the greatest risk for developing lung cancer. As a result of this failure to detect high-risk smokers and the low frequency of early stage detection the five year survival rates for lung cancer (~15%) have not changed appreciably over the past 4–5 decades. Previous screening trials with frequent chest x-rays and sputum cytology have not demonstrated an effect on lung cancer mortality (reviewed by Jett and Midthun et al. (23)). Spiral computerized tomography (CT) scan screening can detect lung tumors at an earlier stage than routine chest x-rays. However, while spiral CT can be highly sensitive it is also non-specific and many newly detected small lesions have proven on resection to be non-malignant scar tissue or old granulomas rather than early lung cancers (2). While final results from large-scale randomized trials using CT scans are still pending, recent work has suggested that this approach does not improve lung cancer mortality (24).
Developing biomarkers that are highly sensitive, specific, and identify smokers at high risk for developing lung cancer or individuals with early stage cancer represents a key approach to improving lung cancer mortality. In order to explore the mechanisms by which individuals respond to the carcinogenic effects of smoking, several groups have used DNA microarrays and SAGE to define the genome-wide impact of smoking and smoking cessation on cytologically normal bronchial airway epithelial cells (25–31) or peripheral blood lymphocytes (32;33) of never, former, and current smokers.
The results of the above studies suggest that it might be possible to detect which smokers the carcinogenic effects of cigarette smoke have resulted in lung cancer. A recent study by Spira et al. used DNA micoarrays to profile the gene expression patterns of cytologically normal large airway epithelial cells in current and former smokers undergoing bronchoscopy for the clinical suspicion of lung cancer (34). An 80-probeset lung cancer-specific biomarker was developed based on a training set of samples (n=77) that could distinguish between smokers with and without lung cancer. The biomarker was both sensitive and specific when tested on an independent test set (n=52) and on an additional prospectively collected set of samples (n=35). This biomarker was also shown to provide information about the likelihood of lung cancer that is independent of clinical risk factors for lung cancer among patients with non-diagnostic bronchoscopies (35). By increasing the diagnostic sensitivity of bronchoscopy, biomarkers such as the one described above, have the potential to expedite more invasive testing and definitive therapy for smokers with lung cancer, and reduce invasive diagnostic procedures for individuals without lung cancer. In addition, if future studies demonstrate that smoking-induced cancer-specific alterations in gene expression precede the development of lung cancer, biomarkers may be useful for indentifying high-risk lung cancer patients.
Molecular Classification and Characterization
Differences in treatment between NSCLC and SCLC make the distinction between these two types of lung cancer important. Within NSCLC, there are potential differences in terms of prognosis and response to newer targeted therapies (36). Accurate molecular classification, therefore, has the potential to identify different molecular subtypes of NSCLC currently not recognized by pathologists that would benefit from subtype-specific therapies. In addition, molecular classification of tumors may augment surgical-pathological staging at surgery, allowing the most appropriate treatment for a given stage of tumor to be used.
Molecular Classification of Lung Cancer
One of the initial applications of high-throughput gene expression technology in the area of lung cancer was to explore whether or not differences in gene expression could be indentified between the different histological subtypes of lung tumors. Two studies in November 2001 began to explore this question using microarray technology and diverse sets of lung tumor samples. The broad goals were to identify gene expression profiles associated with the histological subtypes of lung tumors, identify subclasses of AD where there is frequent disagreement among pathologists, associate gene expression profiles with tumor features such as surgical-pathological stage as well as survival after resection, and identify metastases of non-lung origin.
Garber et al. (37) profiled the gene expression of 67 lung tumors with 5 years of clinical follow-up from 56 patients as well as 5 normal lung samples and 1 fetal lung sample using 24,000 element cDNA microarrays. Hierarchal clustering of samples according to the expression of the most variable genes revealed patterns of gene expression that corresponded to the major morphological classes of lung tumors: AD (n=41), SCC (n=16), LCC (n=5), and SCLC (n=5). The AD tumors were the most heterogeneous and formed 3 distinct clusters. There were differences in survival between the 3 groups, and this was in part associated with tumor grade and lymph node metastases.
In a larger study, Bhattacharjee et al.(38) used Affymetrix U95 microarrays containing 12,600 transcripts to profile gene expression levels of 17 normal lung samples and 186 lung tumors that included 127 ADs, 21 SCC, 20 carcinoids, 6 SCLC, and 12 AD tumors suspected to be of non-lung origin. Using a similar methodology to Garber et al., hierarchal clustering segregated samples based on histological subtype and identified molecular markers associated with each subtype. Both studies found, for example, keratin genes were highly expressed by SCC and genes associated with neuroendocrine differentiation were highly expressed in SCLC. Bhattacharjee et al. examined just the ADs using hierarchal and probabilistic model-based clustering and identified 6 distinct groups. A supervised approach was subsequently used to identify genes strongly associated with each of the 6 clusters. One cluster contained normal lung tissue, another cluster contained tumors suspected to be colon, breast, or liver metastases, and the remaining 4 clusters segregated the ADs based on markers of cell division, proliferation, neuroendocrine origin, and type II alveolar pneumocytes. The clusters were also associated with extent of tumor differentiation, presence of BAC, and patient outcome even when limited to stage I tumors.
