Abstract
Non-enzymatic glycation of peptides and proteins by D-glucose has important implications in the pathogenesis of diabetes mellitus, particularly in the context of development of diabetic complications. The fragmentation behavior of glycated peptides produced from reaction of D-glucose with lysine residues was investigated by electron transfer dissociation (ETD) and collision induced dissociation (CID) tandem mass spectrometry. It was found that high abundance ions corresponding to various degrees of neutral water losses, as well as furylium ion production, dominate the CID spectra, and that the sequence informative b and y ions were rarely observed when Amadori-modified peptides were fragmented. Contrary to what was observed under CID conditions, ions corresponding to neutral losses of water or furylium ion production were not observed in the ETD spectra. Instead, abundant and almost complete series of c and z type ions were observed regardless of whether the modification site was located in the middle of the sequence or close to the N-terminus, greatly facilitating the peptide sequencing. This study strongly suggests that ETD is a better technique for proteomics studies of non-enzymatically glycated peptides and proteins.
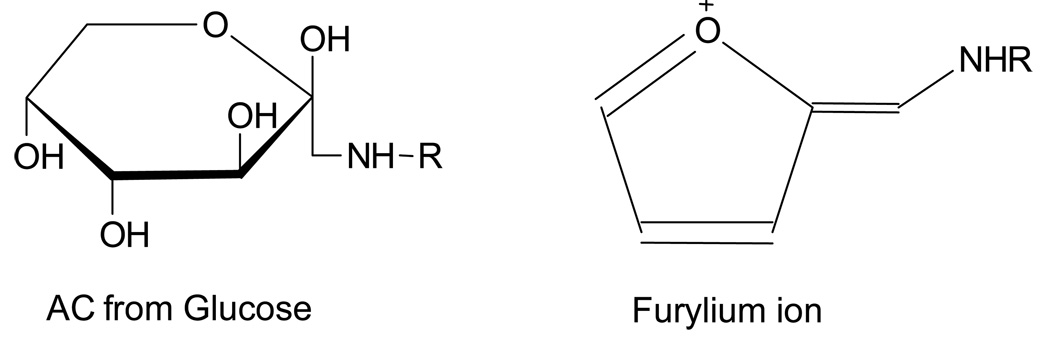
Reducing sugars (glucose, fructose, ribose, etc.) can react with primary amino groups in amino acids and proteins under favorable conditions to form stable, covalent modifications. This non-enzymatic glycosylation or glycation is commonly known as the Maillard or browning reaction, due to the brown pigments typically formed during incubation of reducing sugars with protein for extended periods of time and/or high heat. The Maillard reaction begins with nucleophilic attack by a free amine group of an amino acid or protein on the carbonyl carbon of a reducing sugar to form a carbinolamine, which subsequently undergoes dehydration to form an imine (Schiff base). The Schiff base intermediate slowly rearranges to form a stable ketoamine, called the Amadori compound (Figure 1), resulting from modification by aldose.1 The ketoamines can then undergo further oxidation and rearrangement reactions to form a series of more reactive, colored, and fluorescent compounds, termed advanced glycation end-products (AGEs).2
Figure 1.
Structure of the Amadori compound (AC) resulting from reaction of primary amino groups with glucose, and structure of the furylium ion, which is formed during the collision induced dissociation process by loss of 3 H2O and HCHO.
Glycated hemoglobin (HbA1c) has been used as a clinical diagnostic marker of relatively long-term (~90 days) glucose control in diabetic patients.3 However, the Maillard reaction, and AGEs in general, are believed to play a more pathogenic role in the development of atherosclerosis and diabetic complications. The Maillard hypothesis of diabetic complications proposes that chronic, cumulative chemical modification of proteins by glycation and AGEs alters their turnover, structure, and function.4 Indeed, Maillard-type fluorescence has been shown to be increased in lens crystalline of diabetic patients and to correspond to degree of diabetic retinopathy.5 Similarly, levels of the AGE Nε-carboxymethyllysine (CML) are known to correlate with the severity of both diabetic nephropathy and retinopathy.6 AGEs are also thought to play a role in age-related pathological conditions such as atherosclerosis,7 via their accumulation in tissue collagens, resulting in loss of vascular elasticity and thickening of arterial walls. They have also been proposed to play a role in abnormal amyloid aggregation in age-related neurodegenerative disorders such as Alzheimer’s8, 9 and Parkinson’s10 diseases.
