Abstract
The CTCF paralog BORIS (brother of the regulator of imprinted sites) is an insulator DNA-binding protein thought to play a role in chromatin organization and gene expression. Under normal physiologic conditions, BORIS is predominantly expressed during embryonic male germ cell development; however, it is also expressed in tumors and tumor cell lines and, as such, has been classified as a cancer-germline or cancer-testis gene. It has been suggested that BORIS may be a pro-proliferative factor, whereas CTCF favors antiproliferation. BORIS and CTCF share similar zinc finger DNA-binding domains and seem to bind to identical target sequences. Thus, one critical question is the mechanism governing the DNA-binding specificity of these two proteins when both are present in tumor cells. Chromatin immunoprecipitation (ChIP) in HCT116 cells and their hypermethylated variant showed that BORIS binds to methylated DNA sequences, whereas CTCF binds to unmethylated DNA. Electromobility shift assays, using both whole-cell extracts and in vitro translated CTCF and BORIS protein, and methylation-specific ChIP PCR showed that BORIS is a methylation-independent DNA-binding protein. Finally, experiments in murine hybrid cells containing either the maternal or paternal human chromosome 11 showed that BORIS preferentially binds to the methylated paternal H19 differentially methylated region, suggesting a mechanism in which the affinity of CTCF for the unmethylated maternal allele directs the DNA binding of BORIS toward the paternal allele.
Introduction
Insulator DNA sequences are thought to partition the genome into functional chromosomal domains to regulate gene transcription (1). The compartmentalization of chromatin into distinct regulatory domains seems to alter interactions between target genes and nearby cis-acting enhancer elements (2). CTCF was the first human insulator DNA-binding protein identified that is a ubiquitously expressed, highly conserved, zinc finger protein that has multiple roles in gene regulation (3), including regulation of the imprinted maternal H19 allele (4). CTCF binds to the unmethylated DNA of the differentially methylated region (DMR), limiting access to an enhancer shared between H19 and IGF2 and causing silencing of the maternal IGF2 allele (5).
The CTCF paralog, CTCFL or BORIS (brother of the regulator of imprinted sites), is thought to be predominantly expressed in testis; it has also been detected in >100 cancer cell lines representing all major forms of human tumors (6). BORIS shares an 11 zinc finger domain with CTCF (6); however, CTCF and BORIS differ significantly in their NH2 and COOH termini, suggesting that these regions may interact with different binding partners, altering gene expression by different mechanisms. Indeed, in contrast to CTCF, BORIS seems to activate gene expression (7, 8). These results suggest that CTCF and BORIS use a similar DNA sequence to elicit very different changes in gene expression: one suppresses and the other induces gene expression at the same locus.
It has recently been shown that the human genome contains 13,804 CTCF DNA-binding sites in potential insulators of the human genome, and although these sequences are significantly different fromtranscriptional start sites, their distribution correlated with gene expression (9). Studies have also shown that CTCF is down-regulated in lobular carcinoma in situ of the breast (10), suggesting a potential role of the CTCF DNA-binding target sequences in carcinogenesis. Thus, a model could be proposed whereby the aberrant reexpression of BORIS, which is observed in tumor cells (6), might in turn activate a large number of genes resulting in a cellular phenotype permissive for transformation. However, both CTCF and BORIS seemto be expressed in tumor cells, suggesting a potential competition between these two proteins for CTCF DNA-binding sequences that may subsequently determine the expression of nearby target genes. Thus, one critical question about the activity of these related insulator DNA-binding proteins is the mechanism by which they specifically bind to CTCF target sequences.
Materials and Methods
Cell lines, cell culture, and plasmids
HCT116 cells (human colon carcinoma) and the methyltransferase somatic knockouts DNMT1(-/-)/DNMT3B(-/-) double knockout (DKO) cells were cultured in McCoy's 5A medium containing 10% heat-inactivated fetal bovine serum (FBS). The murine hybrid A911P or A911M cells were grown in DMEM with 10% FBS. Media were supplemented with penicillin (100 units/mL) and streptomycin (100 μg/mL). Primers were used to PCR the DMR region (Supplementary Data) and this fragment was cloned into ptk-Luc (Clontech, Inc.) and transient assay was done as previously described (11).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were done using the Upstate Biotechnology, Inc. kit (12). ChIP was performed using CTCF (C-20; Santa Cruz Biotechnology, Inc.) or BORIS (Supplementary Figs. S1 and S2) antibodies. Purified DNA samples were analyzed by real-time quantitative PCR (QPCR) using SYBR Green PCR Master Mix (Applied Biosystems) with the ABI Prism 7500 Detection System (Applied Biosystems) with primers to the human H19 DMR (Supplementary Materials and Methods). Data were collected and analyzed by comparative CT methods.
