Abstract
Decreased histone acetyltransferase activity and transcriptional dysfunction have been implicated in almost all neurodegenerative conditions. Increasing net histone acetyltransferase activity through inhibition of the histone deacetylases (HDACs) has been shown to be an effective strategy to delay or halt progression of neurological disease in cellular and rodent models. These findings have provided firm rationale for Phase I and Phase II clinical trials of HDAC inhibitors in Huntington’s disease, spinal muscular atrophy, and Freidreich’s ataxia. In this review, we discuss the current findings and promise of HDAC inhibition as a strategy for treating neurological disorders. Despite the fact that HDAC inhibitors are in an advanced stage of development, we suggest other approaches to modulating HDAC function that may be less toxic and more efficacious than the canonical agents developed so far.
Keywords: histone deacetylase inhibitors, histone deacetylases, neurodegeneration, therapeutics, transcription
1. Introduction
Every year, millions of people are diagnosed with neurodegenerative diseases such as stroke, Alzheimer’s disease (AD), amyotrophic lateral sclerosis (ALS), and Parkinson’s disease (PD). At present, there are no effective cures for these diseases. This void creates an emotional strain on the affected individuals and their families, as well as a financial burden on the healthcare system. The onset and severity of these diseases vary considerably because of genetic mutations, environmental factors, and age, making development of treatments difficult; however, there are many other reasons for the absence of effective cures. First, the molecular mechanisms underlying disease onset and progression remain poorly understood. Second, drug developers have traditionally focused on targets that affect a single downstream signalling pathway, thus ignoring the reality that many pathways act serially and in parallel to create the phenotype of a particular disease. Third, therapeutic strategies have focused on targeting a single, well-defined post-transcriptional genetic modification and have avoided engaging endogenous programmes of gene expression that may include tens to hundreds of genes. Given that endogenous adaptive programmes are activated as a consequence of stress, an ideal therapeutic target should affect adaptive gene expression as well as post-translational modifications of pre-existing proteins. The toxicity of agents that act on such targets is minimal, in theory, because the programmes of gene expression active in the target are similar to those activated as part of adaptive responses. Fourth, any drug effects on a target for neurodegenerative diseases must not provide a selective advantage for cancer cells or autoimmune cells. Recently, the histone deacetylase (HDAC) protein family has been recognised as an attractive therapeutic target for the treatment of neurodegenerative diseases because global inhibition of HDACs overcomes all these problems and meets additional challenges. This review summarises current research on and the promise of this novel class of neuroprotective agents.
2. Histone deacetylases
HDACs were first identified as enzymes that catalyze the removal of an acetyl group from the N-terminal tails of histone proteins. This action leads to chromatin compaction and gene expression silencing. Together with histone acetyl transferases (HATs), which catalyze the reverse reaction, HDACs control how accessible chromatin is to transcription factors and thus the level of gene expression. In addition to their role in regulating chromatin structure and gene expression, HDACs deacetylate non-histone proteins such as p53 [1,2], Sp1 [3], E2F [4,5], cAMP response-element-binding protein (CREB) [6], GATA1 [7,8] and tubulin [9].
HDACs are evolutionarily conserved from yeast to man. Human HDACs are classified into four classes. The zinc-dependent enzymes constitute classes I, II and IV, whereas the NAD-dependent enzymes are included in class III (Figure 1). Class I HDACs include HDAC-1, HDAC-2, HDAC-3, and HDAC-8. Class II HDACs include HDAC-4, HDAC-5, HDAC-6, HDAC-7, HDAC-9a and b, and HDAC-10. Class I HDACs are ubiquitously expressed, whereas class II HDACs are primarily expressed in striated muscle, heart and brain. Class III HDACs are referred to as sirtuins because they are homologous to the yeast protein Sir-2. This class includes SIRT1 – 7 (reviewed in references [10,11]). Class IV HDACs include HDAC-11, which is predominantly nuclear and was found to be expressed in the mouse brain [10–12].
Figure 1.
Histone deacetylase classes, expression patterns and sub-cellular localization.
The dynamic state of chromatin and the accessibility of gene promoters to transcription factors and/or transcriptional machinery are dependent on the balance between HAT activity and HDAC activity. This balance is tightly regulated because HATs and HDACs are often found as part of protein complexes that include co-activators or corepressors. Defects in the regulation of the fine interplay between HAT function and HDAC function can lead to many cancers as well as to neurodegenerative disease [13]. For this reason, HDAC inhibitors have been extensively studied in models of neurodegenerative disease [13], and some agents–such as valproic acid (VPA), sodium butyrate (SB) and LBH589–are now being tested in clinical trials in patients with spinal muscular atrophy (SMA), Huntington’s disease (HD), AD and ALS.
3. Histone deacetylase inhibitors
HDAC inhibitors can be divided into several groups on the basis of their chemical structure. The first group includes small-molecule hydroxamates such as trichostatin A (TSA), suberoylanilide hydroxamic acid (SAHA)/vorinostat and scriptaid (Table 1) The second group includes derivatives of aliphatic acid such as SB sodium phenylbutyrate (SPB) and VPA. VPA was widely used as an antiepileptic drug well before its function as a HDAC inhibitor was described [14]. The third group includes cyclic tetrapeptides such as apicidin, trapoxin, depsipeptide (FK-228)/romidepsin. Finally, the last group includes benzamides, such as MS-275/SNDX-275 and Cl-994 [10]. HDAC inhibitors such as the hydroxamates bind to the active site of the HDAC enzyme and function by chelating the Zn2+ ion. These compounds have a ashort half-life, but still exhibits long-term effects [15,16].
Table 1.
Classes and specificities of HDAC inhibitors.
| HDAC inhibitor | Target | |
|---|---|---|
| Hydroxamates | ||
| TSA | Class I and II HDACs |
|
| SAHA/Zolinza/vorinostat | Class I and II HDACs |
|
| LAQ-824 | Class I and II HDACs |
|
| LBH-589/panobinostat | Class I and II HDACs |
|
| PCI 24789 | Class I and II HDACs |
|
| Aliphatic acids | ||
| VPA | Class I and lla HDACs |
|
| Sodium Phenylbutyrate | Class I and lla HDACs |
|
| Sodium Butyrate | Class I and lla HDACs |
|
| Cyclic tetrapeptides | ||
| Apicidin | Class I and III HDACs |
|
| Trapoxin | Class I and IIa HDACs |
|
| Depsipeptide/FK-228/ romidepsin |
Class IHDACs | |
| Benzamides | ||
| M5-275/SNDX-275 | HDAC-l, HDAC-2, HDAC-3 |
|
| MGCD0103 | Class I HDACs |
SAHA: Suberoylanilide hydroxamic acid; SB: Sodium butyrate; TSA: Trichostatin A; VPA: Valproic acid.
3.1 Histone deacetylase inhibitors: mechanism of action
HDAC inhibitors are now in clinical trials to test their capacity to treat a variety of cancers [17–19]. These agents have been shown to be effective in blocking the toxicity of the chemotherapeutic agent cisplatin and are themselves anticancer agents [17–20]. The use of these drugs in combination with chemotherapeutic agents may be a way to abrogate the deleterious effects of chemotherapy, while enhancing its efficacy. Gensert et al. showed that HDAC inhibitors induce the expression of a CD81, a tetraspanin that regulates growth arrest in gliomas and astrocytes. HDAC inhibition thus prevents cell death in the CNS without increasing the likelihood of glia cancers [21].