In order to more fully understand the role of glycation and AGEs in the development of diabetic complications and other disorders, comprehensive proteomic studies are required in order to identify those glycated proteins and sites of glycation whose altered structure may lead to a pathological state. Previous research on protein glycation primarily focused on quantifying the overall protein glycation level by chemical, chromatographic, spectroscopic and radioimmunological methods; however, these methods generally lack specificity and glycation site information.11 Peptide mapping methods, such as LC-ESI-MS or MALDI-MS analysis of enzymatically digested peptides coupled with in-silico digestion of proteins, have been used in glycation site analyses and semi-quantification.12–14 Although certain specific glycation sites have been identified from incubations of model proteins with glucose or ribose, in many cases, identifications of the glycation sites were ambiguous based only on monosaccharide modification-induced mass increases. Therefore, it is difficult to extend these methods to more complex proteome-level glycation analyses. Very recently, tandem mass spectrometry was also applied to glycated peptide sequencing,15 however, these studies were performed with instruments that utilized collision induced dissociation (CID), in which intra-molecular vibrational energy redistribution occurs prior to bond cleavage. Thus, the weakest bonds on the peptide side chain modifications tend to dissociate preferentially, resulting in high abundance ions corresponding to various degrees of water loss. Those ions resulting from neutral losses of water dominate the MS/MS spectrum; very limited and very weak peptide backbone fragmentation is observed, making the confident identification of glycation sites difficult. While certain patterns of neutral losses can hint at the presence of a glycated peptide,15, 16 information leading to a confident identification of the peptide sequence is lacking. Precursor-ion scanning methods based on the Amadori-derived lysine immonium ion at m/z 192.1 were recently used to map glycation sites on glycated bovine serum albumin (BSA) using quadrupole-time-of-flight MS; complete sequence information was obtained for modified BSA peptides, based on singly and doubly charged furylium y ions, as well as doubly charged pyrylium y ions.17 However, due to the low mass cut off limitation of all commercial 3-D or linear ion trap instruments, this method is limited for applications in bottom up proteomics.
Electron capture dissociation (ECD), implemented on FT-ICR mass spectrometers by McLafferty and coworkers,18 represents a significant advance in tandem mass spectrometry. Being non-ergodic in its nature, (i.e. bond dissociation occurs immediately after electron transfer) ECD provides more extensive sequence fragmentation, allowing labile modifications to remain intact. It has been successfully applied to the sequencing of peptides containing labile post translational modifications (PTMs),19–22 including large peptides and intact proteins.23 Very recently, electron transfer dissociation (ETD) fragmentation using a modified linear ion-trap was developed by Hunt and co-workers.24 This technique, being ECD-like but using aromatic anions as an electron vehicle, was demonstrated in sequencing experiments of phosphopeptides; abundant peptide backbone c and z type ions were detected, resulting in almost complete sequence coverage in the ETD fragmentation spectra. It is generally concluded that ETD is particularly well-suited for the characterization of peptides containing PTMs,24 and it has been applied to the analyses of N-linked enzymatically glycosylated peptides.25
In this work, we report the first use of ETD as a fragmentation technique for MS/MS characterization of D-glucose glycated peptides. ETD fragmentation spectra will be compared to those obtained with CID (with a focus on the early glycation product ketoamine), illustrating the much more informative sequence information obtained with ETD.
EXPERIMENTAL
Standard Peptides
Standard glycated peptides with amidated C-termini (AGGK#AAFL-NH2, AK#DSASFL-NH2, and AK#ASASFL-NH2; the # represents the site of glycation with D-glucose) were synthesized and purified according to published solid-phase based post-synthetic approaches.26 Briefly, the peptides were synthesized using 9-fluorenylmethoxycarbonyl/tert-butyl (Fmoc/t-Bu) chemistry, with 4-methyltrityl group protection of the lysine residue to be modified. The resin-supported peptides were then incubated with D-glucose in dimethylformamide at 110°C under nitrogen for 40 min, and the resulting glycated peptides were deprotected with trifluoroacetic acid and purified by HPLC.