Both the Garber et al. and Bhattacharjee et al. studies demonstrated that gene expression patterns could distinguish between the histological subtypes of lung cancer and found that ADs had the greatest heterogeneity. In addition, both studies demonstrated an association between the AD clusters and prognosis. The studies, however, lacked independent test sets to confirm the molecular classifications, however, a study by Hayes et al. demonstrated that the tumor subtypes of AD were reproducible across the two datasets plus an additional dataset (39). The AD specimens also contained a mixture of subtypes that included BACs with known favorable prognoses making it difficult to distinguish between genes related to prognosis or subtype, and the Bhattacharjee study lacked clinical data to confirm metastases from extrapulmonary tumors. Despite these shortcomings, the studies served as a foundation for future lung cancer gene expression studies.
Several smaller studies followed exploring similar questions using different analysis techniques, sample sets, and technologies such as SAGE (40–43). Other studies performed real time PCR and immunohistochemistry to validate gene expression differences between lung tumor subtypes (44), distinguish between primary and metastatic SCC of the lung (45), and explore differences between lung tumors and lung cancer cell lines (46). In addition, other studies have identified molecular markers for pulmonary neuroendocrine tumors using DNA microarrays and linked a subset of these markers to prognosis (47;48). Finally, these studies identified molecular markers for known histological subtypes of lung cancer and suggested refinements to the pathological classification of tumors. Molecular classification of lung tumors may eventually improve prognosis if newly identified subtypes respond differently to current treatments regimens or if they suggest new subtype-specific drug targets.
Molecular Staging of Lung Tumors
In addition to molecular classification of tumors, high-throughput gene expression technologies have been used to characterize tumor stage. A study by Ramaswamy et al. (49) identified a gene expression signature of metastasis that could distinguish between metastatic and primary ADs from multiple tumor types. Stage I and II lung ADs from the Bhattacharjee et al. dataset separated into two groups with significant differences in survival according to the expression of the metastatic gene signature. When the signature was applied to other tumor datasets, tumors expressing the metastatic gene signature consistently had a poor outcome, suggesting that metastatic potential may be encoded in the primary tumor.
Several studies using the primary lung tumor to predict lymph node metastases were subsequently published. Kikuchi et al. (50) and Inamura et al. (51) identified genes associated with lymph node metastasis among primary lung ADs, and Hoang et al. (52) identified genes associated with non-metastatic tumors, those with micrometastases, and those with overt metastasis. Xi et al. (53) used the Bhattacharjee et al. (see above) and the Beer et al. (54) (see Prognosis section below) datasets to examine whether gene expression in primary AD tumors was indicative of lymph node metastases. A 318-gene signature was able to accurately classify node positive patients in the training (Beer et al.) and test (Bhattacharjee et al.) sets, but frequently misclassified node negative patients. The classification as node negative or positive in the node negative patients was associated with survival. These studies suggest that the survival differences observed among stage I ADs in the Garber et al. and Bhattacharjee et al. datasets might be related to the presence of micrometastases or metastatic potential. The use of gene expression for “molecular staging” may enhance the sensitivity of clinical and pathologic methods for staging tumors, improving treatment decisions and ultimately outcomes for lung cancer patients.
miRNA Classification of Lung Cancer
miRNAs are short sequences of RNA about 22 nucleotides long that regulate gene expression by hybridizing to complementary sequences of target mRNA. The binding of miRNAs to mRNAs can result in degradation of the mRNA or repression of mRNA translation into proteins. Recently, expression profiling of miRNAs has contributed to our knowledge of how these short sequences are involved in cancer biology. Yanaihara et al. (55) focused on exploring miRNA expression in normal and cancerous lung tissue. DNA microarrays capable of measuring 352 miRNAs were used to identify 43 miRNAs that were differentially expressed between 104 pairs of normal and lung tumor tissue and 6 miRNAs differentially expressed between AD and SCC.
Prognosis
Thirty to 35% of Stage 1 NSCLC patients relapse following tumor resection(56;57). Clinical trials have indicated a potential survival advantage for early-stage lung cancer patients who receive adjuvant chemotherapy (58). However, it would be useful to identify the subset of these patients who are at low risk for relapse to spare them the side effects of unnecessary treatment. Gene expression profiles have the potential to augment current prognostic indicators such as clincopathological stage, K-ras and p53 mutations, poor differentiation, and high tumor proliferative index.