Many studies have reported that HATs and HDACs are misregulated in neurological disease (reviewed in references [10,22,23]). Accordingly, HDAC inhibitors have been assessed in both cell culture models and rodent models of neurodegenerative disease. Expanded polyglutamine tracts in mutant huntingtin (htt), for example, have been shown to mediate neurodegeneration via inhibition of HAT activity in CREB binding protein (CBP)/p300. Accordingly, Steffan et al. reasoned that HDAC inhibition should alleviate this defect in cellular HAT activity and prevent cell loss [24]. These researchers used molecular and small-molecule suppression of HDAC activity to define a role for HDAC inhibition in neuroprotection. Their seminal observations were rapidly replicated in studies of other polyglutamine expansion diseases [24,25].
A subsequent study suggested that the benefits of HDAC inhibitors extend beyond their effects on the proximate toxicity of polyglutamine expansions by showing that these agents can abrogate oxidative death in cortical neurons [26]. In this study, oxidative stress, a putative mediator of neurodegeneration implicated in almost every neurological condition, was shown to increase acetylation of the protective transcription factor Sp1. TSA treatment increased Sp1 acetylation, binding of Sp1 DNA to its targets, and Sp1-dependent gene expression [3,26]. It also protected cells from oxidative-stress-induced apoptosis both in vitro and in vivo. Sp1 activation was subsequently shown to be necessary for the protective effects of HDAC inhibitors. These data suggest that oxidative stress induces HDAC inhibition as part of an Sp1-dependent adaptive response. Accordingly, augmentation of this adaptive response via a host of structurally distinct class I or class II HDAC inhibitors protected neurons from oxidative death. Recent studies have shown that class II HDACs have conserved cysteine motifs that can be oxidized leading to the translocation of these HDACs from the nucleus to the cytoplasm, the functional equivalent of HDAC inhibition [27].
One of the major challenges in understanding the precise mechanisms by which global HDAC inhibitors protect against oxidative death is the fact that a small, but reproducible, toxicity is observed in cortical neurons exposed continuously to these agents. This toxicity is of particular concern if HDAC inhibitors are to be moved into the clinical setting. Langley et al. observed that pulse treatment with TSA for 2 h was not associated with any toxic effects and maintained all the protective effects of this drug against oxidative-stress-induced cell death [28]. HDAC inhibition correlated with increased p21 gene and protein expression both in cultured neurons and in an in vivo model of permanent ischemia; However, although p21 is sufficient for this protective effect, it is not necessary. This observation is consistent with TSA being a global HDAC inhibitor and potentially functioning by affecting multiple gene expression programmes that together contribute to the protective effect of this drug [28].
The findings of studies by Vecsey et al. [29], Kanai et al. [30], Leng et al. [31], and Politis et al. [32] are consistent with the idea that multiple downstream gene targets contribute to the neuroprotective effects of HDAC inhibitor treatment. Vescey et al. reported that HDAC inhibitors enhance memory by activating genes regulated by the CREB and CBP/HAT complex [29]. Indeed, the authors showed that the CREB/CBP complex mediates hippocampus-dependent memory and hippocampal synaptic plasticity and that TSA does not globally alter gene expression, but rather upregulates the expression of specific genes during memory consolidation [29]. Kanai et al. showed that mature cerebellar granular cells are protected from SYM2081 glutamate-mediated excitotoxicity by VPA, TSA and butyrate [30]. SYM2081 ([2S,4R]-4-methylglutamate) is an inhibitor of excitatory amino-acid transporters and an agonist of low-affinity kainate receptors [30]. Moreover, Kanai and colleagues linked the protective effect of HDAC inhibitors to decreased nuclear accumulation of the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The latter has been reported to influence cytotoxicity by translocating to the nucleus, after being S-nitrosylated, as part of a complex with Siah1 (an E3 ubiquitin ligase). In the nucleus, GAPDH stabilises Siah1, which in turn allows Siah1 to degrade nuclear proteins and contribute to the initiation of apoptosis [33]. A study published by Leng et al., on the other hand, highlights the role of glycogen synthase kinase-3 inhibition and increased β-catenin-dependent and TCF-1/LEF-1-dependent gene expression–through combined treatment with lithium and an HDAC inhibitor (VPA, SPB, SB or TSA)–in protecting aging cerebellar granule cells against glutamate-induced cell death [31]. Finally, Politis et al. showed that HDAC inhibition causes growth arrest and induces differentiation in neuroblastoma cells [32]. The authors identified Cend1, a neuronal lineage-specific cell-cycle-exit molecule, as one of the genes mediating the function of HDAC inhibitors. They showed that knockdown of this gene abolishes the antiproliferative effects and differentiation induced by TSA. Even though these experiments were performed in cell lines, it is important to mention that the aberrant expression of proteins that classically drive cell-cycle progression has been observed in neurodegenerative diseases in humans, such as AD (reviewed in reference [34]).
In addition to affecting the expression and function of transcription factors and cell-cycle-regulating molecules, HDAC inhibition also affects the expression and function of cytoskeletal proteins. Meisel et al. observed that TSA protects cortical neurons from oxygen and glucose deprivation in a model of ischemic cell death [35]. This effect was in part due to upregulation of the actin binding protein gelsolin. Gelsolin induces actin remodelling and filament disassembly by capping the ends of and severing actin microfilaments. TSA failed to protect gelsolin knock-out cells, however. Gelsolin normally protects brain cells from ischemic injury by changing the actin cytoskeleton, reducing Ca2+-induced cytotoxicity [36,37] and stabilising mitochondrial permeability transition [38]. Tubulin is another cytoskeletal target affected by global HDAC inhibition. Acetylation of tubulin is increased via HDAC-6 inhibition, and this process yields a net increase in vesicular transport and release of brain-derived neurotrophic factor (BDNF). Such a modification may be important for diseases where microtubule transport is affected or where release of transcription factors from the cytoskeleton to the nucleus is important, such as HD [39–41] Siddiq et al. (manuscript in preparation)].
In addition to protecting neurons by affecting neuronal gene expression profiles and the acetylation status of neuronal proteins, there is recent evidence to indicate that the neuroprotective effects of HDAC inhibitors may involve astrocytes [42]. One study showed that HDAC inhibitors, such as TSA and SB, increase the expression of glia-cell-line-derived neurotrophic factor (GDNF) and BDNF in astrocytes. This effect was shown to be associated with increased H3 acetylation at the promoters of these genes, as well as with protection of dopaminergic neurons in midbrain neuron-glia cultures [42].
Finally, HDAC inhibition has a role in mediating the plasticity of neuronal development. Lyssiotis et al. demonstrated that following BMP exposure, lineage-committed oligodendrocyte precursor cells can be converted to neural-like stem cells that produce both neurons and glia after BMP exposure [43]. This effect is achieved by the inactivating the repression of 13 genes, including sox-2, that are involved in the maintenance of neuron stem cell fate and the repression of genes involved in oligodendrocyte fate.
