Abstract
Nonenzymatic glycation of peptides and proteins by d-glucose has important implications in the pathogenesis of diabetes mellitus, particularly in the development of diabetic complications. In this work, we report the first proteomics-based characterization of nonenzymatically glycated proteins in human plasma and erythrocyte membranes from individuals with normal glucose tolerance, impaired glucose tolerance, and type 2 diabetes mellitus. Phenylboronate affinity chromatography was used to enrich glycated proteins and glycated tryptic peptides from both human plasma and erythrocyte membranes. The enriched peptides were subsequently analyzed by liquid chromatography coupled with electron transfer dissociation-tandem mass spectrometry, resulting in the confident identification of 76 and 31 proteins from human plasma and erythrocyte membranes, respectively. Although most of the glycated proteins could be identified in samples from individuals with normal glucose tolerance, slightly higher numbers of glycated proteins and more glycation sites were identified in samples from individuals with impaired glucose tolerance and type 2 diabetes mellitus.
Keywords: protein nonenzymatic glycation, Amadori compound, boronate affinity enrichment, electron transfer dissociation tandem mass spectrometry, type 2 diabetes mellitus, human plasma, erythrocyte membrane, comparative proteomics
Introduction
Diabetes mellitus affects approximately 5–10% of the world's population.1,2 Among those affected, noninsulin dependent diabetes mellitus, which is more commonly referred to as type 2 diabetes mellitus (T2DM), represents 90% of all reported cases. In the United States alone, over 12 million individuals have been diagnosed with T2DM, while another estimated 5 million are unaware of their disease.3 There are two key physiological defects in the pathogenesis of T2DM: (1) resistance to the biological action of insulin on peripheral tissues, including skeletal muscle, adipose, and liver tissues, and (2) impaired insulin secretion from pancreatic β-cells, resulting in insulin deficiency.1 Both of these defects lead to hyperglycemia in the untreated state, which has been proposed to contribute to the development of diabetic complications through nonenzymatic chemical modification of tissue proteins by glucose.
Glucose chemically reacts with primary amine groups in proteins to form a stable ketoamine called the Amadori compound.4 This compound is the first meta-stable product of the Maillard reaction and can undergo further oxidation and rearrangement reactions to form a series of more reactive, colored, and/or fluorescent compounds generally referred to as advanced glycation end-products (AGEs). AGEs are believed to play a more pathogenic role in the development of diabetic complications by altering the structure, turnover, and function of tissue proteins.5 For example, tissue AGE levels have been shown to increase in lens crystalline of diabetic patients6 and have been known to correlate with the severity of diabetic neuropathy, nephropathy, and retinopathy.7 AGEs are also thought to play a role in age-related pathological conditions such as atherosclerosis via their accumulation in tissue collagens, which results in loss of vascular elasticity and thickening of arterial walls.8,9 These products have also been proposed to play a role in abnormal amyloid aggregation in age-related neurodegenerative disorders, for example, Alzheimer's10,11 and Parkinson's diseases.12,13 As such, there is great interest in better understanding the role of glycation and AGEs in the development of diabetic complications and other disorders, as well as in discovering more sensitive biomarkers for better monitoring of glycemic control.
Proteomics-based biomarker discovery efforts have recently gained attention due to the power of these approaches for analyzing the complex protein mixtures of clinical samples. Currently, human blood plasma is the most commonly used clinical sample in such analyses, potentially including specific biomarkers for virtually every human disease as a result of either direct or indirect interaction with all the cellular components of the body, for example, tissue- or tumor-specific proteins may be released into the blood stream upon cell damage or cell death.14,15 With an average life span of 21 days,14 the abundant classical plasma proteins are suitable indicators for short to midterm glycemic control. In this respect, glycated albumin, which reflects the status of blood glucose more rapidly and more efficiently than glycated hemoglobin (HbA1c),16 has been suggested as a biomarker for monitoring midterm glycemic control. Conversely, HbA1c, the major protein found in erythrocytes, has been used as a clinical diagnostic marker of relatively long-term (∼90 days) glucose control in diabetic patients since the 1970s.17 With a life span of 120 days, erythrocytes can accumulate relatively high amounts of the Amadori compound, particularly on the cell membrane proteins, which are in close contact with glucose throughout their life cycle. As a result, it is possible that some glycated erythrocyte membrane proteins may provide a more sensitive biomarker of glycemic control.
In this work, we report the first characterization of the glycated proteome in human plasma and erythrocyte membranes from individuals with normal glucose tolerance (NGT), impaired glucose tolerance (IGT), and T2DM. We utilized an approach that addressed two challenges associated with global analysis of glycated proteins: (1) unambiguous identification of the glycation site18 and (2) the under-sampling issue typical in mass spectrometry (MS)-based global proteomics. The glycated proteins and peptides were first specifically enriched using phenylboronate affinity chromatography, after which the enriched, glycated tryptic peptides were separated using reversed-phase liquid chromatography (LC) and analyzed using data-dependent tandem MS with electron transfer dissociation (ETD) fragmentation. ETD fragment the labile Amadori compound can be kept intact during the fragmentation process, thus, facilitating peptide sequencing.19,20
Experimental Procedures
Sample Collection and Pretreatment for Human Plasma and Erythrocytes
All specimens were collected at the School of Medicine of Emory University with standard protocols and made available through the NIDDK Central Repository. Briefly, patient volunteers were recruited with the following exclusion criteria: (1) known to have diabetes; (2) known to have illness that caused them to miss work during the past week (or, if unemployed, would have caused them to miss work had they been employed); (3) known to have had acute infections or illness during the past week (in the opinion of the subject); (4) known to be pregnant; and (5) known to be taking steroid hormones. The volunteers were randomly chosen, without sex, age, race, or life style matches. The oral glucose tolerance test (OGTT) was administered to each volunteer prior to blood draw, and fasting and 2-h plasma glucose levels were used to categorize the patients as having either NGT, IGT, or T2DM according to World Health Organization criteria.21 The details of each patient group are summarized in Table 1. For each individual consenting donor, blood was collected into citrated tubes (catalog no. 363083 or equivalent CPT tubes; Becton Dickinson; Franklin Lakes, NJ) by venipuncture. Immediately after collection, each tube was inverted four times, then either placed in wet ice or refrigerated at 4 °C. The specimens were subsequently centrifuged in a swinging bucket centrifuge at 1500g for 15 min at 4 °C. The resultant plasma from each tube was pooled for each individual into a secondary conical Falcon tube. The buffy coats in the original tubes were removed and stored separately; a 10% volume of dimethylsulfoxide (cryoprotectant) was then added to the remaining erythrocyte pellets, which were transferred to 1.8 mL cryovials for storage at −70 °C. The secondary Falcon tube containing the plasma was recentrifuged at 1500g for 15 min at 4 °C to remove any potentially remaining cellular material, and the top 90% of plasma was then transferred into new tubes and stored at −70 °C. This sample processing was completed within 75 min from time of specimen collection. To reduce biological variation and provide sufficient material for boronate affinity enrichment, samples from each volunteer group (i.e., NGT, IGT, and T2DM) were pooled.