Prognostic gene expression signatures for NSCLC
The Garber et al. and Bhattacharjee et al. studies found correlations between molecular subgroups of lung AD and prognosis. These findings set the stage for the publication of several studies that used supervised approaches to identify genes associated with prognosis among early-stage ADs. The supervised approaches first stratify patients by known outcome, identify genes associated with these outcomes in a set of training samples, and use these genes and an algorithm to predict the outcome of additional test set samples. In 2002, Beer et al. (54) used DNA microarrays to measure gene expression levels in 67 stage I ADs, 19 stage III ADs, and 10 non-neoplastic lung tissues. Stage I and III tumors were divided into training and testing sets and 50 genes associated with survival were identified across the training set using univariate Cox proportional-hazard regression modeling. Expression levels of these genes were combined using a prediction algorithm to calculate a risk index which was then used to stratify patients into low- and high-risk groups. There was a significant difference in survival between test set samples as a whole and the subgroup of stage I test samples predicted to be low- or high-risk. Interestingly, stratifying patients by prognostic markers such as K-ras and p53 mutation status did not identify subgroups with a significant difference in survival. After refining the predictor, it was validated across 84 lung AD samples from Bhattacharjee et al. and patients assigned to the low- and high-risk groups by gene expression varied significantly in survival. Since the publication of this study, several other studies have emerged with gene expression prognostic profiles for early stage NSCLC (59–65).
One such study by Potti et al. (66) analyzed 89 NSCLC patients using DNA microarrays to develop a metagene prediction model capable of predicting disease recurrence. The model had a higher accuracy than models containing clinical data alone (age, sex, tumor diameter, stage of disease, histological subtype, and smoking history) or both clinical and gene expression data. The model was 72% accurate across ACOSOG Z0030 trial samples (n=25), 79% accurate across CALGB 9761 trial samples (n=84), and 80% accurate across an independent set of stage I SCC (n=15). As proposed by Potti et al., a randomized Phase III trial, CALGB 30506, is about to begin to evaluate the metagene predictor to direct adjuvant therapy in high risk stage IA NSCLC patients. While the prediction model was validated on an independent sample set, it remains unclear if the signature is entirely related to differences in prognosis or recognized subtypes of AD (patients with BAC were not identified). In addition, the variables explored in the clinical risk model did not include potentially important prognostic indicators such as tumor grade, histological subtype of AD (67), and the mutational status of cancer-related genes (K-ras, p53). Finally, it is not clear if there were differences in the use of adjuvant chemotherapy treatment among the patients that could effect survival. The trial will hopefully answer several of these questions that were not addressed in the study.
A study by Lu et al. (68), published shortly after the Potti et al. study, performed a meta-analysis of 7 different datasets (10;38;54;69), including a previously unpublished dataset of their own, to identify a gene expression signature that predicts survival in patients with stage I NSCLC. Genes were identified that were common to the microarray platforms used in all of the studies, the datasets were adjusted for systematic bias, and 197 samples with stage I NSCLC from 5 of the 7 datasets were used to identify a gene expression signature of 64 genes predictive of survival. The signature had higher classification power compared to stage, was predictive of survival among ADs and SCCs, and was able to accurately predict survival in the 2 datasets not used to develop the signature. A subset of the 64 genes was also validated using quantitative RT-PCR and immunohistochemistry. This study demonstrates the feasibility of combining different Affymetrix DNA microarrays to increase sample size and predictive power and identify a robust gene expression signature predictive of survival.
Chen et al. (70) recently reported a 5 gene signature capable of predicting survival among patients with NSCLC. Sixteen genes were found to be associated with survival across training and test sets using DNA microarrays measuring 672 previously identified genes (71) associated with invasive activity in invasive NSCLC cell lines. A subset of the sixteen genes (n=5) were correlated with survival using quantitative RT-PCR, and this subset was used to create a decision tree that stratified patients into low- and high-risk for reoccurrence. The predictor was tested on an independent set of 60 patients and on the Beer et al. (54) dataset. The shortcomings of this study include a heterogeneous group of samples that included Stage I, II, and III NSCLC samples and different subtypes of NSCLC. In addition, Chen et al. chose to focus on a set of invasive genes derived from NSCLC cell lines characteristic of the lung tumor, but not the adjacent stromal tissue. The samples used were not microdissected and had both tumor and stromal tissue, and therefore, the analysis may be missing more robust predictive genes.
Given the publication of numerous studies that have identified prognostic gene expression signatures for NSCLC, one important question concerns the comparability of these studies as they have used different microarray platforms, analysis techniques, and samples. The Lu et al. study discussed above as well as other published studies (72–74) have demonstrated the feasibility of combining different datasets to increase the power and robustness of the prognostic signature. In addition to these studies, additional work has been done to determine the feasibility of conducting larger studies involving the participation of multiple laboratories. Recently a large retrospective, multi-site, blinded study by Shedden et al. collected 442 lung ADs with relevant clinical, pathological, and outcome data at 4 institutions from 6 lung cancer treatment sites to characterize the performance of several prognostic models (75). The feasibility of the study was established previously by comparing gene expression data produced on the same microarray platform using a standardized protocol by the 4 participating institutions (76). Eight prognostic classifiers and classifiers based on the work of Potti et al. (66) and Chen et al. (70) were developed and evaluated on designated training and blinded test subsets of the data and produced variable results. The inclusion of clinical covariates improved the performance of most classifiers, more complex classifiers (classifiers that included more genes) had better performance, classifiers trained across samples of all stages performed better across stage I samples, and a small subset of the classifiers performed well across both tests sets (from 2 different institutions).