The studies described above show that many putative targets are involved in the broad effects of HDAC inhibition on neuroprotection and repair. Exactly which targets are crucial for the success of these agents is dependent on a host of factors, including the cell type, the injury stimulus, and the specific HDAC isoforms expressed in a tissue. These studies suggest, however, that it is unlikely that a universal target will be found that can explain the successes of HDAC inhibitors in the treatment of the different disorders of synaptic plasticity and cognition, including neurodegenerative disorders (e.g., HD, PD, ALS, and brain ischemia), neurodevelopmental disorders (Rubinstein–Taybi syndrome, Rett syndrome and fragile X syndrome), mood disorders (depression and anxiety) and motor neuron disease.
3.2 Histone deacetylase inhibitors and diseases
3.2.1 Huntington’s disease
HD is an autosomal, dominant, late-onset neurodegenerative disease characterised by cognitive dysfunction, psychiatric symptoms and movement impairment. HD is caused by a polyglutamine expansion in the 5′-coding region of the htt gene. The presence of the expansion leads to the nuclear translocation and aggregation of the mutant htt and, in turn, to the inhibition of transcription factors such as Sp1 and co-activators such as CBP [26,44–46]. The utility of HDAC inhibition was first observed in a Drosophila model of polyglutamine disease. In this model, SAHA blocked photo-receptor neurodegeneration and increased survival [24,47]. Studies in the R6/2 Huntington mouse model showed that with both SB treatment [45] and SAHA treatment [48] have beneficial effects. To further understand which HDAC is mediating the toxicity in these models and to overcome the absence of specific HDAC inhibitors, experiments using RNA interference were performed in C. elegans neurons expressing a human htt fragment with an expanded polyglutamine tract (Htn-Q150). These studies showed that HDAC-3 acts within neurons in Htn-Q150 to promote degeneration in response [49]. Recently, the beneficial effects of a novel pimelic diphenylamide HDAC inhibitor, HDACi 4b, in an HD mouse model were reported [50].
3.2.2 Parkinson’s disease
PD is a movement disorder characterised by the selective and progressive loss of dopaminergic neurons in the midbrain substantia nigra, which causes muscle rigidity, tremor, and bradykinesia. Even though the molecular mechanisms underlying this disease are still under investigation, α-synuclein is consistently implicated in PD. Kontopoulis et al. observed that nuclear α-synuclein is toxic, whereas its cytoplasmic form is protective in cell cultures and in flies [51]. α-Synuclein in the nucleus was found to be bound to histones, but treatment with two HDAC inhibitors, SB and SAHA, abrogated the toxicity. Furthermore, SPB had protective effects in animals administered 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine, which causes destruction of the dopaminergic nigrostriatal pathway analogous to that observed in PD [52]. VPA was also protective in this setting, inducing the release of neurotrophic factors such as BDNF and GDNF from astrocytes [42,53] or inhibiting of the release of pro-inflammatory factors from microglia [54].
3.2.3 Amyotrophic Lateral sclerosis
ALS is a progressive, lethal neurodegenerative disease characterised by selective loss of both upper and lower motor neurons. So far, the main cause of the sporadic forms of this disease remains elusive, although it is known that both oxidative stress and protein aggregation contribute to the pathology. In addition, altered transcriptional activity leading to defects in many cellular functions has been observed. Sugai et al. reported that VPA can protect spinal motor neurons from glutamate toxicity in organotypic slice cultures [55]. Following this report, another study demonstrated that SPB can significantly extend survival and improve both the clinical and neuropathological phenotypes of G93A transgenic ALS mice [56]. In another study, VPA treatment was shown to induce CBP transcriptional expression in motor neurons and cause delayed disease onset; however, it had no effect on life span [57]. Treatment with HDAC inhibitors has been shown to ameliorate the ALS phenotype, and further synergistic effects are observed on simultaneous treatment with the catalytic antioxidant AEOL 10150 [58]. It is not clear whether this synergy is due to the ability of antioxidants to abrogate the toxic effects of HDAC inhibitors or whether antioxidants act on injury pathways parallel to those ameliorated by HDAC inhibition.
3.2.4 Stroke and ischemia
Stroke is the third leading cause of death in the US and the leading cause of disability. Delayed cell death following a stroke is redox regulated and is thought to be a downstream consequence of oxygen and glucose deprivation. HDAC inhibitors are protective against oxygen and glucose deprivation and against oxidative-stress-induced cell death in vitro, which indicates that HDAC inhibitors are ideal treatment candidates for patients who have experienced a stroke. SAHA and VPA have been tested in a model of focal cerebral ischemia in which cell death was induced by the occlusion of the middle cerebral artery [59,60]. Treatment with SAHA increased histone H3 acetylation within the normal brain and prevented histone deacetylation in the ischemic brain [60]. In addition, it induced the expression of the neuroprotective proteins Hsp70 and Bcl-2 in both control and ischemic brain tissue [60]. Likewise, treatment with VPA also resulted in increased expression of the protective protein Hsp70 [59]. It is important to note that HDAC inhibition also suppressed microglia activation, reduced the number of microglia, and inhibited various inflammatory markers in the ischemic brain. In more-recent studies, particular effort has been devoted to uncovering the specific HDACs involved in stroke and ischemia. In acute coronary syndrome, small interfering RNA knockdown experiments have showed that HDAC-4 reduces infarct size, suggesting a role for this specific isoform [61].
3.2.5 Rubinstein–Taybi syndrome
Rubinstein–Taybi syndrome is an inheritable disorder caused by mutations in the gene encoding the CBP and characterised by mental retardation and skeletal abnormalities [62]. Several different mouse models in which CBP or p300 function is altered have been created [63]. Such mice show deficits in synaptic plasticity and memory that is linked to impairment in histone H2B acetylation. Treatment with SAHA, however, reversed or ameliorated the symptoms [63,64].
3.2.6 Friedreich’s ataxia
Friedreich’s ataxia (FRDA) is caused by a homozygous GAA-repeat expansion mutation within intron 1 of the frataxin (FXN) gene, leading to reduced expression of FXN and its gene product, a highly conserved mitochondrial protein. Repression of FXN gene expression is mediated by increased trimethylation of histone H3 at lysine 9 and by hypoacetylation of histones H3 and H4 [65,66]. Two recent studies demonstrated promising effects of HDAC inhibitors in alleviating symptoms associated with FRDA. The first study showed that a new class of HDAC inhibitors could reverse FXN gene silencing in primary lymphocytes from individuals with FRDA [65]. The second demonstrated that treatment with a novel class I HDAC inhibitor, an analogue of BML-210, could increase histone H3 and H4 acetylation in chromatin near the GAA repeat and restore wild-type FXN levels in the nervous system and heart of KIKI mice, a murine model of FRDA [67].