ETD MS/MS analyses of standard peptides
For LC-MS/MS analyses, an Agilent 6340 Ion Trap LC/MS system (Agilent, Santa Clara, CA, USA) equipped with ETD capability was coupled to an Agilent HPLC-Chip Cube MS interface, well plate sampler, nanoflow pump, and capillary pump.27 Fluoranthene anions from a small negative chemical ionization source were used to transfer electrons in the ETD process. CID and ETD scans were performed alternately. For CID process, the smart fragmentation feature was enabled, and the fragmentation amplitude was 1.3 V with start amplitude of 30% and end amplitude of 200%; for ETD process, reactant accumulation time was 40 ms and reaction time was 80 ms. The fast HPLC separation was carried out on the Chip with an integrated separation column (Zorbax 300SB-C18, 75 µm × 43 mm, 5 µm) and an enrichment column (Zorbax 300SB-C18, 40 nL, 5 µm). The voltage applied to HPLC-Chip capillary was 1725 V. For each separation, 1 µL (0.3 µg) of sample was injected. A short gradient of 3–80% B (Solvent A: 0.1% formic acid in H2O; Solvent B: 0.1% formic acid in 90% CH3>CN) in 7 minutes was used at a flow rate of 600 nL/min.
CID MSn analyses of standard peptides
The fragmentation behavior of peptides glycated with glucose under CID conditions was also investigated using an LCQ-Deca XP (Thermo Electron, San Jose, CA, USA) instrument. Peptides at 20 µM in 50/50 (v/v) CH3CN/H2O were directly infused at 1 µl/min for analyses in positive ionization mode. Electrospray voltage was typically kept between 1.2–2.0 kV, and the inlet capillary was maintained at 150°C. The [M+2H]2+ and [M+H]+ ions were selected as precursors for MS2 or MS3 experiments. For MS2 experiments, the normalized collision energy is typically kept at 22–24% for [M+H]+, while a normalized collision energy of 27–30% was used for MS2 of [M+2H]2+ and MS3 fragmentations.
RESULTS AND DISCUSSION
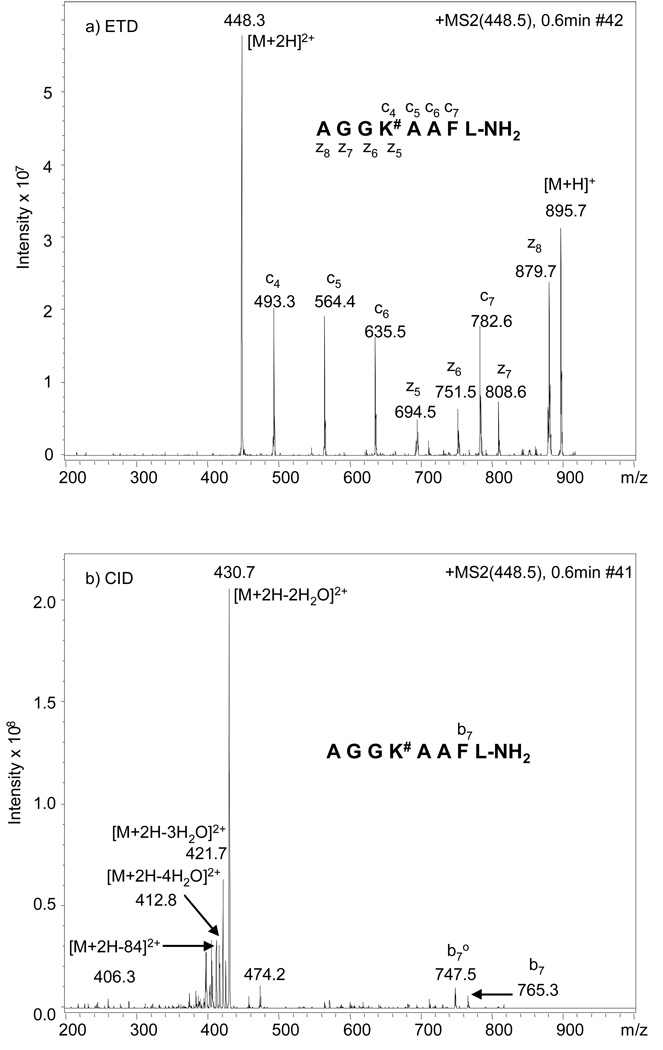
To confine all neutral water losses obtained in the tandem mass spectra of glycated peptides to the sugar moiety and simplify spectral interpretation, the model peptides used in this study were amidated at the C-terminus.17 Figure 2 shows the MS/MS spectra of the [M+2H]2+ ion corresponding to peptide AGGK#AAFL-NH2 under ETD (Figure 2a) and CID (Figure 2b) conditions, respectively. Under CID conditions, the sequence informative b and y ions resulting from peptide back bone cleavage are very rare; only b7 and b7-H2O can be confidently identified, and the most abundant ions represent neutral losses of two to four molecules of H2O. Losses of one to three molecules of water from Amadori-modified peptides during tandem mass spectrometry have been well documented in the literature.15–17, 28 We did not observe the loss of only one molecule of water, which was likely due to the high collision energy used during CID-MS/MS analyses using the Chip-HPLC-MS system. When lower collision energies were used on an independent ion-trap instrument for direct infusion studies of these peptides, the ion corresponding to loss of one molecule of water was highly abundant (data not shown). The loss of four consecutive water molecules has been previously observed and suggested to be produced by loss of four water molecules from five-membered furanose rings.17
Figure 2.