The study illustrates many important points concerning the development of gene expression-based prognostic predictors for early stage lung cancer. While the prognostic classifiers contain different gene sets, there was some concordance between the predictions made by each of the classifiers. This suggests that the power of gene expression to predict prognosis is not restricted to the differential expression of a few genes and that each of the classifiers is measuring aspects of prognosis-related lung AD biology. Similar results have been seen in the setting of breast cancer, where various prognostic classifiers (containing different genes) show high rates of concordance in their outcome predictions of individual samples (77). It is interesting to note that for some lung tumor samples there was complete agreement or disagreement between the classifiers and clinical outcome, while for other samples there was considerable heterogeneity. There are several possible explanations for these discrepancies. Lung ADs have significant histological variation and mixed subtypes, and therefore, it is possible that for some samples, the tissue in the sample may not accurately represent the tumor or the biological process on which a particular classifier depends. In addition, heterogeneity in tissue composition and sample processing or inaccuracies in clinical information may contribute to the variability in the predictions made by the classifiers for a particular sample. There are also potential problems with using overall survival as an endpoint to evaluate prognostic gene expression signatures in subjects with “high risk” tumors that are completely resected or in subjects with “low risk” tumors that develop secondary conditions shortly after diagnosis. The study addresses problems that have plagued past studies such as small number of samples, inconsistent and variable clinical data and sample collection and illustrates many of the remaining challenges associated with developing a prognostic gene expression signature for clinical application.
In addition, the MicroArray Quality Control (MAQC) project led by the FDA evaluated microarray technology for its use in clinical and regulatory settings by examining repeatability of data generated within a particular site, across multiple sites, and between seven different microarray platforms (78). The study observed reproducibility of gene expression measurements between different sites and platforms. The reproducibility of gene expression measurements between sites and across platforms demonstrated by these studies is a critical milestone in the development of gene expression biomarkers that can be routinely used in the clinic.
Prognostic miRNA signatures for NSCLC
Prior to the work of Johnson et al. (79) associating let-7 miRNA and RAS expression in lung cancer, Takamizawa et al. (80) demonstrated that reduced expression of let-7 miRNA in lung cancer was associated with shortened postoperative survival. One-hundred, forty-three lung tissue specimens, predominantly ADs, from stage I, II, and III lung cancers were collected from patients undergoing resection. Let-7 expression was used to dichotomize patients into two groups that had significantly different survival (p = 0.0003) when all samples were analyzed or just ADs. Patients with lower let-7 expression has significantly worse prognosis, independent of disease stage.
Yanihara et al. (55) used microarrays to quantify miRNA expression in lung tumors and found that two miRNAs, mir-155 and let-7a-2, were significantly associated with survival in lung ADs by Kaplan-Meier survival analysis. In a multivariate Cox proportional hazard analysis that included all clinicopathological and molecular factors, increased expression of mir-155 was significantly associated with worse prognosis. Real time RT-PCR across an independent validation set of 32 ADs confirmed a significant relationship between mir-155 expression and survival. A subsequent study by Yu et al. (81) used real-time PCR to measure the expression of 157 miRNAs in 112 NSCLC patients to identify a 5 miRNA signature (let-7a, mir-221, mir-137, mir-372, mir-182) capable of predicting overall and relapse-free survival. Cox proportional hazard regression and risk-score analysis was used to identify the 5 miRNA signature across a training set of samples (n=56). The signature was used to predict the risk (high- or low-) on a test set of samples (n=56) and an independent cohort of NSCLC samples (n=62). There was a statistically significant difference in overall and relapse-free survival between low- and high-risk groups and the signature was a reasonable predictor of survival among subsets of the samples with the same cell type or stage. Yu et al. was also able to show that modulating the levels of 4 out of the 5 miRNAs altered lung cancer cell invasiveness in vitro. The results indicate that miRNA expression profiles can be used as prognostic markers for lung cancer. Future studies profiling both gene and miRNA expression across a large cohort of early stage ADs is needed to determine if an expression signature composed of miRNAs, mRNAs, or both has the greatest diagnostic and prognostic potential in lung cancer.
Pharmacogenomics
Integration of diverse sources of clinical, biological, expression, and sequence information is the promise of personalized medicine and may make it possible to individually tailor treatment regimens for lung cancer. For example, biomarkers may identify chemotherapeutic-specific lung cancer subtypes with the potential to improve prognosis through use of individualized treatments. Work in this direction is already starting to yield promising results.
Staunton et al. (82) used DNA microarrays to measure gene expression in the NCI-60 panel (a collection of 60 human cancer cell lines (83;84)). By combining the untreated gene expression profile of each cell line together with information about each cell lines’ chemosensitivity profile, they were able to predict drug sensitivity in an independent test set of cell lines. A subsequent study by Potti et al. (85) repeated and built upon Staunton’s work. They showed that the drug sensitivity predictors derived from the NCI-60 data were capable of accurately predicting patient response to various chemotherapeutic agents, and were further able to predict that lung cancer patients sensitive to docetaxel were likely to be resistant to etoposide – both front-line chemotherapy options. The work by Potti et al. also connected patterns of chemotherapy sensitivity with deregulation of known oncogenic pathways. For example, a relationship between docetaxel resistance and deregulation of the PI3-kinase pathway was observed. Using a panel of 17 NSCLC cell lines a significant association was found between docetaxel resistance and sensitivity to a PI3-kinase inhibitor (LY-294002), suggesting its use as a second-line therapy.