3.2.7 Fragile X syndrome
Mutations in the FMR1 gene result in fragile X syndrome and mental retardation. The most common mutation is expansion of a CGG repeat tract at the 5′ end of this gene. This expansion leads to cytosine methylation and transcriptional silencing. Coffee et al. observed that normal individuals have acetylated H3 and H4 associated with the FMR1 locus, whereas individuals with fragile X syndrome do not [68]. When this group examined cells from patients with fragile X syndrome, they observed that inhibitors of DNA methylation completely restored H3 and H4 acetylation and gene expression at the FMR1 locus. On the other hand, TSA only restored H4 acetylation and could not restore FMR1 expression. A study published by Chiurazzi et al., however, reported a consistent but mild reactivation of the FMR1 gene when lymphoblastoid cell lines of non-mosaic full mutation patients were treated with 4-phenylbutyrate, SB or TSA [69]. Moreover, they showed that combining these HDAC inhibitors with 5-azadC leads to a marked increase in FMR1 expression, suggesting a synergistic effect of histone hyperacetylation and DNA demethylation. The above studies did not test whether sirtuins are involved in reactivating the FMR1 locus; however, a more-recent study reported that inhibition of SIRT1 can induce histone H3 and H4 acetylation and reactivate expression of the FMR1 gene [70].
3.2.8 Mood and anxiety disorders
Therapies for mood and anxiety disorders mainly target the regulation of synaptic transmission. The prevalent working hypothesis is that increased levels of neurotransmitters lead to increased expression of specific genes, whose products act to alter neural function and behaviour. Several recent papers have uncovered the role of chromatin remodelling in these diseases. Tsankova et al. have demonstrated that BDNF expression is reduced in mice subjected to chronic social defeat and that treatment with the tricyclic antidepressant imipramine can restore normal behaviour through increased BDNF gene expression [71]. This increase in expression was associated with increased acetylation of the BDNF promoter through downregulation of HDAC-5 activity. Microarray studies have indicated that gene expression profiles are highly altered in the hippocampus of adult rats depending on early maternal care, a model of anxiety disorder, and that altered glucocorticoid receptor gene expression is, in part, responsible for the long-lasting changes seen in behaviour [72]. In these studies, treatment with TSA was shown to reverse the repression of the glucocorticoid receptor gene and decrease anxiety. Epigenetic changes have also been found to be associated with specific mood disorders, such as schizophrenia and those provoked by drug addiction. The efficacy of HDAC inhibition in ameliorating symptoms in animal models of these conditions has been reported in several studies published the last year [73–77].
3.2.9 Motor neuron diseases
SMA is a neurodegenerative disease of the motor neurons that results in progressive muscle weakness. It is also the leading hereditary cause of infant mortality. In humans, there are two survival motor neuron genes, SMN1 and SMN2, which arise from a recent duplication. Homozygous loss of SMN1 causes SMA, and the expression and accuracy of splicing of SMN2 appears to modulate the severity of the disease. Increasing the expression and splicing of the SMN2 gene and product, respectively, by HDAC inhibition is one of the therapeutic approaches being investigated at present [78]. In this regard, particular emphasis has been placed on HDAC-2 after the observation that this molecule influences the expression of SMN2 [79]. In a detailed study investigating the efficacy of different HDAC inhibitors in ameliorating SMA pathology, SAHA was found to be the best therapeutic agent in in vitro and in ex vivo models of this disease: SAMA selectively increased SMN2 activity and had the lowest toxicity [80]. Indeed, both SAHA and FK-228 were able to bypass the silencing of the SMN2 gene by DNA methylation characteristic of SMA patients, whereas other inhibitors such as SPB and VPA could not [81]. Positive results have also been reported with the benzamide M344 in fibroblast cells derived from patients with SMA [82]. Interestingly the aliphatic acid HDAC inhibitor SPB shows poor efficacy in lymphoblastoid cell lines, despite its ability to inhibit class I HDACs (and in particular HDAC-2) [83]. Moreover, MS-275, a potent HDAC-2 inhibitor, is unable to increase SMN2 expression [79]. Given the discrepancy observed between these systems, more-detailed studies investigating the remaining zinc-dependent HDACs and aiming at understanding the role of HDAC inhibition in SMA are warranted [79].
Spinal and bulbar muscular atrophy (SBMA) is a recessive autosomal disease that leads to muscular paralysis and atrophy. It is characterised by the presence of a poly Q expansion within the androgen receptor. The mutant protein that causes SBMA is thought to inhibit HAT activity, resulting in transcriptional dysfunction and subsequent neuronal degeneration. To date, the only study of HDAC and SBMA reported in the literature is the investigation of SB and its therapeutic effects in a transgenic mouse model of SBMA. In this study, SB treatment ameliorated many of the neurological phenotypes associated with SBMA, but this effect was dose-dependent and seen only over a narrow range. The high toxicity observed at higher doses probably discouraged further studies [84].
Even though HDAC inhibitors have shown promise in cellular and rodent models of neurodegeneration, many issues need to be addressed before we can use these drugs in the clinic.
4. Expert opinion
4.1 HDAC inhibitor specificity
Most of the HDAC inhibitors that are at present being studied or used in clinical trials are global HDAC inhibitors. As such, HDAC inhibition affects 2 – 5% of all genes [85,86]. It is probable that affecting this many genes generates desirable gene expression (that which is associated with survival and repair, or restores gene expression in a transcription-dysfunctional state), but also undesirable gene expression (that which is associated with death and the toxicity). For example, HDAC inhibition is associated with toxicity in cell culture models of oxidative stress [28] and in clinical trials. In a recent issue of Neuron, Kim et al. reported that HDAC-1 inactivation by p25/cdk-5 induced aberrant cellcycle activity and double-stranded DNA breaks, which usually precede neuronal apoptosis [87]. Thus, more studies aiming at understanding the molecular mechanisms underlying diseases and at validating the targets through which HDAC inhibition is functioning are both necessary and critical. Determining which HDAC isoforms are bona fide therapeutic targets in neurodegenerative diseases and designing specific inhibitors for these targets remain the most crucial challenges for investigators and for the success of these drugs in the clinic. Indeed, it may be that while a single HDAC isoform is a therapeutic target at one gene locus, it may not be at another locus; thus, investigators may have to consider strategies that not only target a single isoform, but do so in a gene-specific manner. Many factors have contributed to the difficulty in synthesizing specific HDAC inhibitors. First, until the recent publication of the crystal structure of human HDAC-8 in complex with structurally diverse hydroxamic acid inhibitors, most of the design for synthetic inhibitors was based on the crystal structure of an HDAC-homolog found in the bacterium Aquifex aeolicus [88,89]. The availability of these structures, as well as the identification of an increasing number of non-histone targets specific to HDACs, may help in designing more-effective and more-specific inhibitors. Second, the close homology between the catalytic sites of these enzymes and their presence as part of protein complexes also contributes to the difficulty in designing specific inhibitors. The latter makes purifying HDACs tedious, and in many cases there is loss of enzymatic activity, even with high purifications [90].