Table 1.
Brief Summary of Clinical Data for the Three Different Volunteer Groupsa
| group | number | fast glucose | 2 h glucose | cholesterols | HDL | LDL | HbA1c | ALB/CR |
|---|---|---|---|---|---|---|---|---|
| NGT | 12 | 87.8 (88.5) | 80.3 (79.5) | 173.2 (170.5) | 59.8 (57.0) | 101.3 (95.5) | 5.0 (5.1) | 8.8 (6.0) |
| IGT | 11 | 100.2 (98.0) | 167.6 (170.0) | 219.4 (208.0) | 51.1 (49.0) | 145.6 (127.0) | 5.3 (5.3) | 11.1 (8.0) |
| T2DM | 8 | 127.8 (126.0) | 230.1 (221.0) | 180.6 (178.5) | 34.0 (33.5) | 116.1 (112.0) | 6.2 (6.1) | 10.5 (9.0) |
For each group, only the average and median values (number in parenthesis) are given. All data shown were from the 2nd visit. HDL, high density lipoprotein; LDL, low density lipoprotein; ALB/CR, ratio of albumin to creatinine in urine. Glucose concentration is reported as mg/dL and HbA1c levels as %.
Chemicals and Materials
All chemicals and glucose assay kits were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise stated. Micro-BCA assay kits were purchased from Pierce (Rockford, IL), and Amicon Ultra-15 5KDa cutoff concentrators were purchased from Millipore (Billerica, MA). Sequencing-grade modified trypsin was purchased from Promega (Madison, WI). An empty Tricorn high performance LC column (5–100 mm) was purchased from GE Healthcare, while Glycogel II boronate affinity gel (Pierce, Rockford, IL) was a gift from Dr. Bart Haigh of the Institute for Bioanalytics (Branford, CT).
Immunodepletion of Plasma To Remove High-Abundance Plasma Proteins
Twelve high-abundance plasma proteins (albumin, IgG, α1-antitrypsin, IgA, IgM, transferrin, haptoglobin, α1-acid glycoprotein, α2-macroglobulin, apolipoprotein A-I, apolipoprotein A-II, and fibrinogen) that constitute ∼96% of the total protein mass of human plasma were simultaneously separated from other proteins using a ProteomeLab IgY-12 LC10 (125 μL plasma loading capacity) affinity LC column (Beckman Coulter, Fullerton, CA) with an Agilent 1100 series HPLC system (Agilent, Palo Alto, CA). All IgY-12 separations were performed based on the manufacturer's instructions regarding column usage and loading capacity. The following three buffers were used in a separation scheme that consisted of sample loading-washing-stripping-neutralization followed by re-equilibration for a total cycle time of 48 min: (1) 10 mM Tris-HCl, 150 mM NaCl, pH 7.4 (TBS) for dilution/washing; (2) 100 mM glycine, pH 2.5, for stripping; and (3) 100 mM Tris-HCl, pH 8.0, for neutralizing.
To generate sufficient material for subsequent boronate affinity enrichment of glycated proteins/peptides, plasma aliquots from 12 NGT, 11 IGT, and 8 T2DM donors were pooled to create approximately 5 mL of material per group, which was passed through the depletion column. The corresponding flow-through fractions were collected, pooled, and individually concentrated in Amicon Ultra-15 concentrators that have an Ultracel regenerated cellulose membrane with 5 kDa molecular weight cutoff (Millipore), followed by a buffer exchange to 50 mM NH4HCO3 in the same unit according to the manufacturer's instructions. Protein concentration was then measured using the BCA protein assay.
Preparation of Erythrocyte Membrane Proteins
Aliquots of erythrocytes from individual donors were pooled to create uniform samples corresponding to NGT, IGT, and T2DM. Approximately 2 mL of erythrocyte suspension was diluted in 6 vol of cold 5 mM sodium phosphate buffer, pH 8, and incubated at room temperature for 15 min. The cell lysate was then centrifuged at 200g for 6 min to remove any remaining whole cells. The pellet was washed once with sodium phosphate buffer and centrifuged at 200g for 6 min, and the two supernatants were combined and centrifuged at 14 000g for 8 min to separate membranes from supernatant (soluble proteins). Membranes were washed five times with sodium phosphate buffer until the supernatant became colorless and the membranes retained little color. The membranes were then reconstituted in 50 mM NH4HCO3, and protein concentration was measured by BCA assay prior to trypsin digestion.
Protein Digestion
Plasma proteins or erythrocyte membranes (10 mg/mL) were dissolved in 8 M urea/100 mM NH4HCO3 (pH 8.2) and reduced with 5 mM dithiothreitol (DTT) for1hat37 °C; free sulfhydryl groups were then alkylated with 20 mM iodoacetamide at room temperature for1hinthe dark. Samples were subsequently diluted with 50 mM NH4HCO3 (pH 8.0) to reduce the urea concentration to below 1 M, and CaCl2 was added to a final concentration of 1.5 mM prior to the addition of sequencing-grade modified trypsin at a ratio of 1:40 (w/w, enzyme/protein). Samples were digested at 37 °C for 12 h. The final digestion mixture was passed through C18 SPE cartridges for desalting, and eluted peptide solutions were concentrated by a Speed-Vac (Thermo Savant, Milford, MA) before being processed further.
Boronate Affinity Chromatography
The empty chromatography column was slurry-packed with boronate affinity gel under gravity flow. Prior to affinity fractionation, free glucose that remained in human plasma samples was removed with Amicon concentrators to avoid competition of boronate binding sites; the completeness of glucose removal was determined by micro glucose assay (Sigma-Aldrich). Proteins or postdigestion peptide samples (up to 0.75 mg) were then dissolved in 500 μL of buffer A (50 mM MgCl2, 250 mM NH4OAc, pH 8.1) and injected on column using an Agilent 1100 series HPLC system equipped with a fraction collector. The flow rate was set at 1.0 mL/min, and the following gradient was used to fractionate, regenerate, and equilibrate the column: 0–10 min, 100% A; 10.1–20 min, 100% B (0.1 M HOAc); 20.1–30 min, 100% A. The LC effluent was monitored at 280 nm with a UV detector. The percentage of glycated proteins or peptides was calculated as the peak area that corresponded to bound proteins or peptides divided by the sum of the peak areas corresponding to bound and unbound proteins or peptides. Eluted glycated proteins were concentrated and desalted with Amicon concentrators prior to tryptic digestion, while eluted glycated peptides were concentrated by Speed-Vac and subsequently desalted with C18 SPE cartridges.