Following the above work, Hsu et al. (86) developed predictors of cisplatin (a first line agent) and pemetrexed (a second line agent) sensitivity using the NCI-60 data and data from Gyorffy et al. (87). They found that docetaxel, abraxane, and pemetrexed sensitivity was significantly inversely correlated with sensitivity to cisplatin (p<0.01) suggesting their use in ciplatin-resistant patients. Another study by Gemma et al. (88) coupled gene expression data generated using 10 human lung cancer cell lines and drug sensitivity data across 8 anti-cancer drugs used in lung cancer chemotherapy (docetaxel, paclitaxel, gemcitabine, vinorelbine, 5-FU, SN38, CDDP, and CBDCA) to demonstrate sensitivity to gemcitabine was uncorrelated with sensitivity to the other agents, suggesting that combination therapy regimens that include gemcitabine might be interesting to pursue clinically.
Many of the studies described earlier in this review profiled gene expression in primary human tumors to identify gene expression predictors of clinical and pathological variables. An exciting aspect of the studies described above is that they use gene expression information from cell lines and demonstrate that this information can lead to clinically relevant predictors of drug sensitivity in lung cancer patients. These results, while tantalizing, are preliminary and need to be validated in larger longitudinal cohorts of lung cancer patients being treated with various chemotherapeutic regimens and followed for measures of disease outcome.
Conclusions and Future Directions
The studies described in this review demonstrate the potential for gene expression signatures to impact lung cancer management; however, numerous obstacles remain to the routine application of these profiles in the clinic. Further work on computational approaches for merging datasets across platforms is needed to effectively leverage the collective data being generated. In addition, large longitudinal studies measuring gene expression as well as routine clinical, biochemical, and pathologic measures are needed to demonstrate that gene expression is a better predictor of outcome than more routine measures. This could be accomplished by leveraging existing large-scale prospective clinical trials or epidemiologic studies and collecting biological samples for gene expression studies from those subjects. Additionally, integrating high-throughput gene expression measurements with other forms of molecular data (SNPs, methylation, proteomics) may give a more complete picture and result in the identification of the most robust diagnostic, prognostic, and predictive markers. However, the ultimate barrier to adoption of these markers in the clinic is the need for more of them to be validated in prospective multicenter studies to demonstrate their reproducibility and accuracy across multiple sites and operators. Physicians and other health care providers will need to be trained in the proper handling and storage of biological specimens for gene expression studies given RNA’s inherent instability. While the FDA has begun to address some of the regulatory issues surrounding multivariate gene expression assays, additional guidance is needed from physicians, third-party payers, and regulatory bodies if these tests are to be translated into clinical benefit for lung cancer patients.
Acknowledgments
Support: This work was supported by NIH/NCI R01CA124640 (AS, MEL, and JB) and the National Institute of Environmental Health Sciences (NIEHS)/NIH U01 ES016035.
Abbreviations
- NSCLC
non-small cell lung cancer
- SCLC
small cell lung cancer
- AD
adenocarcinoma
- SCC
squamous cell carcinoma
- LCC
large cell carcinoma
- BAC
bronchioloalveolar carcinoma
- DNA
deoxyribonucleic acid
- cDNA
complementary DNA
- RNA
ribonucleic acid
- mRNA
messenger RNA
- miRNA
microRNA
- SAGE
serial analysis of gene expression
- CT
computerized tomography
Footnotes
Disclosure: M.E.L. and A.S. have equity in Allegro Dx Inc.