4.2 HDAC inhibitor doses and delivery
As with other drugs, dosing is one of the most critical issues in the use of HDAC inhibitors in a clinical setting. The proper balance between obtaining effective results, and at the same time overcoming the toxicity and side effects associated with global inhibitors [91,92] will be essential. For example, even though HDAC inhibitors are protective against oxidative-stress-induced cortical neuronal cell death, they show high levels of toxicity when cortical neurons are treated for prolonged periods; however, pulse treatment with these same HDAC inhibitors alleviates the toxicity while maintaining the protective effect [28]. Studies suggest that the half-life of an HDAC inhibitor might dictate the associated toxicity and undesirable side affects in vivo. In addition to being administered at an appropriate dose, HDAC inhibitors also need to be able to cross the blood–brain barrier (BBB) intact in order to be effective in treating neurodegenerative diseases. There is some evidence that SAHA, SB and VPA all cross the BBB, but, in general, studies of BBB transport have either not been done or have been done poorly [93,94]. In cancer trials, some cardiovascular issues have arisen following the use of these drugs. It remains unclear whether this effect is common to all HDAC inhibitors or whether it is specific to a particular class of inhibitors, but this issue emphasizes the urgency to design more-specific HDAC inhibitors and new approaches to modulating HDAC activity.
Expert opinion
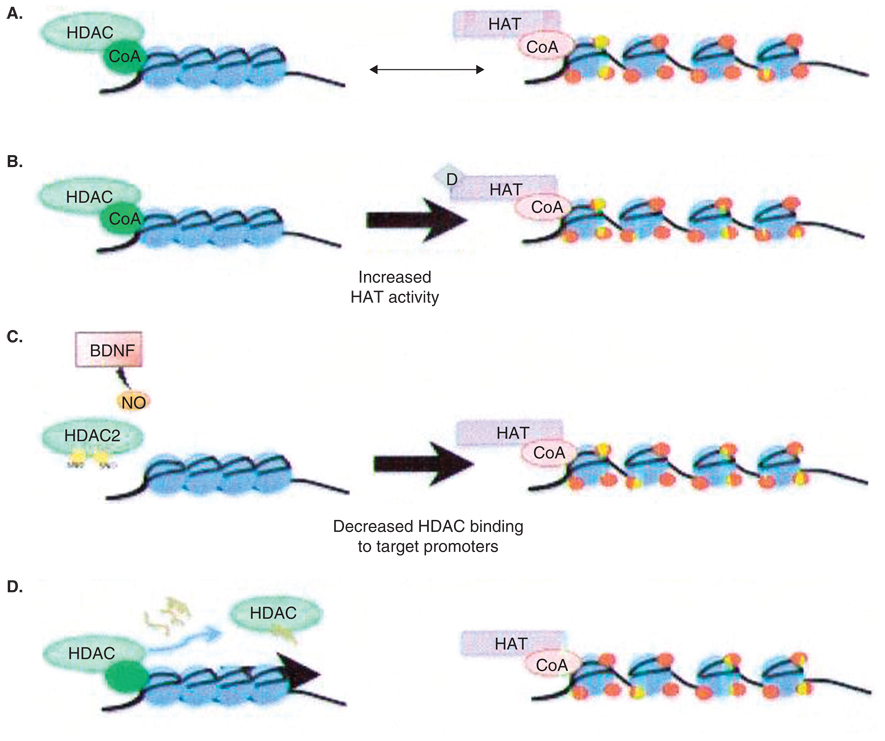
In conclusion, data indicate that loss of HAT function contributes to neurodegeneration. In cellular and rodent models of neurodegenerative disease, HDAC inhibition has been shown to be effective at restoring proper acetylation levels and thus indirectly increasing HAT activity, and thus is a promising therapeutic strategy (Figure 2). Nevertheless, the HDAC inhibitors that are available at present lack specificity with regard to individual HDAC isoforms. As such, more-selective inhibitors are needed in order to avoid the adverse effects of this approach. In addition, the inhibitors available at present function by inhibiting HDAC catalytic activity. It would be useful to design inhibitors or peptides that can interfere with the domains required for corepressor interaction or HDAC promoter recruitment (Figure 2D). This approach requires a good understanding of the crystal structure of the complexes as well as of the nature of the interaction. Such a strategy may cede additional specificity to having isoform specific inhibitors, however, as some interactions may target a specific HDAC to some promoters and not others. This effect could in turn further alleviate some of the off-target effects associated with toxicity. Other strategies to decrease HDAC function include modulating the ability of HDAC to interact with promoters and regulating HDAC subcellular localisation. For example, a recent study showed that BDNF induces NO synthesis and S-nitrosylation of two cysteine residues (Cys262 and Cys274) in HDAC-2 [95]. This nitrosylation does not affect the catalytic activity of HDAC-2, but rather its ability to bind to CREB-dependent promoters (Figure 2C). Another recent study showed that reduction of two cysteine residues (Cys667 and Cys669) that are usually oxidized in response to reactive oxygen species prevents HDAC-4 entry into the nucleus [27] (Figure 2E). Targeting these and similar signalling pathways may be a more-effective way of inhibiting HDAC function than targeting an HDACs catalytic activity. Finally, neuroprotection can be achieved in an autonomous manner through non-neuronal cells, as there is some evidence that HDAC inhibition in astrocytes can protect neurons through the release of BDNF and GDNF [21,42,53] (Figure 2F). Many questions and issues remain as to how HDAC inhibition is neuroprotective and what is the best way to achieve inhibition of this enzyme. Nevertheless, this strategy has been shown to be effective in most models of neurodegeneration. The challenge remains in determining the most appropriate and effective way of administering these drugs in the clinic.
Figure 2. Strategies to inhibit histone deacetylase activity.
A. Recruitment of histone deacetylases (HDACs) to gene promoters followed by histone deacetylation leads to DNA compaction and repression of gene expression, whereas recruitment of histone acetyltransferases (HATs) leads to an open DNA conformation and activation of gene expression. B–E. Therapeutic strategies that involve either increasing HAT activity directly or increasing HAT activity indirectly through HDAC inhibition. B. Targeting HATs and directly increasing their activity. C. Targeting signaling pathways that induce post-translational modifications that lead to reduced target promoter binding. D. Designing peptides that can interfere with the interaction between the HDAC and the relevant corepressors and prevent recruitment of HDAC to target promoters. E. Targeting the redox state of HDACs in order to control its subcellular localization. F. Targeting HDAC activity in glia as a mechanism to protect the neurons.
BDNF: Brain-derived neurotrophic factor; CoA: Coenzyme A; CoR: ; NO: Nitric oxide.
Acknowledgements
We would like to thank A Siddiq for her critical reading of this review and also MA Rivieccio and C Brochier for their suggestions. We acknowledge the support of the Adelson Foundation (CO #190772), the New York State Department of Health, NIA PO1, the Burke Foundation and the Goldsmith Foundation.
Footnotes
Declaration of interest
The authors state no conflict of interest and have received no payment in preparation of this manuscript.
Contributor Information
Sama F Sleiman, Email: ssleiman@burke.org.