LC–MS/MS Analyses of Peptides
An Agilent 6340 Ion Trap LC-MS system equipped with ETD capability was coupled with an Agilent HPLC-Chip Cube MS interface, well plate sampler, nanoflow pump, and capillary pump for LC–MS/MS analyses. Fluoranthene anions from a small negative chemical ionization source were used to transfer electrons in the ETD fragmentation process. Four ETD MS/MS scans were performed after each MS survey scan. If the precursor ions had been fragmented twice, then a dynamic exclusion window of 1 min was used to discriminate against previously analyzed ions. During the ETD process, the reactant accumulation time and reaction time were 40 and 80 ms, respectively. The HPLC separation was carried out on the Chip with an integrated separation column (Zorbax 300SB-C18, 75 μm × 150 mm, 5 μm), and an enrichment column (Zorbax 300SB-C18, 160 nL, 5 μm). The voltage applied to the HPLC-Chip capillary was 1725 V and the LC solvents were (A) 0.1% formic acid in H2O and (B) 0.1% formic acid in 90% CH3CN/10% H2O. Each sample was analyzed in duplicate, and 2.0 μg of sample was injected for each analysis. A gradient separation of 3–43% B over 80 min, 43–90% B over 5 min, and 90% B for 5 min was used at a flow rate of 300 nL/min. MS/MS data were acquired over a mass range of 300–2200 m/z.
Data Analysis
Spectrum Mill MS Proteomics Workbench Rev A.03.03.072 (Agilent Technologies) was used to identify peptides by searching LC–MS/MS data obtained in ETD fragmentation mode against the human International Protein Index (IPI) database (Version 3.20 consisting of 61 225 protein entries; available online at www.ebi.ac.uk/IPI). The minimum matched peak intensity was set to 40%, with a precursor ion mass tolerance of ±2.5 Da and a product ion mass tolerance of ±0.7 Da; a sequence tag length >1 was also specified. A parameter file incorporating the following mass modifications was used in the search for glycated peptides: static modification for cysteine residues (carbamidomethylation, 57.02 Da), dynamic modification for methionine residues (oxidation, 16.00 Da), and dynamic modification for lysine residues (AC, 162.05 Da). Additionally, the following charge-dependent criteria that were modified from Molina et al.22 were used to filter raw search results: 5+, score >14; 4+, score >12; 3+, score >9; 2+, score >8; a percentage scored peak intensity (%SPI) >60 and Delta Rank1 − Rank2 >1 were used for all charge states. Further, each data set was searched against the same IPI database with all protein sequences reversed in order to estimate the peptide false discovery rate. With these filtering criteria, the false discovery rate was <1%. For semiquantitation of glycated peptides, only those glycated peptides that were identified at least two times more frequently in IGT and T2DM states than in NGT, or glycated peptides that were identified at least two times in either IGT or T2DM groups only were retained for semiquantitative data comparison.
Results
BAC Enrichment of Glycated Peptides
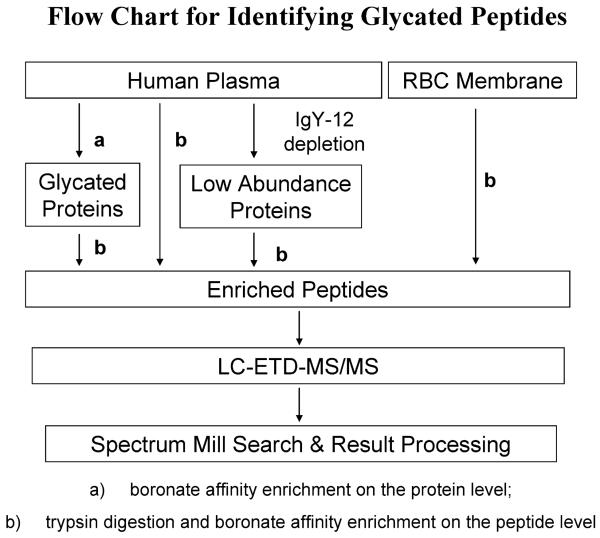
Our prior investigation of in vitro glycated human serum demonstrated that boronate affinity chromatography with Glycogel II boronate affinity gel is a powerful technique for specific enrichment of glycated proteins in the presence of enzymatically glycosylated proteins. The technique is based on the fact that the boronate anion has significantly higher affinity for the coplanar cis-diols that exist in the Amadori compound than the equatorial cisdiols present on mannose glycans.20,23 Furthermore, electro-static repulsion prevents the negatively charged sialic acids present on the outside of glycan chains in glycosylated proteins from getting close to boronate anions. We also demonstrated that, compared to dual enrichment on both protein and peptide levels, affinity enrichment on the peptide level is more applicable when a limited amount of sample is available.20 Further, glycated species were observed to be present in only bound fractions, which reduced the number of samples requiring LC–MS/MS analysis.20 Therefore, because of the limited amount of sample (∼500 μg of total protein obtained) in the current study, erythrocyte membranes and the low-abundance portion of plasma proteins (∼4% of whole plasma protein weight) were affinity-enriched at only the peptide level (Figure 1). Alternatively, whole plasma proteins were processed using dual enrichment, first at the protein level and then at the peptide level. In a parallel experiment, whole plasma was also enriched at only the peptide level.
Figure 1.
Flowchart depicting glycated peptide identification from blood plasma and erythrocyte membranes of NGT, IGT, and T2DM individuals. Pooled samples from each patient group were processed and analyzed individually as outlined in the chart.
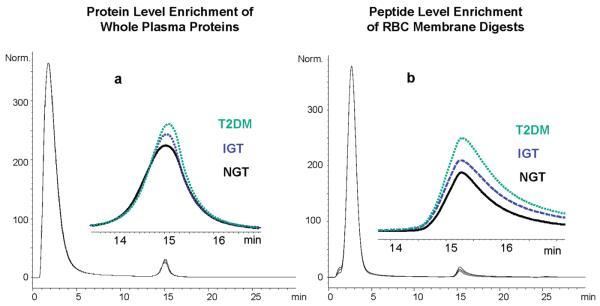
Chromatographic differences among the three different sample types (NGT, IGT, and T2DM) are shown in Figure 2. For whole plasma samples (Figure 2a), the bound portion accounted for only ∼4.5% of the total sample amount. While the difference in peak areas of bound glycated proteins is very small among the three different volunteer groups, the overall abundance of glycated species is positively correlated with fasting and 2-h OGTT glucose concentrations (T2DM > IGT > NGT). This trend is more apparent for the glycated proteins from whole plasma than for the glycated proteins from the low-abundance protein fraction of plasma (data not shown). A possible explanation for this observation is that low-abundance proteins, which have higher turn-over rates compared with the classical plasma proteins and account for less than 4% of the plasma protein weight,15 do not accumulate significant amounts of the Amadori compound before in vivo proteolytic digestion. Similarly, the observed quantitative difference (based on HPLCUV) in levels of enriched glycated plasma peptides was not as readily distinguished among the three sample groups as the enrichment on the protein level (data not shown). In contrast, the observed difference in the levels of glycated peptides is more profound for the erythrocyte membrane samples (Figure 2b) and can be attributed to the much longer life span of this cell type (∼120 days) that results in more effective accumulation of the Amadori compound.
Figure 2.