References
- 1.Hirsch FR, Merrick DT, Franklin WA. Role of biomarkers for early detection of lung cancer and chemoprevention. Eur Respir J. 2002;19:1151–1158. doi: 10.1183/09031936.02.00294102. [DOI] [PubMed] [Google Scholar]
- 2.Jett JR. Limitations of screening for lung cancer with low-dose spiral computed tomography. Clin Cancer Res. 2005;11:4988s–4992s. doi: 10.1158/1078-0432.CCR-05-9000. [DOI] [PubMed] [Google Scholar]
- 3.MacRedmond R, McVey G, Lee M, et al. Screening for lung cancer using low dose CT scanning: results of 2 year follow up. Thorax. 2006;61:54–56. doi: 10.1136/thx.2004.037580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 5.van dV, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 6.Ma XJ, Wang Z, Ryan PD, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 7.Marchionni L, Wilson RF, Wolff AC, et al. Systematic Review: Gene Expression Profiling Assays in Early-Stage Breast Cancer. Ann Intern Med. 2008 doi: 10.7326/0003-4819-148-5-200803040-00208. [DOI] [PubMed] [Google Scholar]
- 8.Sweet-Cordero A, Mukherjee S, Subramanian A, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- 9.Borczuk AC, Gorenstein L, Walter KL, et al. Non-small-cell lung cancer molecular signatures recapitulate lung developmental pathways. Am J Pathol. 2003;163:1949–1960. doi: 10.1016/S0002-9440(10)63553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 11.Fan JB, Chee MS, Gunderson KL. Highly parallel genomic assays. Nat Rev Genet. 2006;7:632–644. doi: 10.1038/nrg1901. [DOI] [PubMed] [Google Scholar]
- 12.Thomas RK, Weir B, Meyerson M. Genomic approaches to lung cancer. Clin Cancer Res. 2006;12:4384s–4391s. doi: 10.1158/1078-0432.CCR-06-0098. [DOI] [PubMed] [Google Scholar]
- 13.Granville CA, Dennis PA. An overview of lung cancer genomics and proteomics. Am J Respir Cell Mol Biol. 2005;32:169–176. doi: 10.1165/rcmb.F290. [DOI] [PubMed] [Google Scholar]
- 14.Risch A, Plass C. Lung cancer epigenetics and genetics. Int J Cancer. 2008;123:1–7. doi: 10.1002/ijc.23605. [DOI] [PubMed] [Google Scholar]
- 15.Emmert-Buck MR, Bonner RF, Smith PD, et al. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 16.Schena M, Shalon D, Davis RW, et al. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 17.Lockhart DJ, Dong H, Byrne MC, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 18.Velculescu VE, Zhang L, Vogelstein B, et al. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 19.Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 20.Cloonan N, Forrest AR, Kolle G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 21.Morin RD, O’Connor MD, Griffith M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shields PG. Molecular epidemiology of lung cancer. Ann Oncol. 1999;10 (Suppl 5):S7–11. doi: 10.1093/annonc/10.suppl_5.s7. [DOI] [PubMed] [Google Scholar]
- 23.Jett JR, Midthun DE. Screening for lung cancer: current status and future directions: Thomas A. Neff lecture Chest. 2004;125:158S–162S. doi: 10.1378/chest.125.5_suppl.158s. [DOI] [PubMed] [Google Scholar]
- 24.Bach PB, Jett JR, Pastorino U, et al. Computed tomography screening and lung cancer outcomes. JAMA. 2007;297:953–961. doi: 10.1001/jama.297.9.953. [DOI] [PubMed] [Google Scholar]
- 25.Spira A, Beane J, Shah V, et al. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci USA. 2004;101:10143–10148. doi: 10.1073/pnas.0401422101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beane J, Sebastiani P, Liu G, et al. Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biol. 2007;8:R201. doi: 10.1186/gb-2007-8-9-r201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lonergan KM, Chari R, Deleeuw RJ, et al. Identification of novel lung genes in bronchial epithelium by serial analysis of gene expression. Am J Respir Cell Mol Biol. 2006;35:651–661. doi: 10.1165/rcmb.2006-0056OC. [DOI] [PubMed] [Google Scholar]
- 28.Chari R, Lonergan KM, Ng RT, et al. Effect of active smoking on the human bronchial epithelium transcriptome. BMC Genomics. 2007;8:297. doi: 10.1186/1471-2164-8-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hackett NR, Heguy A, Harvey BG, et al. Variability of antioxidant-related gene expression in the airway epithelium of cigarette smokers. Am J Respir Cell Mol Biol. 2003;29:331–343. doi: 10.1165/rcmb.2002-0321OC. [DOI] [PubMed] [Google Scholar]
- 30.Carolan BJ, Heguy A, Harvey BG, et al. Up-regulation of expression of the ubiquitin carboxyl-terminal hydrolase L1 gene in human airway epithelium of cigarette smokers. Cancer Res. 2006;66:10729–10740. doi: 10.1158/0008-5472.CAN-06-2224. [DOI] [PubMed] [Google Scholar]
- 31.Harvey BG, Heguy A, Leopold PL, et al. Modification of gene expression of the small airway epithelium in response to cigarette smoking. J Mol Med. 2007;85:39–53. doi: 10.1007/s00109-006-0103-z. [DOI] [PubMed] [Google Scholar]
- 32.Lampe JW, Stepaniants SB, Mao M, et al. Signatures of environmental exposures using peripheral leukocyte gene expression: tobacco smoke. Cancer Epidemiol Biomarkers Prev. 2004;13:445–453. [PubMed] [Google Scholar]
- 33.van Leeuwen DM, van AE, Gottschalk RW, et al. Cigarette smoke-induced differential gene expression in blood cells from monozygotic twin pairs. Carcinogenesis. 2007;28:691–697. doi: 10.1093/carcin/bgl199. [DOI] [PubMed] [Google Scholar]