Rajiv R Ratan, Email: rrr2001@med.cornell.edu.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Liu L, Scolnick DM, Trievel RC, et al. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19(2):1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90(4):595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 3.Ryu H, Lee J, Zaman K, et al. Sp1 and Sp3 are oxidative stress-inducible, antideath transcription factors in cortical neurons. J Neurosci. 2003;23(9):3597–3606. doi: 10.1523/JNEUROSCI.23-09-03597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez-Balbas MA, Bauer UM, Nielsen SJ, et al. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19(4):662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marzio G, Wagener C, Gutierrez MI, et al. E2F family members are differentially regulated by reversible acetylation. J Biol Chem. 2000;275(15):10887–10892. doi: 10.1074/jbc.275.15.10887. [DOI] [PubMed] [Google Scholar]
- 6.Lu Q, Hutchins AE, Doyle CM, et al. Acetylation of cAMP-responsive element-binding protein (CREB) by CREB-binding protein enhances CREB-dependent transcription. J Biol Chem. 2003;278(18):15727–15734. doi: 10.1074/jbc.M300546200. [DOI] [PubMed] [Google Scholar]
- 7.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396(6711):594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 8.Hung HL, Lau J, Kim AY, et al. CREB-Binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol Cell Biol. 1999;19(5):3496–3505. doi: 10.1128/mcb.19.5.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Li N, Caron C, et al. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 2003;22(5):1168–1179. doi: 10.1093/emboj/cdg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langley B, Gensert JM, Beal MF, Ratan RR. Remodeling chromatin and stress resistance in the central nervous system: histone deacetylase inhibitors as novel and broadly effective neuroprotective agents. Curr Drug Targets CNS Neurol Disord. 2005;4(1):41–50. doi: 10.2174/1568007053005091. [DOI] [PubMed] [Google Scholar]
- 11.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7(10):854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 12.Voelter-Mahlknecht S, Ho AD, Mahlknecht U. Chromosomal organization and localization of the novel class IV human histone deacetylase 11 gene. Int J Mol Med. 2005;16(4):589–598. [PubMed] [Google Scholar]
- 13.Hahnen E, Hauke J, Trankle C, et al. Histone deacetylase inhibitors: possible implications for neurodegenerative disorders. Expert Opin Investig Drugs. 2008;17(2):169–184. doi: 10.1517/13543784.17.2.169. [DOI] [PubMed] [Google Scholar]
- 14.Gottlicher M. Valproic acid: an old drug newly discovered as inhibitor of histone deacetylases. Ann Hematol. 2004;83(Suppl 1):S91–S92. doi: 10.1007/s00277-004-0850-2. [DOI] [PubMed] [Google Scholar]
- 15.Plumb JA, Finn PW, Williams RJ, et al. Pharmacodynamic response and inhibition of growth of human tumor xenografts by the novel histone deacetylase inhibitor PXD101. Mol Cancer Ther. 2003;2(8):721–728. [PubMed] [Google Scholar]
- 16.McLaughlin F, La Thangue NB. Histone deacetylase inhibitors open new doors in cancer therapy. Biochem Pharmacol. 2004;68(6):1139–1144. doi: 10.1016/j.bcp.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 17.Carew JS, Giles FJ, Nawrocki ST. Histone deacetylase inhibitors: mechanisms of cell death and promise in combination cancer therapy. Cancer Lett. 2008;269(1):7–17. doi: 10.1016/j.canlet.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 18.Moradei O, Vaisburg A, Martell RE. Histone deacetylase inhibitors in cancer therapy: new compounds and clinical update of benzamide-type agents. Curr Top Med Chem. 2008;8(10):841–858. doi: 10.2174/156802608784911581. [DOI] [PubMed] [Google Scholar]
- 19.Shankar S, Srivastava RK. Histone deacetylase inhibitors: mechanisms and clinical significance in cancer: HDAC inhibitor-induced apoptosis. Adv Exp Med Biol. 2008;615:261–298. doi: 10.1007/978-1-4020-6554-5_13. [DOI] [PubMed] [Google Scholar]
- 20.Drottar M, Liberman MC, Ratan RR, Roberson DW. The histone deacetylase inhibitor sodium butyrate protects against cisplatin-induced hearing loss in guinea pigs. Laryngoscope. 2006;116(2):292–296. doi: 10.1097/01.mlg.0000197630.85208.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gensert JM, Baranova OV, Weinstein DE, Ratan RR. CD81, a cell cycle regulator, is a novel target for histone deacetylase inhibition in glioma cells. Neurobiol Dis. 2007;26(3):671–680. doi: 10.1016/j.nbd.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Saha RN, Pahan K. HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death Differ. 2006;13(4):539–550. doi: 10.1038/sj.cdd.4401769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rouaux C, Loeffler JP, Boutillier AL. Targeting CREB-binding protein (CBP) loss of function as a therapeutic strategy in neurological disorders. Biochem Pharmacol. 2004;68(6):1157–1164. doi: 10.1016/j.bcp.2004.05.035. [DOI] [PubMed] [Google Scholar]
- 24.Steffan JS, Bodai L, Pallos J, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413(6857):739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 25.La Spada AR, Morrison RS. The power of the dark side: Huntington’s disease protein and p53 form a deadly alliance. Neuron. 2005;47(1):1–3. doi: 10.1016/j.neuron.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 26.Ryu H, Lee J, Olofsson BA, et al. Histone deacetylase inhibitors prevent oxidative neuronal death independent of expanded polyglutamine repeats via an Sp1-dependent pathway. Proc Natl Acad Sci USA. 2003;100(7):4281–4286. doi: 10.1073/pnas.0737363100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ago T, Liu T, Zhai P, et al. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 2008;133(6):978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 28.Langley B, D’annibale MA, Suh K, et al. Pulse inhibition of histone deacetylases induces complete resistance to oxidative death in cortical neurons without toxicity and reveals a role for cytoplasmic p21(waf1/cip1) in cell cycle-independent neuroprotection. J Neurosci. 2008;28(1):163–176. doi: 10.1523/JNEUROSCI.3200-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vecsey CG, Hawk JD, Lattal KM, et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27(23):6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanai H, Sawa A, Chen RW, et al. Valproic acid inhibits histone deacetylase activity and suppresses excitotoxicity-induced GAPDH nuclear accumulation and apoptotic death in neurons. Pharmacogenomics J. 2004;4(5):336–344. doi: 10.1038/sj.tpj.6500269. [DOI] [PubMed] [Google Scholar]
- 31.Leng Y, Liang MH, Ren M, et al. Synergistic neuroprotective effects of lithium and valproic acid or other histone deacetylase inhibitors in neurons: roles of glycogen synthase kinase-3 inhibition. J Neurosci. 2008;28(10):2576–2588. doi: 10.1523/JNEUROSCI.5467-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Politis PK, Akrivou S, Hurel C, et al. BM88/Cend1 is involved in histone deacetylase inhibition-mediated growth arrest and differentiation of neuroblastoma cells. FEBS Lett. 2008;582(5):741–748. doi: 10.1016/j.febslet.2008.01.052. [DOI] [PubMed] [Google Scholar]