ETD (a) and CID (b) MS/MS spectra of the [M+2H]2+ ion of peptide AGGK#AAFL-NH2, in which # represents glucose adduct to lysine. The spectra were acquired with alternating ETD and CID scanning.
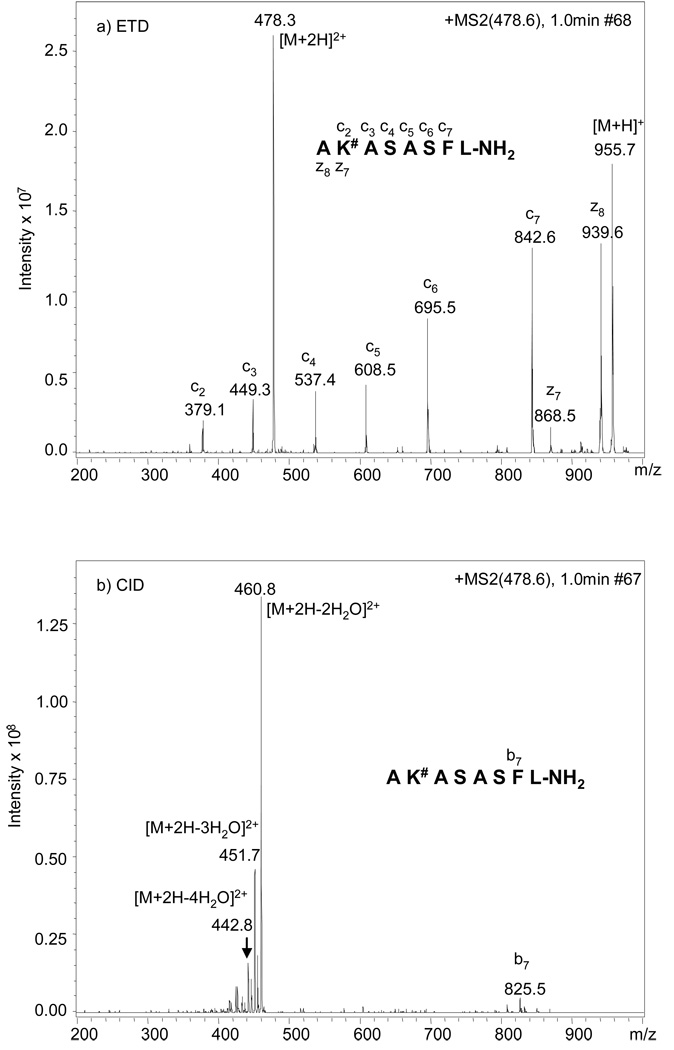
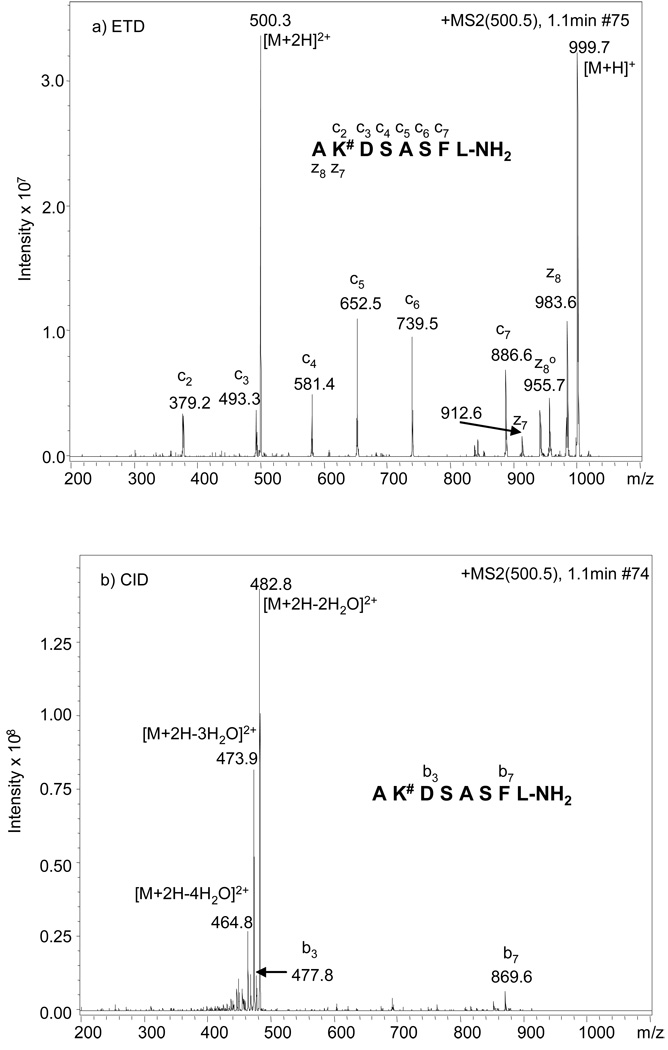
In addition to the water loss, the loss of 84 Da (3 H2O + HCHO) to form a furylium ion (as shown in Figure 1), although not very abundant, was also observed (Figure 2b). The furylium ion has been observed in MS/MS studies of the model compounds Nε-fructose-lysine and Nα-formyl-Nε-fructoselysine (Amadori compound formed between glucose and lysine, data not shown), as well as in glucose modified dipeptides16 and glucose modified oligopeptides.15, 17 This neutral loss pattern of various degrees of water and formation of furylium ion is generally charge independent, as similar results were observed when the [M+H]+ was used as the precursor for MS/MS (data not shown). Similar water loss features were also obtained with peptide AK#ASASFL-NH2 (Figure 3b) and AK#DSASFL-NH2 (Figure 4b) under CID conditions, where the glucose modification is close to the N-terminus in both peptides; however, compared to the MS/MS spectra for peptide AGGK#AAFL-NH2, the [M+2H-84]2+ ions (corresponding to furylium ion production) are almost undetectable for the latter two peptides. In addition, the number of b and y ions generated in the CID-MS/MS spectra of these two peptides are also rare when the modified lysine is present in the middle of the sequence, i.e. only b3 and b7 ions were observed for AK#DSASFL-NH2 and b7 is the only observable b or y ion for AK#ASASFL-NH2. Any attempt to identify the sequence of these peptides or the glycation site with such information poor fragmentation spectra would be extremely difficult.
Figure 3.
ETD (a) and CID (b) MS/MS spectra of the [M+2H]2+ ion of peptide AK#ASASFL-NH2, in which # represents glucose adduct to lysine. The spectra were acquired with alternating ETD and CID scanning.
Figure 4.
ETD (a) and CID (b) MS/MS spectra of the [M+2H]2+ of peptide AK#DSASFL-NH2, in which # represents glucose adduct to lysine. The spectra were acquired with alternating ETD and CID scanning.