- 34.Spira A, Beane JE, Shah V, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–366. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 35.Beane J, Sebastiani P, Whitfield TH, et al. A Prediction Model for Lung Cancer Diagnosis that Integrates Genomic and Clinical Features. Cancer Prevention Research. 2008;1:65–76. doi: 10.1158/1940-6207.CAPR-08-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 37.Garber ME, Troyanskaya OG, Schluens K, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayes DN, Monti S, Parmigiani G, et al. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J Clin Oncol. 2006;24:5079–5090. doi: 10.1200/JCO.2005.05.1748. [DOI] [PubMed] [Google Scholar]
- 40.Hibi K, Liu Q, Beaudry GA, et al. Serial analysis of gene expression in non-small cell lung cancer. Cancer Res. 1998;58:5690–5694. [PubMed] [Google Scholar]
- 41.Nacht M, Dracheva T, Gao Y, et al. Molecular characteristics of non-small cell lung cancer. Proc Natl Acad Sci USA. 2001;98:15203–15208. doi: 10.1073/pnas.261414598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamagata N, Shyr Y, Yanagisawa K, et al. A training-testing approach to the molecular classification of resected non-small cell lung cancer. Clin Cancer Res. 2003;9:4695–4704. [PubMed] [Google Scholar]
- 43.Kim B, Lee HJ, Choi HY, et al. Clinical validity of the lung cancer biomarkers identified by bioinformatics analysis of public expression data. Cancer Res. 2007;67:7431–7438. doi: 10.1158/0008-5472.CAN-07-0003. [DOI] [PubMed] [Google Scholar]
- 44.Sugita M, Geraci M, Gao B, et al. Combined use of oligonucleotide and tissue microarrays identifies cancer/testis antigens as biomarkers in lung carcinoma. Cancer Res. 2002;62:3971–3979. [PubMed] [Google Scholar]
- 45.Talbot SG, Estilo C, Maghami E, et al. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res. 2005;65:3063–3071. doi: 10.1158/0008-5472.CAN-04-1985. [DOI] [PubMed] [Google Scholar]
- 46.Virtanen C, Ishikawa Y, Honjoh D, et al. Integrated classification of lung tumors and cell lines by expression profiling. Proc Natl Acad Sci USA. 2002;99:12357–12362. doi: 10.1073/pnas.192240599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones MH, Virtanen C, Honjoh D, et al. Two prognostically significant subtypes of high-grade lung neuroendocrine tumours independent of small-cell and large-cell neuroendocrine carcinomas identified by gene expression profiles. Lancet. 2004;363:775–781. doi: 10.1016/S0140-6736(04)15693-6. [DOI] [PubMed] [Google Scholar]
- 48.He P, Varticovski L, Bowman ED, et al. Identification of carboxypeptidase E and gamma-glutamyl hydrolase as biomarkers for pulmonary neuroendocrine tumors by cDNA microarray. Hum Pathol. 2004;35:1196–1209. doi: 10.1016/j.humpath.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 49.Ramaswamy S, Ross KN, Lander ES, et al. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 50.Kikuchi T, Daigo Y, Katagiri T, et al. Expression profiles of non-small cell lung cancers on cDNA microarrays: identification of genes for prediction of lymph-node metastasis and sensitivity to anti-cancer drugs. Oncogene. 2003;22:2192–2205. doi: 10.1038/sj.onc.1206288. [DOI] [PubMed] [Google Scholar]
- 51.Inamura K, Shimoji T, Ninomiya H, et al. A metastatic signature in entire lung adenocarcinomas irrespective of morphological heterogeneity. Hum Pathol. 2007;38:702–709. doi: 10.1016/j.humpath.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 52.Hoang CD, D’Cunha J, Tawfic SH, et al. Expression profiling of non-small cell lung carcinoma identifies metastatic genotypes based on lymph node tumor burden. J Thorac Cardiovasc Surg. 2004;127:1332–1341. doi: 10.1016/j.jtcvs.2003.11.060. [DOI] [PubMed] [Google Scholar]
- 53.Xi L, Lyons-Weiler J, Coello MC, et al. Prediction of lymph node metastasis by analysis of gene expression profiles in primary lung adenocarcinomas. Clin Cancer Res. 2005;11:4128–4135. doi: 10.1158/1078-0432.CCR-04-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 55.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 56.Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet. 2000;355:479–485. doi: 10.1016/S0140-6736(00)82038-3. [DOI] [PubMed] [Google Scholar]
- 57.Nesbitt JC, Putnam JB, Jr, Walsh GL, et al. Survival in early-stage non-small cell lung cancer. Ann Thorac Surg. 1995;60:466–472. doi: 10.1016/0003-4975(95)00169-l. [DOI] [PubMed] [Google Scholar]
- 58.Booth CM, Shepherd FA. Adjuvant chemotherapy for resected non-small cell lung cancer. J Thorac Oncol. 2006;1:180–187. [PubMed] [Google Scholar]
- 59.Miura K, Bowman ED, Simon R, et al. Laser capture microdissection and microarray expression analysis of lung adenocarcinoma reveals tobacco smoking-and prognosis-related molecular profiles. Cancer Res. 2002;62:3244–3250. [PubMed] [Google Scholar]
- 60.Endoh H, Tomida S, Yatabe Y, et al. Prognostic model of pulmonary adenocarcinoma by expression profiling of eight genes as determined by quantitative real-time reverse transcriptase polymerase chain reaction. J Clin Oncol. 2004;22:811–819. doi: 10.1200/JCO.2004.04.109. [DOI] [PubMed] [Google Scholar]