(a) Boronate affinity chromatograms of whole plasma from different volunteer groups. The inset is a zoomed view of binding fractions, which are labeled with the respective sample name. Peaks at approximately 2 min are nonglycated proteins, and glycated proteins elute near 15 min. (b) Boronate affinity chromatograms showing the enrichment of glycated peptides from tryptic digests of erythrocyte membranes. The glycated peptides elute slightly after 15 min, and the nonglycated peptides are washed out near 2 min. The chromatograms in both (a) and (b) were normalized based on the most intense peak in each trace, that is, the peak corresponding to the flow-through fraction.
Overview of Glycated Peptides and Proteins Identified by LC-ETD-MS/MS
Glycated peptides obtained from boronate affinity enrichment were analyzed by LC-ETD-MS/MS. In our previous studies, we utilized an instrument capable of alternating ETD and CID scans, which provided complementary information regarding the Amadori modification; ETD provided high quality glycated peptide sequence information, while CID provided additional confirmation of the presence of the Amadori compound based on a characteristic series of neutral losses.19,20 However, we chose to use only ETD fragmentation in the current study in order to increase the duty cycle of the MS/MS analyses.
Samples were analyzed in duplicate and included (1) glycated peptides from erythrocyte membranes, (2) glycated peptides from whole plasma enriched first at the protein and then at the peptide level, (3) glycated peptides from whole plasma enriched only at the peptide level, and (4) glycated peptides from low-abundance plasma proteins enriched only at the peptide level. We were able to confidently identify 3272 spectra from 24 LC–MS/MS data sets (using the rigorous filtering criteria described in Experimental Procedures and providing an estimated false discovery rate of <1%); 1610 (49.2%) of these spectra contained Amadori-modified lysines. More specifically, we confidently identified 75 unique glycated peptides (31 unique glycated proteins) from erythrocyte membranes and 260 unique glycated peptides (76 unique glycated proteins) from plasma of which 114 unique glycated peptides (39 unique glycated proteins) were from nonimmunodepleted human plasma and 156 unique glycated peptides (46 unique glycated proteins) were from the low-abundance protein fraction of human plasma.
The list of identified glycated proteins includes proteins already known to be glycated, as well as many new proteins that were identified as glycated for the first time. Note the glycated proteins, glycated peptides, and glycation sites identified from whole plasma, immunodepleted plasma, and erythrocyte membranes are listed in Tables S1–S3 (Supporting Information). On the basis of our results, the most abundant plasma protein human serum albumin (HSA), which has 59 lysines, is heavily glycated in vivo as evidenced by the 31 glycation sites identified from 38 unique glycated peptides. K525 was the most frequent glycation site based on spectra count and represents ∼20% of the glycated peptides, similar to previous reports.24,25 Although many of the HSA glycation sites have been previously observed in studies of HSA glycated in vitro,20,24,25 the fact that many of these glycation sites occur in vivo is encouraging. In addition to HSA, other high-abundance plasma proteins identified as glycated include serotransferrin, alpha-1-antitrypsin, alpha-2-macroglobulin, apolipoprotein A-I and A-II, fibrinogen, and alpha-1-acid glycoprotein. From the immunodepleted plasma fraction, we were also able to identify several moderately abundant glycated proteins, such as ceruloplasmin, hemopexin, complement C3, C4A and C5 precursors, heparin cofactor 2 precursor, kininogen-1 precursor, vitamin D-binding protein precursor, apolipoprotein B-100, afamin, and lumican precursors. Some of these proteins have been previously identified as Amadori-modified in plasma from type 2 diabetic patients.26
From erythrocyte membranes, we identified ankyrin, flotillin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), spectrin, erythrocyte membrane protein bands 4.1 and 4.2, as well as the hemoglobin alpha and beta subunits, which were not completely removed even after rigorous washing of the erythrocyte membranes during sample preparation. Ankyrin, spectrin, and erythrocyte membrane protein band 4.2 are major structural proteins. These proteins were reported to be heavily glycated in erythrocyte membranes of diabetic patients, and observed membrane abnormalities in these individuals were largely attributed to oxidation originating from glycation.27 GADPH plays an important role in regulating multiple pathways that relate to the development of diabetic complications, and its modification by glycation was reported to result in lower enzymatic activity.28 Flotillin-1 and Flotillin-2 were recently identified as the major integral proteins of erythrocyte lipid rafts,29 and the former was shown to be dysregulated in diabetes,30 possibly due to glycation.
Semiquantitative Comparison of Glycated Proteins among Different Subject Groups
A majority of the identified glycated proteins appear in all three subject groups, with little variation in terms of the numbers of glycated peptides or glycation sites. However, based on MS/MS spectrum counting, a number of glycated peptides were observed more frequently in both IGT and T2DM groups compared to the NGT group. Additionally, some glycated peptides were only observed in either IGT or T2DM groups. Only those glycated peptides that were identified at least two times more frequently in IGT and T2DM states than in NGT, or glycated peptides that were identified at least two times in either IGT or T2DM groups only, were retained for semiquantitative data comparison. In total, there were 14 and 50 unique glycated peptides considered as up-regulated in IGT and T2DM groups relative to the NGT group in erythrocyte membrane (Table 2) and plasma samples (Table 3), respectively. Of special note are previously unreported glycated peptides that were identified solely in the T2DM group, such as (K)VVAGVANALAHkYH(−) from either hemoglobin subunit delta or beta; (R)LEALkENGGAR(L) from apolipoprotein A-I precursor; and (K)SVIPSDGPSVACVkK(A) from serotransferrin precursor. These glycated peptides may serve as potential targets for further evaluation in the diagnosis of type 2 diabetes mellitus.
Table 2.