- 33.Hara MR, Thomas B, Cascio MB, et al. Neuroprotection by pharmacologic blockade of the GAPDH death cascade. Proc Natl Acad Sci USA. 2006;103(10):3887–3889. doi: 10.1073/pnas.0511321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin LJ. DNA damage and repair: relevance to mechanisms of neurodegeneration. J Neuropathol Exp Neurol. 2008;67(5):377–387. doi: 10.1097/NEN.0b013e31816ff780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meisel A, Harms C, Yildirim F, et al. Inhibition of histone deacetylation protects wild-type but not gelsolin-deficient neurons from oxygen/glucose deprivation. J Neurochem. 2006;98(4):1019–1031. doi: 10.1111/j.1471-4159.2006.04016.x. [DOI] [PubMed] [Google Scholar]
- 36.Furukawa K, Fu W, Li Y, et al. The actin-severing protein gelsolin modulates calcium channel and NMDA receptor activities and vulnerability to excitotoxicity in hippocampal neurons. J Neurosci. 1997;17(21):8178–8186. doi: 10.1523/JNEUROSCI.17-21-08178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Endres M, Fink K, Zhu J, et al. Neuroprotective effects of gelsolin during murine stroke. J Clin Invest. 1999;103(3):347–354. doi: 10.1172/JCI4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harms C, Bosel J, Lautenschlager M, et al. Neuronal gelsolin prevents apoptosis by enhancing actin depolymerization. Mol Cell Neurosci. 2004;25(1):69–82. doi: 10.1016/j.mcn.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 39.Giannakakou P, Nakano M, Nicolaou KC, et al. Enhanced microtubule-dependent trafficking and p53 nuclear accumulation by suppression of microtubule dynamics. Proc Natl Acad Sci USA. 2002;99(16):10855–10860. doi: 10.1073/pnas.132275599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mabjeesh NJ, Escuin D, LaVallee TM, et al. 2ME2 inhibits tumor growth and angiogenesis by disrupting microtubules and dysregulating HIF. Cancer Cell. 2003;3(4):363–375. doi: 10.1016/s1535-6108(03)00077-1. [DOI] [PubMed] [Google Scholar]
- 41.Dompierre JP, Godin JD, Charrin BC, et al. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington’s disease by increasing tubulin acetylation. J Neurosci. 2007;27(13):3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu X, Chen PS, Dallas S, et al. Histone deacetylase inhibitors up-regulate astrocyte GDNF and BDNF gene transcription and protect dopaminergic neurons. Int J Neuropsychopharmacol. 2008;11(8):1123–1134. doi: 10.1017/S1461145708009024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lyssiotis CA, Walker J, Wu C, et al. Inhibition of histone deacetylase activity induces developmental plasticity in oligodendrocyte precursor cells. Proc Natl Acad Sci USA. 2007;104(38):14982–14987. doi: 10.1073/pnas.0707044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunah AW, Jeong H, Griffin A, et al. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington’s disease. Science. 2002;296(5576):2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 45.Ferrante RJ, Kubilus JK, Lee J, et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington’s disease mice. J Neurosci. 2003;23(28):9418–9427. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steffan JS, Kazantsev A, Spasic-Boskovic O, et al. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci USA. 2000;97(12):6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pallos J, Bodai L, Lukacsovich T, et al. Inhibition of specific HDACs and sirtuins suppresses pathogenesis in a Drosophila model of Huntington’s disease. Hum Mol Genet. 2008;17(23):3767–3775. doi: 10.1093/hmg/ddn273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hockly E, Richon VM, Woodman B, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington’s disease. Proc Natl Acad Sci USA. 2003;100(4):2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bates EA, Victor M, Jones AK, et al. Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J Neurosci. 2006;26(10):2830–2838. doi: 10.1523/JNEUROSCI.3344-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas EA, Coppola G, Desplats PA, et al. The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington’s disease transgenic mice. Proc Natl Acad Sci USA. 2008;105(40):15564–15569. doi: 10.1073/pnas.0804249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kontopoulos E, Parvin JD, Feany MB. Alpha-synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum Mol Genet. 2006;15(20):3012–3023. doi: 10.1093/hmg/ddl243. [DOI] [PubMed] [Google Scholar]
- 52.Gardian G, Yang L, Cleren C, et al. Neuroprotective effects of phenylbutyrate against MPTP neurotoxicity. Neuromolecular Med. 2004;5(3):235–241. doi: 10.1385/NMM:5:3:235. [DOI] [PubMed] [Google Scholar]
- 53.Chen PS, Peng GS, Li G, et al. Valproate protects dopaminergic neurons in midbrain neuron/glia cultures by stimulating the release of neurotrophic factors from astrocytes. Mol Psychiatry. 2006;11(12):1116–1125. doi: 10.1038/sj.mp.4001893. [DOI] [PubMed] [Google Scholar]
- 54.Chen PS, Wang CC, Bortner CD, et al. Valproic acid and other histone deacetylase inhibitors induce microglial apoptosis and attenuate lipopolysaccharide-induced dopaminergic neurotoxicity. Neuroscience. 2007;149(1):203–212. doi: 10.1016/j.neuroscience.2007.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sugai F, Yamamoto Y, Miyaguchi K, et al. Benefit of valproic acid in suppressing disease progression of ALS model mice. Eur J Neurosci. 2004;20(11):3179–3183. doi: 10.1111/j.1460-9568.2004.03765.x. [DOI] [PubMed] [Google Scholar]
- 56.Ryu H, Smith K, Camelo SI, et al. Sodium phenylbutyrate prolongs survival and regulates expression of anti-apoptotic genes in transgenic amyotrophic lateral sclerosis mice. J Neurochem. 2005;93(5):1087–1098. doi: 10.1111/j.1471-4159.2005.03077.x. [DOI] [PubMed] [Google Scholar]
- 57.Rouaux C, Panteleeva I, Rene F, et al. Sodium valproate exerts neuroprotective effects in vivo through CREB-binding protein-dependent mechanisms but does not improve survival in an amyotrophic lateral sclerosis mouse model. J Neurosci. 2007;27(21):5535–5545. doi: 10.1523/JNEUROSCI.1139-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petri S, Kiaei M, Kipiani K, et al. Additive neuroprotective effects of a histone deacetylase inhibitor and a catalytic antioxidant in a transgenic mouse model of amyotrophic lateral sclerosis. Neurobiol Dis. 2006;22(1):40–49. doi: 10.1016/j.nbd.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 59.Ren M, Leng Y, Jeong M, et al. Valproic acid reduces brain damage induced by transient focal cerebral ischemia in rats: potential roles of histone deacetylase inhibition and heat shock protein induction. J Neurochem. 2004;89(6):1358–1367. doi: 10.1111/j.1471-4159.2004.02406.x. [DOI] [PubMed] [Google Scholar]
- 60.Faraco G, Pancani T, Formentini L, et al. Pharmacological inhibition of histone deacetylases by suberoylanilide hydroxamic acid specifically alters gene expression and reduces ischemic injury in the mouse brain. Mol Pharmacol. 2006;70(6):1876–1884. doi: 10.1124/mol.106.027912. [DOI] [PubMed] [Google Scholar]