- 61.Tomida S, Koshikawa K, Yatabe Y, et al. Gene expression-based, individualized outcome prediction for surgically treated lung cancer patients. Oncogene. 2004;23:5360–5370. doi: 10.1038/sj.onc.1207697. [DOI] [PubMed] [Google Scholar]
- 62.charoenrat P, Rusch V, Talbot SG, et al. Casein kinase II alpha subunit and C1-inhibitor are independent predictors of outcome in patients with squamous cell carcinoma of the lung. Clin Cancer Res. 2004;10:5792–5803. doi: 10.1158/1078-0432.CCR-03-0317. [DOI] [PubMed] [Google Scholar]
- 63.Raponi M, Zhang Y, Yu J, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66:7466–7472. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 64.Seike M, Yanaihara N, Bowman ED, et al. Use of a cytokine gene expression signature in lung adenocarcinoma and the surrounding tissue as a prognostic classifier. J Natl Cancer Inst. 2007;99:1257–1269. doi: 10.1093/jnci/djm083. [DOI] [PubMed] [Google Scholar]
- 65.Larsen JE, Pavey SJ, Passmore LH, et al. Gene expression signature predicts recurrence in lung adenocarcinoma. Clin Cancer Res. 2007;13:2946–2954. doi: 10.1158/1078-0432.CCR-06-2525. [DOI] [PubMed] [Google Scholar]
- 66.Potti A, Mukherjee S, Petersen R, et al. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med. 2006;355:570–580. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- 67.Sun Z, Yang P, Aubry MC, et al. Can gene expression profiling predict survival for patients with squamous cell carcinoma of the lung? Mol Cancer. 2004;3:35. doi: 10.1186/1476-4598-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu Y, Lemon W, Liu PY, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med. 2006;3:e467. doi: 10.1371/journal.pmed.0030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Borczuk AC, Shah L, Pearson GD, et al. Molecular signatures in biopsy specimens of lung cancer. Am J Respir Crit Care Med. 2004;170:167–174. doi: 10.1164/rccm.200401-066OC. [DOI] [PubMed] [Google Scholar]
- 70.Chen HY, Yu SL, Chen CH, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 71.Chen JJ, Peck K, Hong TM, et al. Global analysis of gene expression in invasion by a lung cancer model. Cancer Res. 2001;61:5223–5230. [PubMed] [Google Scholar]
- 72.Jiang H, Deng Y, Chen HS, et al. Joint analysis of two microarray gene-expression data sets to select lung adenocarcinoma marker genes. BMC Bioinformatics. 2004;5:81. doi: 10.1186/1471-2105-5-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parmigiani G, Garrett-Mayer ES, Anbazhagan R, et al. A cross-study comparison of gene expression studies for the molecular classification of lung cancer. Clin Cancer Res. 2004;10:2922–2927. doi: 10.1158/1078-0432.ccr-03-0490. [DOI] [PubMed] [Google Scholar]
- 74.Tamayo P, Scanfeld D, Ebert BL, et al. Metagene projection for cross-platform, cross-species characterization of global transcriptional states. Proc Natl Acad Sci USA. 2007;104:5959–5964. doi: 10.1073/pnas.0701068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shedden K, Taylor JM, Enkemann SA, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008 doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dobbin KK, Beer DG, Meyerson M, et al. Interlaboratory comparability study of cancer gene expression analysis using oligonucleotide microarrays. Clin Cancer Res. 2005;11:565–572. [PubMed] [Google Scholar]
- 77.Fan C, Oh DS, Wessels L, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 78.Shi L, Reid LH, Jones WD, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 80.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 81.Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 82.Staunton JE, Slonim DK, Coller HA, et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci USA. 2001;98:10787–10792. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grever MR, Schepartz SA, Chabner BA. The National Cancer Institute: cancer drug discovery and development program. Semin Oncol. 1992;19:622–638. [PubMed] [Google Scholar]
- 84.Stinson SF, Alley MC, Kopp WC, et al. Morphological and immunocytochemical characteristics of human tumor cell lines for use in a disease-oriented anticancer drug screen. Anticancer Res. 1992;12:1035–1053. [PubMed] [Google Scholar]
- 85.Potti A, Dressman HK, Bild A, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–1300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 86.Hsu DS, Balakumaran BS, Acharya CR, et al. Pharmacogenomic strategies provide a rational approach to the treatment of cisplatin-resistant patients with advanced cancer. J Clin Oncol. 2007;25:4350–4357. doi: 10.1200/JCO.2007.11.0593. [DOI] [PubMed] [Google Scholar]
- 87.Gyorffy B, Surowiak P, Kiesslich O, et al. Gene expression profiling of 30 cancer cell lines predicts resistance towards 11 anticancer drugs at clinically achieved concentrations. Int J Cancer. 2006;118:1699–1712. doi: 10.1002/ijc.21570. [DOI] [PubMed] [Google Scholar]
- 88.Gemma A, Li C, Sugiyama Y, et al. Anticancer drug clustering in lung cancer based on gene expression profiles and sensitivity database. BMC Cancer. 2006;6:174. doi: 10.1186/1471-2407-6-174. [DOI] [PMC free article] [PubMed] [Google Scholar]