Summary of Unique Glycated Peptides Considered as Up-Regulated in IGT and T2DM Groups from Erythrocyte Membrane Samplesa
| gene | accession no. | protein description | peptide sequence | glycation site |
R. T. (min) |
S. C. NGT |
S. C. IGT |
S. C. T2DM |
|---|---|---|---|---|---|---|---|---|
| ANK1 | IPI00216697.2 | ankyrin 1 isoform 1 | (R)AGHTEVAkYLLQNK(A) | K454 | 26.7 | 2 | 5 | 4 |
| ANK1 | IPI00216697.2 | ankyrin 1 isoform 1 | (R)VAkVLLDK(G) | K355 | 16.9 | 0 | 2 | 2 |
| EPB41 | IPI00003921.2 | isoform 1 of protein 4.1 | (K)HAkGQDLLK(R) | K231 | 8.8 | 0 | 0 | 2 |
| GAPDH | IPI00219018.6 | glyceraldehyde-3-phosphate dehydrogenase | (K)AGAHLQGGAkR(V) | K116 | 2.9 | 1 | 2 | 2 |
| GAPDH | IPI00219018.6 | glyceraldehyde-3-phosphate dehydrogenase | (K)AVGkVIPELNGK(L) | K218 | 26.4 | 1 | 2 | 2 |
| GAPDH | IPI00219018.6 | glyceraldehyde-3-phosphate dehydrogenase | (K)FHGTVkAENGK(L) | K60 | 5.3 | 2 | 6 | 5 |
| GAPDH | IPI00219018.6 | glyceraldehyde-3-phosphate dehydrogenase | (K)VkVGVNGFGR(I) | K4 | 23.3 | 0 | 1 | 1 |
| HBD | IPI00473011.2 | hemoglobin subunit delta | (K)VVAGVANALAHkYH(−) | K144 | 27.5 | 0 | 0 | 4 |
| MPP1 | IPI00215610.2 | 55 kDa erythrocyte membrane protein | (R)SHIkNALLSQNPEK(F) | K299 | 20.1 | 0 | 2 | 1 |
| SPTA1 | IPI00220741.4 | spectrin alpha chain, erythrocyte | (K)ALkAQLIDER(T) | K1486 | 23.5 | 1 | 2 | 3 |
| SPTB | IPI00216704.6 | isoform 2 of spectrin beta chain, erythrocyte | (K)EkVQLIEDR(H) | K1247 | 19.9 | 1 | 2 | 2 |
| SPTB | IPI00216704.6 | isoform 2 of spectrin beta chain, erythrocyte | (K)NNEkAQEASVLLR(D) | K1261 | 22.9 | 1 | 2 | 2 |
| SPTB | IPI00216704.6 | isoform 2 of spectrin beta chain, erythrocyte | (R)LQGQVDKHYAGLkDVAEER(K) | K1675 | 24.3 | 0 | 1 | 2 |
| SPTB | IPI00216704.6 | isoform 2 of spectrin beta chain, erythrocyte | (R)TQLVDTADkFR(F) | K1904 | 25.4 | 1 | 2 | 1 |
Modified amino acids are shown in lower case; k, Amadori modified lysine; m, oxidized methionine. R. T. stands for LC retention time; S. C. stands for scan counts in MS/MS analyses.
Table 3.
Summary of Unique Glycated Peptides Considered as Up-Regulated in IGT and T2DM Groups from Blood Plasma Samplesa
| gene | accession no. | protein description | peptide sequence | glycation sites | R. T. (min) |
S. C. NGT |
S. C. IGT |
S. C. T2DM |
|---|---|---|---|---|---|---|---|---|
| A2M | IPI00478003.1 | alpha-2-macroglobulin precursor | (R)LVDGkGVPIPNK(V) | K375 | 19.2 | 0 | 2 | 0 |
| ALB | IPI00022434.2 | ALB protein | (K)AACLLPkLDELRDEGK(A) | K181 | 33.8 | 1 | 3 | 1 |
| ALB | IPI00022434.2 | ALB protein | (K)AEFAEVSkLVTDLTK(V) | K233 | 47.6 | 19 | 31 | 22 |
| ALB | IPI00022434.2 | ALB protein | (K)DVCkNYAEAK(D) | K317 | 8.4 | 0 | 3 | 2 |
| ALB | IPI00022434.2 | ALB protein | (K)LDELRDEGkASSAK(Q) | K190 | 9.2 | 9 | 7 | 15 |
| ALB | IPI00022434.2 | ALB protein | (K)TCVADESAENCDkSLHTLFGDK(L) | K64 | 31.0 | 0 | 3 | 1 |
| ALB | IPI00022434.2 | ALB protein | (R)DAHkSEVAHR(F) | K4 | 2.3 | 1 | 2 | 2 |
| ALB | IPI00022434.2 | ALB protein | (R)LkCASLQK(F) | K199 | 7.5 | 1 | 0 | 3 |
| ALB | IPI00022434.2 | ALB protein | (R)NLGkVGSkCCK(H) | K432; K436 | 5.0 | 0 | 2 | 2 |
| ALB | IPI00384697.1 | ALB protein | (K)HKPkATKEQLK(A) | K562 | 3.1 | 1 | 3 | 3 |
| APOA1 | IPI00021841.1 | apolipoprotein A-I precursor | (R)LEALkENGGAR(L) | K206 | 8.8 | 0 | 0 | 3 |
| APOA2 | IPI00021854.1 | apolipoprotein A-II precursor | (K)SYFEKSkEQLTPLIK(K) | K67 | 28.7 | 0 | 2 | 0 |
| APOB | IPI00022229.1 | apolipoprotein B-100 precursor | (K)LTIFkTELR(V) | K4040 | 32.8 | 0 | 2 | 1 |
| APOB | IPI00022229.1 | apolipoprotein B-100 precursor | (K)QAEAVLkTLQELK(K) | K327 | 39.5 | 0 | 1 | 1 |
| APOB | IPI00022229.1 | apolipoprotein B-100 precursor | (K)TkNSEEFAAAmSR(Y) | K117 | 12.3 | 0 | 1 | 1 |
| APOB | IPI00022229.1 | apolipoprotein B-100 precursor | (R)FLkNIILPVYDK(S) | K3673 | 44.8 | 0 | 1 | 3 |
| C3 | IPI00164623.4 | 187 kDa protein | (K)LTQSkIWDVVEK(A) | K615 | 33.1 | 0 | 1 | 1 |
| C3 | IPI00164623.4 | 187 kDa protein | (K)VTIkPAPETEK(R) | K1370 | 13.8 | 1 | 2 | 2 |
| C3 | IPI00164623.4 | 187 kDa protein | (R)IFTVNHkLLPVGR(T) | K155 | 35.6 | 0 | 4 | 4 |
| C3 | IPI00164623.4 | 187 kDa protein | (R)YISKYELDkAFSDR(N) | K1438 | 30.6 | 0 | 4 | 1 |
| C4A | IPI00032258.4 | complement C4-A precursor | (R)ADLEkLTSLSDR(Y) | K1503 | 26.6 | 0 | 1 | 1 |
| C4A | IPI00032258.4 | complement C4-A precursor | (R)GLESQTkLVNGQSHISLSK(A) | K304 | 24.9 | 0 | 2 | 0 |
| C4A | IPI00032258.4 | complement C4-A precursor | (R)YIYGkPVQGVAYVR(F) | K274 | 26.4 | 0 | 1 | 2 |
| CFB | IPI00019591.1 | isoform 1 of complement factor B precursor (fragment) | (K)QPWQAkISVIRPSK(G) | K498 | 31.7 | 0 | 0 | 2 |
| CFH | IPI00029739.4 | isoform 1 of complement factor H precursor | (K)DQYkVGEVLK(F) | K588 | 22.3 | 0 | 2 | 0 |
| CP | IPI00017601.1 | ceruloplasmin precursor | (R)kAEEEHLGILGPQLHADVGDK(V) | K799 | 33.1 | 1 | 0 | 2 |
| CP | IPI00017601.1 | ceruloplasmin precursor | (R)RPYLkVFNPR(R) | K906 | 19.6 | 0 | 2 | 2 |
| CP | IPI00017601.1 | ceruloplasmin precursor | (R)VTFHNkGAYPLSIEPIGVR(F) | K468 | 34.9 | 0 | 2 | 0 |
| FGA | IPI00021885.1 | isoform 1 of fibrinogen alpha chain precursor | (K)mkPVPDLVPGNFK(S) | K227 | 28.5 | 0 | 2 | 3 |
| FGA | IPI00021885.1 | isoform 1 of fibrinogen alpha chain precursor | (K)VIEkVQHIQLLQK(N) | K148 | 27.6 | 1 | 3 | 7 |
| FGA | IPI00021885.1 | isoform 1 of fibrinogen alpha chain precursor | (R)KVIEkVQHIQLLQK(N) | K148 | 21.1 | 2 | 5 | 3 |
| FGB | IPI00298497.3 | fibrinogen beta chain precursor | (R)kGGETSEmYLIQPDSSVKPYR(V) | K247 | 26.5 | 0 | 2 | 0 |
| FRAS1 | IPI00329327.3 | isoform 2 of extracellular matrix protein fras1 precursor | (R)LVIEFKTHAk(F) | K3547 | 23.6 | 0 | 1 | 2 |
| GC | IPI00555812.4 | vitamin D-binding protein precursor | (K)VmDkYTFELSR(R) | K345 | 26.3 | 0 | 2 | 0 |