When the [M+2H]2+ ions from the same peptides were selected as precursors for analysis by ETD, a series of abundant c and z type ions were observed (Figure 2a, Figure 3a and Figure 4a). Contrary to what was observed under CID conditions, neither furylium ions nor ions corresponding to neutral losses of water from the glucose modification site were observed, clearly demonstrating that the Amadori moiety is stable under ETD conditions and bond cleavage is less dependent on peptide composition and side chain modification(s). The observation of more c ions than z ions in the fragmentation spectra of peptides with the glucose modification close to the N-terminus is due to the charge localizations in these peptides. Each doubly charged peptide contains one charge at the N-terminal alpha-amine as well as one charge at the side-chain amine of the modified lysine residue. During the ETD process, the charge at the N-terminal alpha-amine is neutralized via electron transfer. Therefore, only those peptide fragments containing the modified lysine residue will be detected. It has been reported that for non-tryptic peptides, triply charged precursors exhibit more extensive fragmentation than doubly charged precursors under ETD.29 However, this is more likely due to charge localization in doubly charge peptides, resulting in some fragments retaining one charge and others being undetected as neutral species. Regardless, this should not reduce the utility of ETD for sequencing of doubly charged glycated peptides. In addition, while non-tryptic peptides were used in this study in order to confine all water losses obtained in the CID process to the Amadori moiety and to simplify interpretation of the resultant tandem mass spectra, complete sequence coverage for tryptic peptides containing the Amadori modification is expected when analyzed by ETD, as this approach has been successfully applied in the analysis of tryptic peptides containing phosphorylated residues.24
In a typical ETD fragmentation reaction, the charge states of multiply charged cation precursors are reduced by collisions with anions to form so-called radical cation species. Subsequently, the N-Cα bonds of the radical cations fragment and form a series of c and z ions. This charge reduction process prevents ETD from being applied to singly charged precursors; however, this is less of a concern for proteomics, in general, because the vast majority of peptides resulting from enzymatic digestion are multiply charged when using ESI. The MS/MS spectra shown in this report were obtained from doubly charged precursors, and it is clearly demonstrated this technique is well suited for glycated peptide analyses by identifying the sequences without ambiguity. It is reported that higher charge states are more efficient for the ETD/ECD process,29, 30 which is amenable to analyses of peptide digests from such complex samples as cell lysates and plasma. An additional advantage of ETD/ECD analyses of tryptic digests of glycated proteins is that these techniques are more powerful for sequencing longer peptides. In this respect, it has been reported that enzymatic activity of trypsin is inhibited by lysine glycation,12, 15 resulting in longer peptides containing one or more missed cleavage sites. With traditional CID fragmentation, longer peptides are difficult to sequence even when not containing post translational modifications. In addition, the ETD spectra shown in this report were all from single scans, without any spectrum averaging. This highlights the efficiency of the ETD process, which is important to consider when utilizing ETD to identify peptides eluting from high-efficiency capillary LC separations of complex samples. It is important to note the lower sensitivity of ETD compared to CID (approximately 30% on the basis of ion current ratio between ETD and CID), which may be a concern when utilizing ETD for the identification of low abundance glycated proteins from complex samples. However, the better sequence coverage provided by ETD results in higher database searching scores for modified peptides24 and likely compensates for the relatively lower sensitivity of the technique.
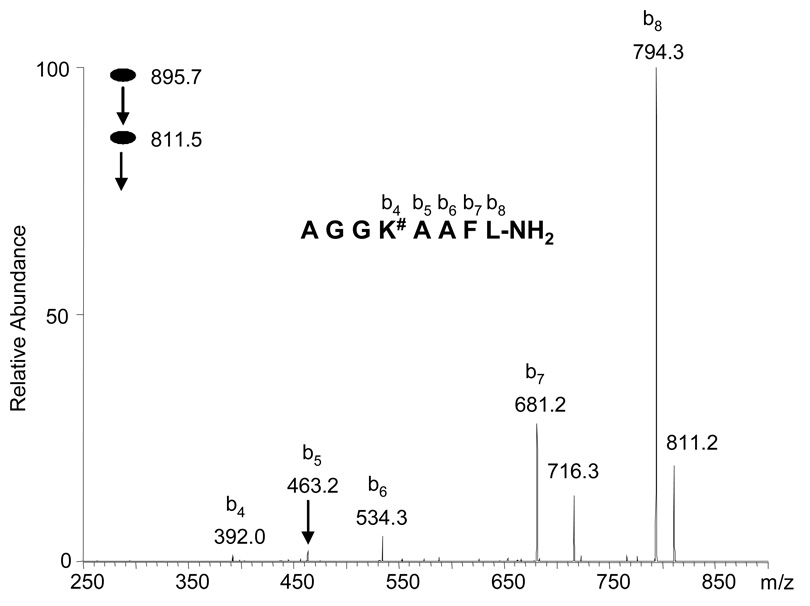
The pattern of various degrees of water losses in conjunction with formation of the unique furylium ion under CID conditions may serve as a marker for glycated peptides. This will complement peptide sequence information obtained from ETD analyses and provide an additional level of confidence to further reduce false positive identifications (it is important to note that ETD and CID were performed alternately on the instrument used in this study). The furylium ion has been used to trigger MS3 fragmentation for peptide sequencing under CID conditions.31 However, this can be problematic when it is implemented to complex samples as the furylium ion is not often very abundant (Figure 3b, Figure 4b and other unpublished data), particularly when multiply charged species were selected for MS/MS, which will result in even lower quality fragment ions in MS3. Even if the furylium ion is abundant, the CID process, owing to its limitations (i.e. charge localization and residue effect32) may still preferentially cleave certain amide bonds over others on the peptide backbone. Figure 5 illustrates this point, as no fragmentation comparable to that obtained by ETD is observed when the [M+H-84]+ of peptide AGGK*AAFL-NH2 was further fragmented. Furthermore, when coupled with HPLC for complex sample analyses, neutral-loss triggered events will require a longer instrument duty cycle, resulting in under sampling.