Bisulfite pyrosequencing and methylation-specific ChIP PCR
DNA (10 μg) was treated with sodium bisulfite according to established methods (12). Treated DNA was resuspended in 40 mL of distilled water for PCR. All primers are listed in Supplementary Materials and Methods. PCR products were used directly for pyrosequencing according to the manufacturer's instruction (Biotage). Methylation-specific ChIP PCR (MS-ChIP-PCR) was carried out in HCT116 cells. Following ChIP with an anti-BORIS or anti-CTCF antibody, the pulled-down chromatin was subjected to bisulfite conversion using the EZ DNA Methylation Gold kit (Zymo Research). Primer sets were designed using Methyl Primer Express software (Applied Biosystems). Each reaction contained 10 μL Fast Cycling Taq Master mix (Qiagen), 5 μL of converted DNA, and 1 AL each of 0.3 μmol/L forward and reverse primer. QPCR and PCR were done as described above.
Preparation of nuclear extracts and electromobility shift assay
Nuclear extracts were prepared as previously described method (11). In vitro BORIS and CTCF protein was prepared using Expressway In Vitro Protein Synthesis System(Invitrogen). For the electromobility shift assay (EMSA) methylation analysis of BORIS and CTCF DNA binding, 1 μg of PCR product was used. Each reaction consisted of 30 μL total volume containing 1 Ag DNA, 4 units of SssI methylase enzyme, and buffer, and reactions were carried out at 37°C for 1 h. The same procedure was performed for unmethylated DNA. The DNA reactions were gel purified, and 50 ng of DNA were used for labeling with [32P]ATP. For each EMSA, 10,000 cpmof labeled DNA were used with in vitro transcribed CTCF or BORIS protein.
Results
BORIS binds to the H19 DMR in cultured cells
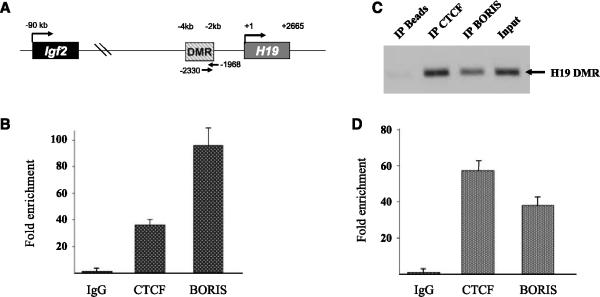
We initially determined whether BORIS, like CTCF (3), binds to the DMR of the H19 locus (Fig. 1A). ChIP analysis was done using either an anti-CTCF (Santa Cruz Biotechnology) or an anti-BORIS (Supplementary Fig. S2) antibody. Primers were used that overlap the primary H19 CTCF DNA-binding site that has been shown to regulate IGF2/H19 gene expression (2). ChIP of Tera-1 testicular tumor or HCT116 colon cancer cells followed by PCR or QPCR yielded a significant enrichment of the H19 DMR with both CTCF and BORIS antibodies (Fig. 1B-D). As a negative control, an H19 genomic region without a CTCF consensus site was amplified (data not shown) using primers that have been previously described (14). These results show that, like CTCF, BORIS binds to the H19 DMR (15).
Figure 1.
BORIS is an H19 DMR DNA-binding protein. A, diagram of the H19 region and the location of the primers used for ChIP analysis. B, BORIS binds to the H19 DMR in testicular tumor cells. Tera-1 testicular tumor cells were fixed with 1% formaldehyde to cross-link protein-DNA interactions and then sonicated, and fixed cells were immunoprecipitated with either an anti-CTCF or anti-BORIS (Supplementary Fig. S2) antibody. DNA was eluted and purified before analysis by real-time QPCR with primers that cover the CTCF-binding site in the H19 DMR region. BORIS binds to the H19 DMR in HCT116 colon tumor cells as measured by PCR (C) and QPCR (D). HCT116 cells were harvested and CTCF and BORIS DNA binding was measured as described above. All ChIP experiments were done in triplicate. Columns, mean; bars, SD.