| GC | IPI00555812.4 | vitamin D-binding protein precursor | (R)kFPSGTFEQVSQLVK(E) | K51 | 44.3 | 3 | 4 | 6 |
| HP | IPI00431645.1 | HP protein | (R)HYEGSTVPEkK(T) | K143 | 6.0 | 1 | 2 | 4 |
| HP | IPI00478493.3 | 38 kDa protein | (R)YQCkNYYK(L) | K53 | 7.1 | 0 | 0 | 2 |
| IGHM | IPI00168728.1 | FLJ00385 protein (fragment) | (K)ALPAPIEkTISK(T) | K327 | 27.8 | 0 | 1 | 3 |
| IGL@ | IPI00154742.4 | IGLC1 protein | (K)AGVETTTPSkQSNNK(Y) | K190 | 5.8 | 1 | 3 | 6 |
| IGL@ | IPI00448800.4 | 25 kDa protein | (K)ADGSPVkAGVETTKPSK(Q) | K183 | 10.4 | 2 | 3 | 5 |
| ITIH1 | IPI00292530.1 | inter-alpha-trypsin inhibitor heavy chain H1 precursor | (K)LDAQASFLPkELAAQTIK(K) | K214 | 45.7 | 0 | 1 | 2 |
| KNG1 | IPI00032328.1 | isoform HMW of kininogen-1 precursor | (R)ITEATkTVGSDTFYSFK(Y) | K64 | 34.0 | 0 | 2 | 2 |
| LUM | IPI00020986.2 | lumican precursor | (K)ILGPLSYSKIk(H) | K307 | 28.4 | 1 | 1 | 2 |
| LUM | IPI00020986.2 | lumican precursor | (R)LKEDAVSAAFkGLK(S) | K181 | 31.8 | 1 | 1 | 3 |
| ORM2 | IPI00020091.1 | alpha-1-acid glycoprotein 2 precursor | (K)DKCEPLEkQHEK(E) | K188 | 5.1 | 1 | 2 | 4 |
| SERPINA1 | IPI00553177.1 | alpha-1-antitrypsin precursor | (K)GTQGkIVDLVK(E) | K192 | 16.4 | 0 | 1 | 1 |
| SERPINA1 | IPI00553177.1 | alpha-1-antitrypsin precursor | (K)QINDYVEkGTQGK(I) | K187 | 12.5 | 0 | 1 | 3 |
| TF | IPI00022463.1 | serotransferrin precursor | (K)CLkDGAGDVAFVK(H) | K215 | 22.3 | 0 | 1 | 2 |
| TF | IPI00022463.1 | serotransferrin precursor | (K)GDVAFVkHQTVPQNTGGK(N) | K553 | 14.8 | 2 | 1 | 5 |
| TF | IPI00022463.1 | serotransferrin precursor | (K)SVIPSDGPSVACVkK(A) | K61 | 23.3 | 0 | 0 | 2 |
Modified amino acids are shown in lower case; k, Amadori modified lysine; m, oxidized methionine. R. T. stands for LC retention time; S. C. stands for scan counts in MS/MS analyses.
Discussion
Human plasma represents the largest and most challenging human proteome available for study, due to the extremely high dynamic range (>109) of its composite proteins.14,15 While albumin, the most abundant plasma protein, accounts for ∼60% of the plasma protein weight, this number increases to 96%whenthe12mostabundantplasmaproteinsareconsidered.14,15 Thus, the discovery of very low abundance glycated proteins represents the proverbial “needle in a haystack”. Since immunodepletion to remove high-abundance plasma proteins has been reported to greatly improve the dynamic range of proteomic measurements,15,31 we utilized an IgY-12 immunodepletion column to deplete the 12 most abundant plasma proteins and enhance our ability to identify low level glycated proteins. As a result, we were able to identify many proteins present in the moderate- to low-abundance range in the plasma proteome. In contrast, we were only able to identify the proteins that are known to be highly abundant in whole plasma. However, even with immunodepletion, we were only able to identify 76 plasma proteins as glycated. Utilizing a multidimensional separation, such as strong cation exchange chromatography coupled with reversed-phase chromatography, could further improve the peptide measurement dynamic range. For example, Washburn et al. utilized multidimensional protein identification technology (MudPIT) for large scale proteome analysis of Saccharomyces cerevisiae,32 and, more recently, Liu et al. reported more than 300 confidently identified plasma proteins after coupling 2D-LC-MS/MS with immunodepletion.31
The measurement of HbA1c reflects the level of nonenzymatic glycation inside the erythrocyte; however, it can also reflect differences in blood glucose levels over time, as well as the individual effects of biological factors that influence cellular glucose transport and the nonenzymatic protein glycation/ deglycation cycle.33 Glycation of the major plasma proteins and erythrocyte membrane proteins more often reflects glycation in the extracellular compartment, which is more directly associated with mean blood glucose over a given time period. While HbA1c is generally correlated linearly to mean blood glucose,33 this correlation can vary widely in populations; some have attributed this variability to individual susceptibility to hemoglobin glycation (the high and low glycator hypothesis).34-37 In addition, while the volunteers were classified into three different groups based on OGTT results, their levels of HbA1c were not as dramatically different as the 2 h glucose measurements (Table 1). This indicates that the volunteers have significantly different plasma glucose responses immediately following a glucose challenge (and likely after meals, as well), but have more modest differences in long-term glucose control as measured by levels of HbA1c. Furthermore, the variability of HbA1c among individuals might well be greater than that caused by hyperglycemia, particularly when comparing individuals with NGT to those with abnormal glucose tolerance (prediabetes and previously unrecognized diabetes, as in this study) who have been diagnosed very early in their disease.34,35,37,38 Thus, the observation that protein glycation (based on boronate affinity chromatography) in the T2DM group was not significantly higher than in the NGT group is not unexpected. Further, low-abundance plasma proteins that have higher turn-over rates compared with the classical plasma proteins may not accumulate significant amounts of the Amadori compound before in vivo proteolytic digestion. These points may also explain the observation that a majority of the glycated proteins were identified in all three subject groups, with few major quantitative variations. However, the background of glycated proteins is extensive, that is, almost every major protein identified contained at least one glycation site.