- 61.Granger A, Abdullah I, Huebner F, et al. Histone deacetylase inhibition reduces myocardial ischemia-reperfusion injury in mice. FASEB J. 2008;22(10):3549–3560. doi: 10.1096/fj.08-108548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petrij F, Giles RH, Dauwerse HG, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376(6538):348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 63.Hallam TM, Bourtchouladze R. Rubinstein-Taybi syndrome: molecular findings and therapeutic approaches to improve cognitive dysfunction. Cell Mol Life Sci. 2006;63(15):1725–1735. doi: 10.1007/s00018-005-5555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alarcon JM, Malleret G, Touzani K, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42(6):947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 65.Herman D, Jenssen K, Burnett R, et al. Histone deacetylase inhibitors reverse gene silencing in Friedreich’s ataxia. Nat Chem Biol. 2006;2(10):551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 66.Greene E, Mahishi L, Entezam A, et al. Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res. 2007;35(10):3383–3390. doi: 10.1093/nar/gkm271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rai M, Soragni E, Jenssen K, et al. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLoS ONE. 2008;3(4):e1958. doi: 10.1371/journal.pone.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coffee B, Zhang F, Warren ST, Reines D. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat Genet. 1999;22(1):98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- 69.Chiurazzi P, Pomponi MG, Pietrobono R, et al. Synergistic effect of histone hyperacetylation and DNA demethylation in the reactivation of the FMR1 gene. Hum Mol Genet. 1999;8(12):2317–2323. doi: 10.1093/hmg/8.12.2317. [DOI] [PubMed] [Google Scholar]
- 70.Biacsi R, Kumari D, Usdin K. SIRT1 inhibition alleviates gene silencing in Fragile X mental retardation syndrome. PLoS Genet. 2008;4(3):e1000017. doi: 10.1371/journal.pgen.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsankova NM, Berton O, Renthal W, et al. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9(4):519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 72.Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci USA. 2006;103(9):3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sharma RP, Grayson DR, Gavin DP. Histone deactylase 1 expression is increased in the prefrontal cortex of schizophrenia subjects: analysis of the National Brain Databank microarray collection. Schizophr Res. 2008;98(1–3):111–117. doi: 10.1016/j.schres.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Romieu P, Host L, Gobaille S, et al. Histone deacetylase inhibitors decrease cocaine but not sucrose self-administration in rats. J Neurosci. 2008;28(38):9342–9348. doi: 10.1523/JNEUROSCI.0379-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pandey SC, Ugale R, Zhang H, et al. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008;28(14):3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Giorgini F, Moller T, Kwan W, et al. Histone deacetylase inhibition modulates kynurenine pathway activation in yeast, microglia, and mice expressing a mutant huntingtin fragment. J Biol Chem. 2008;283(12):7390–7400. doi: 10.1074/jbc.M708192200. [DOI] [PubMed] [Google Scholar]
- 77.Gavin DP, Kartan S, Chase K, et al. Reduced baseline acetylated histone 3 levels, and a blunted response to HDAC inhibition in lymphocyte cultures from schizophrenia subjects. Schizophr Res. 2008;103(1–3):330–332. doi: 10.1016/j.schres.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Echaniz-Laguna A, Bousiges O, Loeffler JP, Boutillier AL. Histone deacetylase inhibitors: therapeutic agents and research tools for deciphering motor neuron diseases. Curr Med Chem. 2008;15(13):1263–1273. doi: 10.2174/092986708784534974. [DOI] [PubMed] [Google Scholar]
- 79.Kernochan LE, Russo ML, Woodling NS, et al. The role of histone acetylation in SMN gene expression. Hum Mol Genet. 2005;14(9):1171–1182. doi: 10.1093/hmg/ddi130. [DOI] [PubMed] [Google Scholar]
- 80.Hahnen E, Eyupoglu IY, Brichta L, et al. In vitro and ex vivo evaluation of second-generation histone deacetylase inhibitors for the treatment of spinal muscular atrophy. J Neurochem. 2006;98(1):193–202. doi: 10.1111/j.1471-4159.2006.03868.x. [DOI] [PubMed] [Google Scholar]
- 81.Hauke J, Riessland M, Lunke S, et al. Survival motor neuron gene 2 silencing by DNA methylation correlates with spinal muscular atrophy disease severity and can be bypassed by histone deacetylase inhibition. Hum Mol Genet. 2009;18(2):304–317. doi: 10.1093/hmg/ddn357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Riessland M, Brichta L, Hahnen E, Wirth B. The benzamide M344, a novel histone deacetylase inhibitor, significantly increases SMN2 RNA/protein levels in spinal muscular atrophy cells. Hum Genet. 2006;120(1):101–110. doi: 10.1007/s00439-006-0186-1. [DOI] [PubMed] [Google Scholar]
- 83.Dayangac-Erden D, Topaloglu H, Erdem-Yurter H. A preliminary report on spinal muscular atrophy lymphoblastoid cell lines: Are they an appropriate tool for drug screening? Adv Ther. 2008;25(3):274–279. doi: 10.1007/s12325-008-0030-1. [DOI] [PubMed] [Google Scholar]
- 84.Minamiyama M, Katsuno M, Adachi H, et al. Sodium butyrate ameliorates phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2004;13(11):1183–1192. doi: 10.1093/hmg/ddh131. [DOI] [PubMed] [Google Scholar]
- 85.Van Lint C, Emiliani S, Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 1996;5(4–5):245–253. [PMC free article] [PubMed] [Google Scholar]
- 86.Glaser KB, Li J, Staver MJ, et al. Role of class I and class II histone deacetylases in carcinoma cells using siRNA. Biochem Biophys Res Commun. 2003;310(2):529–536. doi: 10.1016/j.bbrc.2003.09.043. [DOI] [PubMed] [Google Scholar]
- 87.Kim D, Frank CL, Dobbin MM, et al. Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron. 2008;60(5):803–817. doi: 10.1016/j.neuron.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Somoza JR, Skene RJ, Katz BA, et al. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure. 2004;12(7):1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 89.Vannini A, Volpari C, Filocamo G, et al. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc Natl Acad Sci USA. 2004;101(42):15064–15069. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Curtin M, Glaser K. Histone deacetylase inhibitors: the Abbott experience. Curr Med Chem. 2003;10(22):2373–2392. doi: 10.2174/0929867033456576. [DOI] [PubMed] [Google Scholar]
- 91.Boutillier AL, Trinh E, Loeffler JP. Selective E2F-dependent gene transcription is controlled by histone deacetylase activity during neuronal apoptosis. J Neurochem. 2003;84(4):814–828. doi: 10.1046/j.1471-4159.2003.01581.x. [DOI] [PubMed] [Google Scholar]
- 92.Salminen A, Tapiola T, Korhonen P, Suuronen T. Neuronal apoptosis induced by histone deacetylase inhibitors. Brain Res Mol Brain Res. 1998;61(1–2):203–206. doi: 10.1016/s0169-328x(98)00210-1. [DOI] [PubMed] [Google Scholar]
- 93.Yin D, Ong JM, Hu J, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor: effects on gene expression and growth of glioma cells in vitro and in vivo. Clin Cancer Res. 2007;13(3):1045–1052. doi: 10.1158/1078-0432.CCR-06-1261. [DOI] [PubMed] [Google Scholar]
- 94.Lucke A, Mayer T, Altrup U, et al. Simultaneous and continuous measurement of free concentration of valproate in blood and extracellular space of rat cerebral cortex. Epilepsia. 1994;35(5):922–926. doi: 10.1111/j.1528-1157.1994.tb02535.x. [DOI] [PubMed] [Google Scholar]
- 95.Nott A, Watson PM, Robinson JD, et al. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455(7211):411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]