BORIS preferentially binds to the methylated H19 DMR
Although BORIS and CTCF both seemto bind to the H19 DMR, the mechanism governing DNA binding is unknown, and it seemed reasonable to suggest that methylation might determine the specificity of binding. To address this idea, two cell lines were used, (a) HCT116 cells and (b) HCT116 somatic DKO cells, which have both DNMT1 and DNMT3B deleted. These cells have been previously characterized; the methylation is decreased ~95% in the DKO cells (13) compared with the parental HCT116 cells.
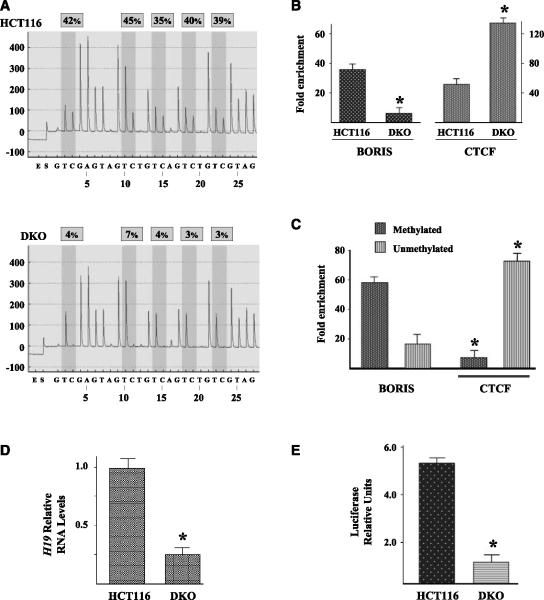
The methylation status of the H19 DMR was determined by bisulfite pyrosequencing (12) and clearly showed hypomethylation of the DMR in the DKO compared with HCT116 cells (Fig. 2A). ChIP analysis with an anti-BORIS antibody showed that BORIS binding was highest in the HCT116 cells and lowest in the unmethylated DKO cells (Fig. 2B, left). In contrast, CTCF DNA binding is highest in the unmethylated DKO cells and lowest in the HCT116 cells (Fig. 2B, right). Western blot analysis showed similar BORIS and CTCF protein levels in all HCT116 cell lines, ruling out that DNA binding is a function of total BORIS or CTCF levels (Supplementary Fig. S3). These experiments suggest that methylation may be at least one mechanism governing the DNA binding of CTCF or BORIS.
Figure 2.
BORIS preferentiallybinds to methylated H19 DMR. A, the H19 DMR is hypomethylated in somatic DNMT1/DNMT3B DKO cell lines. HCT116 (top) and the HCT116-derived DNMT1(-/-)/DNMT3B(-/-) somatic DKOs (bottom) cells were analyzed by bisulfite pyrosequencing at the H19 (CTCF-binding site 1) DMR. B, BORIS and CTCF bind to the H19 DMR depending on cellular DNMT status. HCT116 and DKO cells were examined by ChIP analysis with either an anti-BORIS (left) or anti-CTCF (right) antibodyto determine relative levels of CTCF and BORIS binding to the H19 DMR. C, BORIS preferentially binds to methylated DNA. MS-ChIP-PCR was done using HCT116 cells. In these experiments, cells were harvested followed by ChIP with either an anti-BORIS or anti-CTCF antibody. The immunoprecipitated DNA was bisulfite treated and subjected to PCR with either methylated or unmethylated primer sets complementary to the H19 DMR. All ChIP experiments were done in triplicate. D, H19 expression is decreased in DKO cells. HCT116 and DKO cells were harvested and total RNA was isolated and purity checked, and levels of H19 RNA were determined using real-time PCR with Taqman probes. All results are the mean of at least three separate experiments. Results are presented as H19 relative RNA levels where 1.0 represents H19 RNA level in control HCT116 cells. All experiments were normalized to glyceraldehyde-3-phosphate dehydrogenase. E, transfection of a luciferase reporter plasmids contains the H19 DMR cloned upstream of the ptk-Luc (DMR-tk-Luc) into HCT116 or DKO cells. Cells were also transfected with cytomegalovirus-β-gal and results are presented as luciferase/β-galactosidase relative units. 1.0 represents luciferase activity in DKO cells transfected with DMR-tk-Luc. All results are the mean of at least three experiments, all done in duplicate. The results of individual transfections varied by <25%. Columns, mean; bars, SD. Statistical significance was established by Student's t test. *, P < 0.05.