In summary, the utility of ETD-MS/MS for characterizing in vivo glycated peptides has been further demonstrated. ETD provided high quality spectral information for glycated peptides while allowing the labile Amadori modification to remain intact. As a result, several peptides were identified as doubly glycated in the current study (listed in Supporting Information Tables S1–S3). Because of the challenges associated with peptide-centric proteomics, a more accurate quantitative characterization of the potential dysregulation of the identified glycated proteins will benefit from future quantitative studies based on the accurate mass and time tag approach39,40 to discover potential biomarkers.
Conclusions
Using a bottom-up proteomics approach and phenylboronate affinity chromatography, we were able to identify 76 and 31 nonenzymatically glycated proteins from plasma and erythrocyte membranes, respectively. To the best of our knowledge, this is the most comprehensive report of the glycated human plasma and erythrocyte membrane proteomes to date. The associated 260 and 75 unique glycated plasma and erythrocyte membrane peptides provide a foundation for subsequent investigation of glycation and the development of diabetic complications. Although the semiquantitative data presented here indicates increased levels of some glycated peptides, and therefore proteins, in the IGT and T2DM groups versus the NGT group, caution should be taken as this data was produced from a limited number of pooled samples. Further studies with a large population of individuals with NGT, IGT, and T2DM are required in order to further validate the significance of these glycated peptides and proteins and their potential use as biomarkers of IGT and T2DM.
Supplementary Material
Acknowledgment
The authors thank Dr. Bart Haigh of the Institute for Bioanalytics for kindly providing the Glycogel II boronate affinity gel, and Drs. Matthew E. Monroe and Konstantinos Petritis of Pacific Northwest National Laboratory (PNNL) for their help with the Spectrum Mill software. The authors also thank Professors John W. Baynes of the University of South Carolina and Robert M. Cohen of the University of Cincinnati for helpful discussions. Portions of this research were supported by NIH NIDDK (DK071283 and DK066204), NCRR (RR00039 and RR018522), and the Robert W. Woodruff Health Sciences Center Fund. Portions of this research were performed at the Environmental Molecular Science Laboratory, a national scientific user facility located at PNNL and sponsored by the U.S. Department of Energy (DOE) Office of Biological and Environmental Research. PNNL is operated by Battelle for the DOE under Contract No. DE-AC06-76RLO-1830.
Footnotes
Supporting Information Available
Tables summarizing the unique glycated peptides and proteins identified from whole human blood plasma, the unique glycated peptides and proteins identified from immuno-depleted human blood plasma with IgY-12 column, and the unique glycated peptides and proteins identified from human erythrocyte membrane. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kahn B. Type 2 diabetes: when insulin secretion fails to compensate for insulin resistance. Cell. 1998;92(5):593–596. doi: 10.1016/s0092-8674(00)81125-3. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein B. Insulin resistance: from benign to type 2 diabetes mellitus. Rev. Cardiovasc. Med. 2003;4(Suppl 6):S3–S10. [PubMed] [Google Scholar]
- 3.Skyler J, Oddo C. Diabetes trends in the USA. Diabetes/Metab. Res. Rev. 2002;18(Suppl 3):S21–S26. doi: 10.1002/dmrr.289. [DOI] [PubMed] [Google Scholar]
- 4.Baynes JW, Watkins NG, Fisher CI, Hull CJ, Patrick JS, Ahmed MU, Dunn JA, Thorpe SR. The Amadori product on protein: structure and reactions. Prog. Clin. Biol. Res. 1989;304:43–67. [PubMed] [Google Scholar]
- 5.Baynes JW. The role of AGEs in aging: causation or correlation. Exp. Gerontol. 2001;36(9):1527–1537. doi: 10.1016/s0531-5565(01)00138-3. [DOI] [PubMed] [Google Scholar]
- 6.Franke S, Dawczynski J, Strobel J, Niwa T, Stahl P, Stein G. Increased levels of advanced glycation end products in human cataractous lenses. J. Cataract Refract. Surg. 2003;29(5):998–1004. doi: 10.1016/s0886-3350(02)01841-2. [DOI] [PubMed] [Google Scholar]
- 7.Ahmed N. Advanced glycation endproducts--role in pathology of diabetic complications. Diabetes Res. Clin. Pract. 2005;67(1):3–21. doi: 10.1016/j.diabres.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Pennathur S, Heinecke JW. Mechanisms for oxidative stress in diabetic cardiovascular disease. Antioxid. Redox Signaling. 2007;9(7):955–969. doi: 10.1089/ars.2007.1595. [DOI] [PubMed] [Google Scholar]
- 9.Brownlee M. Advanced protein glycosylation in diabetes and aging. Annu. Rev. Med. 1995;46:223–234. doi: 10.1146/annurev.med.46.1.223. [DOI] [PubMed] [Google Scholar]
- 10.Sato T, Shimogaito N, Wu X, Kikuchi S, Yamagishi S, Takeuchi M. Toxic advanced glycation end products (TAGE) theory in Alzheimer's disease. Am. J. Alzheimer's Dis. Other Demen. 2006;21(3):197–208. doi: 10.1177/1533317506289277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeuchi M, Kikuchi S, Sasaki N, Suzuki T, Watai T, Iwaki M, Bucala R, Yamagishi S. Involvement of advanced glycation end-products (AGEs) in Alzheimer's disease. Curr. Alzheimer Res. 2004;1(1):39–46. doi: 10.2174/1567205043480582. [DOI] [PubMed] [Google Scholar]
- 12.Munch G, Luth HJ, Wong A, Arendt T, Hirsch E, Ravid R, Riederer P. Crosslinking of alpha-synuclein by advanced glycation endproducts--an early pathophysiological step in Lewy body formation. J, Chem, Neuroanat. 2000;20(3–4):253–257. doi: 10.1016/s0891-0618(00)00096-x. [DOI] [PubMed] [Google Scholar]
- 13.Sasaki N, Fukatsu R, Tsuzuki K, Hayashi Y, Yoshida T, Fujii N, Koike T, Wakayama I, Yanagihara R, Garruto R, Amano N, Makita Z. Advanced glycation end products in Alzheimer's disease and other neurodegenerative diseases. Am. J. Pathol. 1998;153(4):1149–1155. doi: 10.1016/S0002-9440(10)65659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 15.Qian WJ, Jacobs JM, Liu T, Camp DG, II, Smith RD. Advances and challenges in liquid chromatography-mass spectrometry-based proteomics profiling for clinical applications. Mol. Cell. Proteomics. 2006;5(10):1727–1744. doi: 10.1074/mcp.M600162-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi S, Uchino H, Shimizu T, Kanazawa A, Tamura Y, Sakai K, Watada H, Hirose T, Kawamori R, Tanaka Y. Comparison of glycated albumin (GA) and glycated hemoglobin (HbA1c) in type 2 diabetic patients: usefulness of GA for evaluation of short-term changes in glycemic control. Endocr. J. 2007;54(1):139–144. doi: 10.1507/endocrj.k06-103. [DOI] [PubMed] [Google Scholar]