Figure 5.
MS3 spectrum of the [M+H]+ ion of peptide AGGK#AAFL-NH2 under CID conditions, in which # represents glucose adduct to lysine. Shown is the MS/MS of [M+H-84]+ generated in the first stage MS/MS of the [M+H]+ ion.
Conclusions
Three different glycated peptides were analyzed by ETD MS2 and CID MS2 and MS3. For all of these peptides, ETD analysis, owing to its unique fragmentation mechanism, provided almost complete series of fragment ions. In contrast, the CID process preferentially cleaved the weakest bonds on the Amadori moiety, resulting in high abundant neutral losses of water in conjunction with relatively moderate to low abundant furylium ion formation. ETD clearly is better suited for glycated peptide analyses, as the Amadori compound contains several labile hydroxyl groups. It is highly anticipated that this technique will greatly facilitate the identification of novel glycated proteins and sites of glycation and may lead to new biomarkers for diabetes mellitus and its related complications. Using ETD for large-scale glycated proteomics analyses of plasma samples from patients with type 2 diabetes mellitus is currently underway in our laboratory.
Acknowledgments
The authors would like to thank Prof. John W. Baynes at the University of South Carolina for providing Nα-formyl-Nε-fructoselysine. This research was supported by NIH grant (5R21DK071283) to R.D.S. (PI) and T.O.M. (co-PI); portions of this research were supported through the National Center for Research Resources (RR018522) and performed at the Environmental Molecular Science Laboratory, a national scientific user facility located at Pacific Northwest National Laboratory (PNNL) and sponsored by the U. S. Department of Energy (DOE) Office of Biological and Environmental Research. PNNL is operated by Battelle for the DOE under Contract No. DE-AC06-76RLO-1830. R.H. and A.F. gratefully acknowledge financial support by the Deutsche Forschungsgemeinschaft (Graduiertenkolleg 378).
References
- 1.Baynes JW, Watkins NG, Fisher CI, Hull CJ, Patrick JS, Ahmed MU, Dunn JA, Thorpe SR. Prog. Clin. Biol. Res. 1989;304:43. [PubMed] [Google Scholar]
- 2.Ahmed N, Thornalley PJ. Biochem. Soc. Trans. 2003;31:1417. doi: 10.1042/bst0311417. [DOI] [PubMed] [Google Scholar]
- 3.Krishnamurti U, Steffes MW. Clin. Chem. 2001;47:1157. [PubMed] [Google Scholar]
- 4.Thorpe SR, Baynes JW. Drugs & Aging. 1996;9:69. doi: 10.2165/00002512-199609020-00001. [DOI] [PubMed] [Google Scholar]
- 5.Mosier MA, Occhipinti JR, Burstein NL. Arch. Opthalmol. 1986;104:1340. doi: 10.1001/archopht.1986.01050210094032. [DOI] [PubMed] [Google Scholar]
- 6.McCance DR, Dyer DG, Dunn JA, Bailie KE, Thorpe SR, Baynes JW. J. Clin. Invest. 1993;91:2470. doi: 10.1172/JCI116482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baynes JW. Exp. Gerontol. 2001;36:1527. doi: 10.1016/s0531-5565(01)00138-3. [DOI] [PubMed] [Google Scholar]
- 8.Nicolls MR. Curr. Alzheimer Res. 2004;1:47. doi: 10.2174/1567205043480555. [DOI] [PubMed] [Google Scholar]
- 9.Moreira PI, Smith MA, Zhu X, Nunomura A, Castellani RJ, Perry G. Ann. N. Y. Acad. Sci. 2005;1043:545. doi: 10.1196/annals.1333.062. [DOI] [PubMed] [Google Scholar]