Although these experiments suggest that methylation may regulate CTCF and BORIS DNA binding, they do not a priori reveal the binding characteristics in a tissue culture model system. To confirmthe role of methylation in DNA binding, MS-ChIP-PCR was done in HCT116 cells. In these experiments, ChIP was performed with either an anti-BORIS or an anti-CTCF antibody and the pulled-down chromatin was then subjected to bisulfite conversion. These samples were subsequently used for methylation-specific PCR with primers (Methyl Primer Express software) designed to detect unmethylated versus methylated DNA. These experiments showed that BORIS preferentially binds to methylated DNA (Fig. 2C, left). In contrast, CTCF seems to preferentially bind to unmethylated chromatin (Fig. 2C, right).
H19 gene expression is decreased in HCT-derived DKO cells
The results above show that BORIS preferentially binds to the H19 DMR in HCT116 cells, whereas CTCF preferentially binds to the H19 DMR in DKO cells (Fig. 2B). Thus, it seemed logical to suggest that the expression of H19 would correlate with the levels of BORIS or CTCF binding to the DMR. As such, HCT116 and DKO cells were harvested and H19 RNA levels were determined by RT-PCR. These experiments showed a roughly 70% decrease in H19 RNA levels in DKO compared with HCT116 cells (Fig. 2D). The H19 DMR was subsequently cloned upstreamof ptk-Luc and this plasmid (DMR-tk-Luc) was transfected into both HCT116 and DKO cells. A roughly 5-fold in increase in luciferase activity was observed in HCT116 cells transfected with DMR-tk-Luc compared with DKO cells (Fig. 2E).
BORIS binding is methylation independent
EMSAs and supershift experiments were done using HCT116 nuclear lysates mixed with a 32P-labeled SssI-methylated oligonucleotide containing the H19 DMR CTCF sequence. Supershift experiments with either an anti-BORIS (Fig. 3A, lanes 3 and 4) or an anti-CTCF (lanes 7 and 8) antibody showed that BORIS, but not CTCF, binds to methylated DNA. In contrast, supershift experiments using an unmethylated 32P-labeled oligonucleotide showed preferential CTCF binding (Supplementary Fig. S4A) with some minimal binding of BORIS (Supplementary Fig. S4B, lane 2 versus lane 4). This was our first indication that BORIS might also bind to unmethylated DNA.
Figure 3.
BORIS, but not CTCF, DNA binding is methylation independent. A, BORIS, but not CTCF, binds to a methylated H19 DMR CTCF DNA-binding sequence. HCT116 cells were harvested and nuclear cell extracts were used for EMSA with a 32P-labeled SssI-methylated oligonucleotide containing the H19 CTCF-binding sequence. Lanes 1 and 5, probe alone; lanes 2 to 4 and 6 to 8, incubated with nuclear extract. Supershift assays were done by preincubating extracts with increasing concentrations (+ or ++) of either an anti-BORIS (lanes 3 and 4) or an anti-CTCF antibody (lanes 7 and 8). Arrows, position of the protein-DNA complex and free probe; small arrows, shifted bands. B, BORIS, but not CTCF, DNA binding in vitro is independent on methylation. EMSAs were done with in vitro translated BORIS (left) or CTCF (right) with either an unmethylated oligonucleotide containing a CTCF-binding site (lanes 1, 2, 5, and 6) or an identical SssI-methylated oligonucleotide (lanes 3, 4, 7, and 8). All gel and supershift experiments were done in triplicate. Sections of fluorograms from native gels using a Typhoon phosphorimager are shown. Arrows, the supershift as well as the protein-CTCF-DNA complex and free unbound oligonucleotide probe. C and D, siRNA knockdown of CTCF decreases CTCF and increases BORIS binding to the H19 DMR. HCT116 cells were transfected with either control (Cont)or CTCF siRNA (see Supplementary Fig. S5 for decreased CTCF levels). ChIP was done followed by either (C) QPCR with primers that cover the H19 DMR or (D) MS-ChIP-QPCR via bisulfite treatment and QPCR with either methylated or unmethylated primer sets to the H19 DMR. All ChIP experiments were done in triplicate. Columns, mean; bars, SD. Statistical significance was established by Student's t test. *, P < 0.05.
Follow-up experiments were done using in vitro translated BORIS and CTCF with either an SssI-methylated or an unmethylated oligonucleotide containing a CTCF DNA-binding consensus sequence. EMSA analysis showed that the DNA binding of BORIS was unaltered by the methylation status of the oligo (Fig. 3B, left, lane 2 versus lane 4). In contrast, CTCF bound to the 32P-labeled unmethylated oligo (Fig. 3B, right, lane 6) but not to the SssI-methylated oligo (lane 8). The results of these experiments imply that, at least in vitro, BORIS seems to bind equally well to both methylated and unmethylated DNA, suggesting that BORIS, in contrast to CTCF, is a methylation-independent DNA-binding protein.