- 17.Goldstein DE, Wiedmeyer HM, England JD, Little RR, Parker KM. Recent advances in glycosylated hemoglobin measurements. Crit. Rev. Clin. Lab Sci. 1984;21(3):187–228. doi: 10.3109/10408368409165782. [DOI] [PubMed] [Google Scholar]
- 18.Lapolla A, Fedele D, Reitano R, Arico NC, Seraglia R, Traldi P, Marotta E, Tonani R. Enzymatic digestion and mass spectrometry in the study of advanced glycation end products/ peptides. J. Am. Soc. Mass Spectrom. 2004;15(4):496–509. doi: 10.1016/j.jasms.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, Frolov A, Tang N, Hoffmann R, van de Goor T, Metz TO, Smith RD. Application of electron transfer dissociation mass spectrometry in analyses of non-enzymatically glycated peptides. Rapid Commun. Mass Spectrom. 2007;21(5):661–666. doi: 10.1002/rcm.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Tang N, Brock JW, Mottaz HM, Ames JM, Baynes JW, Smith RD, Metz TO. Enrichment and analysis of nonenzymatically glycated peptides: boronate affinity chromatography coupled with electron-transfer dissociation mass spectrometry. J. Proteome Res. 2007;6(6):2323–2330. doi: 10.1021/pr070112q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabetic Med. 1998;15(7):539–553. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 22.Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 2007;104(7):2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barnes HH, Ishimaru CA. Purification of catechol siderophores by boronate affinity chromatography: identification of chrysobactin from Erwinia carotovora subsp. carotovora. Biometals. 1999;12(1):83–87. doi: 10.1023/a:1009223615607. [DOI] [PubMed] [Google Scholar]
- 24.Iberg N, Fluckiger R. Nonenzymatic glycosylation of albumin in vivo. Identification of multiple glycosylated sites. J. Biol. Chem. 1986;261(29):13542–13545. [PubMed] [Google Scholar]
- 25.Gadgil HS, Bondarenko PV, Treuheit MJ, Ren D. Screening and sequencing of glycated proteins by neutral loss scan LC/MS/ MS method. Anal. Chem. 2007;79(15):5991–5999. doi: 10.1021/ac070619k. [DOI] [PubMed] [Google Scholar]
- 26.Jaleel A, Halvatsiotis P, Williamson B, Juhasz P, Martin S, Nair KS. Identification of Amadori-modified plasma proteins in type 2 diabetes and the effect of short-term intensive insulin treatment. Diabetes Care. 2005;28(3):645–652. doi: 10.2337/diacare.28.3.645. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz RS, Madsen JW, Rybicki AC, Nagel RL. Oxidation of spectrin and deformability defects in diabetic erythrocytes. Diabetes. 1991;40(6):701–708. doi: 10.2337/diab.40.6.701. [DOI] [PubMed] [Google Scholar]
- 28.Lee HJ, Howell SK, Sanford RJ, Beisswenger PJ. Methylglyoxal can modify GAPDH activity and structure. Ann. N.Y. Acad. Sci. 2005;1043:135–145. doi: 10.1196/annals.1333.017. [DOI] [PubMed] [Google Scholar]
- 29.Salzer U, Prohaska R. Stomatin, flotillin-1, and flotillin-2 are major integral proteins of erythrocyte lipid rafts. Blood. 2001;97(4):1141–1143. doi: 10.1182/blood.v97.4.1141. [DOI] [PubMed] [Google Scholar]
- 30.Jiang M, Jia L, Jiang W, Hu X, Zhou H, Gao X, Lu Z, Zhang Z. Protein disregulation in red blood cell membranes of type 2 diabetic patients. Biochem. Biophys. Res. Commun. 2003;309(1):196–200. doi: 10.1016/s0006-291x(03)01559-6. [DOI] [PubMed] [Google Scholar]
- 31.Liu T, Qian WJ, Mottaz HM, Gritsenko MA, Norbeck AD, Moore RJ, Purvine SO, Camp DG, II, Smith RD. Evaluation of multiprotein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol. Cell. Proteomics. 2006;5(11):2167–2174. doi: 10.1074/mcp.T600039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Washburn MP, Wolters D, Yates JR., III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19(3):242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 33.Hempe JM, Gomez R, McCarter RJ, Jr., Chalew SA. High and low hemoglobin glycation phenotypes in type 1 diabetes: a challenge for interpretation of glycemic control. J. Diabetes Complications. 2002;16(5):313–320. doi: 10.1016/s1056-8727(01)00227-6. [DOI] [PubMed] [Google Scholar]
- 34.Cohen RM. A1C: does one size fit all. Diabetes Care. 2007;30(10):2756–2758. doi: 10.2337/dc07-1301. [DOI] [PubMed] [Google Scholar]
- 35.Cohen RM, Snieder H, Lindsell CJ, Beyan H, Hawa MI, Blinko S, Edwards R, Spector TD, Leslie RD. Evidence for independent heritability of the glycation gap (glycosylation gap) fraction of HbA1c in nondiabetic twins. Diabetes Care. 2006;29(8):1739–1743. doi: 10.2337/dc06-0286. [DOI] [PubMed] [Google Scholar]
- 36.Herman WH, Ma Y, Uwaifo G, Haffner S, Kahn SE, Horton ES, Lachin JM, Montez MG, Brenneman T, Barrett-Connor E. Differences in A1C by race and ethnicity among patients with impaired glucose tolerance in the Diabetes Prevention Program. Diabetes Care. 2007;30(10):2453–2457. doi: 10.2337/dc06-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yudkin JS, Forrest RD, Jackson CA, Ryle AJ, Davie S, Gould BJ. Unexplained variability of glycated haemoglobin in non-diabetic subjects not related to glycaemia. Diabetologia. 1990;33(4):208–215. doi: 10.1007/BF00404798. [DOI] [PubMed] [Google Scholar]
- 38.Lachin JM, Genuth S, Nathan DM, Rutledge BN. The hemoglobin glycation index is not an independent predictor of the risk of microvascular complications in the Diabetes Control and Complications Trial. Diabetes. 2007;56(7):1913–1921. doi: 10.2337/db07-0028. [DOI] [PubMed] [Google Scholar]
- 39.Zimmer JS, Monroe ME, Qian WJ, Smith RD. Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom. Rev. 2006;25(3):450–482. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qian WJ, Monroe ME, Liu T, Jacobs JM, Anderson GA, Shen Y, Moore RJ, Anderson DJ, Zhang R, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Camp DG, II, Smith RD. Quantitative proteome analysis of human plasma following in vivo lipopolysaccharide administration using 16O/18O labeling and the accurate mass and time tag approach. Mol. Cell. Proteomics. 2005;4(5):700–709. doi: 10.1074/mcp.M500045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.