- 10.Munch G, Gerlach M, Sian J, Wong A, Riederer P. Ann. Neurol. 1998;44:S85. doi: 10.1002/ana.410440713. [DOI] [PubMed] [Google Scholar]
- 11.Furth AJ. Anal. Biochem. 1988;175:347. doi: 10.1016/0003-2697(88)90558-1. [DOI] [PubMed] [Google Scholar]
- 12.Brock JW, Hinton DJ, Cotham WE, Metz TO, Thorpe SR, Baynes JW, Ames JM. J. Proteome Res. 2003;2:506. doi: 10.1021/pr0340173. [DOI] [PubMed] [Google Scholar]
- 13.Lapolla A, Fedele D, Reitano R, Bonfante L, Guizzo M, Seraglia R, Tubaro M, Traldi P. J. Mass Spectrom. 2005;40:969. doi: 10.1002/jms.842. [DOI] [PubMed] [Google Scholar]
- 14.Ames JM. Ann. N. Y. Acad. Sci. 2005;1043:225. doi: 10.1196/annals.1333.028. [DOI] [PubMed] [Google Scholar]
- 15.Lapolla A, Fedele D, Reitano R, Arico NC, Seraglia R, Traldi P, Marotta E, Tonani R. J. Am. Soc. Mass Spectrom. 2004;15:496. doi: 10.1016/j.jasms.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 16.Mennella C, Visciano M, Napolitano A, Del Castillo MD, Fogliano V. J. Pept. Sci. 2006;12:291. doi: 10.1002/psc.722. [DOI] [PubMed] [Google Scholar]
- 17.Frolov A, Hoffmann P, Hoffmann R. J. Mass Spectrom. 2006;41:1117. doi: 10.1002/jms.1117. in press. [DOI] [PubMed] [Google Scholar]
- 18.Zubarev RA, Kelleher N, McLafferty FW. J. Am. Chem. Soc. 1998;120:3265. [Google Scholar]
- 19.Stensballe A, Jensen ON, Olsen JV, Haselmann KF, Zubarev RA. Rapid Commun. Mass Spectrom. 2000;14:1793. doi: 10.1002/1097-0231(20001015)14:19<1793::AID-RCM95>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 20.Bakhtiar R, Guan Z. Biochem. Biophys. Res. Commun. 2005;334:1. doi: 10.1016/j.bbrc.2005.05.138. [DOI] [PubMed] [Google Scholar]
- 21.Mirgorodskaya E, Roepstorff P, Zubarev RA. Anal. Chem. 1999;71:4431. doi: 10.1021/ac990578v. [DOI] [PubMed] [Google Scholar]
- 22.Hakansson K, Chalmers MJ, Quinn JP, McFarland MA, Hendrickson CL, Marshall AG. Anal. Chem. 2003;75:3256. doi: 10.1021/ac030015q. [DOI] [PubMed] [Google Scholar]
- 23.Bakhtiar R, Guan Z. Biotechnol. Lett. 2006;28:1047. doi: 10.1007/s10529-006-9065-z. [DOI] [PubMed] [Google Scholar]
- 24.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9528. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogan JM, Pitteri SJ, Chrisman PA, McLuckey SA. J. Proteome Res. 2005;4:628. doi: 10.1021/pr049770q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frolov A, Singer D, Hoffmann R. J. Pept. Sci. 2006;12:389. doi: 10.1002/psc.739. [DOI] [PubMed] [Google Scholar]
- 27.Tang N, Goodley P. Agilent Application Notes. 2006 [Google Scholar]
- 28.Jeric I, Versluis C, Horvat S, Heck AJ. J. Mass Spectrom. 2002;37:803. doi: 10.1002/jms.337. [DOI] [PubMed] [Google Scholar]
- 29.Pitteri SJ, Chrisman PA, Hogan JM, McLuckey SA. Anal. Chem. 2005;77:1831. doi: 10.1021/ac0483872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitteri SJ, Chrisman PA, McLuckey SA. Anal. Chem. 2005;77:5662. doi: 10.1021/ac050666h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaler TA, Wang W, Blackshear PJ, Becker CH. 54th ASMS Conference on Mass Spectrometry and Allied Topics; Seattle. 2006. p. MP402. [Google Scholar]
- 32.Huang Y, Triscari JM, Tseng GC, Pasa-Tolic L, Lipton MS, Smith RD, Wysocki VH. Anal. Chem. 2005;77:5800. doi: 10.1021/ac0480949. [DOI] [PMC free article] [PubMed] [Google Scholar]