Small interfering RNA knockdown of CTCF increases BORIS binding to the H19 DMR
Because the results above suggested that BORIS can bind to unoccupied unmethylated CTCF DNAbinding sites in a cell-free system, we hypothesized that BORIS may be excluded from binding to unmethylated CTCF DNA-binding sites simply due to the competitive occupancy of CTCF. To investigate this idea, HCT116 cells were treated with either control or CTCF small interfering RNA (siRNA; Supplementary Fig. S5) followed by ChIP with either an anti-CTCF or an anti-BORIS antibody. These results clearly showed a decrease in CTCF (Fig. 3C, left) and an increase in BORIS (right) binding to the H19 DMR in the absence of normal CTCF protein (Supplementary Fig. S5). MS-ChIP-PCR (described above) confirmed that siRNA knockdown of CTCF significantly increased BORIS binding to unmethylated DNA (Fig. 3D, 3rd versus 4th column) with little change to methylated DNA (1st versus 2nd column). siRNA knockdown of CTCF proportionally decreased CTCF DNA binding (Supplementary Fig. S6). These results suggest that CTCF may preferentially bind to unmethylated DNA, and if these sites become unoccupied, then BORIS can subsequently bind.
BORIS binding is limited to the paternal H19 DMR
It is well established that the H19 DMR from the maternal allele is hypomethylated, whereas the paternal allele is methylated and that this plays a key role in the regulation of IGF2 imprinting (14, 15). To address whether BORIS binds to the H19 DMR of either the maternal or paternal origin, two murine hybrid cell lines that contain human chromosome 11 of either paternal (A911P) or maternal (A911M) origin were used (16). ChIP analysis with an anti-BORIS antibody showed preferential binding to the paternal allele (Fig. 4A, left) with minimal binding to the maternal allele (right). In contrast, ChIP analysis with an anti-CTCF antibody showed preferential binding of CTCF to the maternal allele (right) as has been previously shown (16) with very little binding to the paternal allele (left). Methylation-specific PCR (Fig. 4B) and bisulfite pyrosequencing (Fig. 4C) of these two murine hybrid cell lines clearly showed that the maternal H19 DMR is hypomethylated, whereas the paternal H19 DMR is methylated.
Figure 4.
BORIS preferentially binds to the paternal H19 DMR. A, BORIS binds preferentially to the paternal allele. A911M and A911P mouse hybrid cells that contain human chromosome 11 of either the paternal (left) or maternal (right) origin, respectively, were analyzed via ChIP using either an anti-CTCF or an anti-BORIS antibodywith primers to the H19 DMR. DNA was eluted and analyzed by QPCR with primers to the human H19 DMR (Fig. 1A). All ChIP experiments were done in triplicate. Columns, mean; bars, SD. B and C, methylation-specific analysis of the paternal and maternal H19 DMR in the murine hybrid cell lines. A911M and A911P mouse hybrid cells were harvested, bisulfite treated, and subjected to QPCR with either methylated or unmethylated primer sets to the human H19 DMR (B) or analyzed by bisulfite pyrosequencing (C).
Discussion
BORIS and CTCF share similar zinc finger DNA-binding domains and bind to the same cis-acting DNA-binding element; however, their NH2-terminal and carboxy amino acid sequences are distinct, suggesting that these two insulator DNA-binding proteins may have divergent cellular functions (6, 17). Whereas CTCF plays a central role in IGF2/H19 imprinting (4), the intracellular function of BORIS is unclear. However, it is well established that BORIS is expressed in malignancies, and in this setting, both CTCF and BORIS are present in tumor cells (6). Thus, the mechanism governing either CTCF or BORIS DNA binding to CTCF DNA-binding target sequences is very likely to provide insight into the cellular role of these two insulator DNA-binding proteins and the subsequent expression of downstream genes, some of which may be prosurvival or pro-proliferative.
In this work, we found that BORIS DNA-binding activity is largely independent of the methylation status of its target sequence. In contrast, CTCF DNA binding is limited to unmethylated DNA. We also showed that BORIS preferentially binds to the paternal H19 DMR using matched murine hybrid cells that contain human chromosome 11 of either maternal or paternal origin. In contrast, CTCF seems to preferentially bind to the maternal H19 DMR. These results are surprising because these two proteins contain a significant degree of homology in their zinc finger DNAbinding domains and both bind to identical cis-acting DNA regulator elements (6). siRNA knockdown of CTCF decreased CTCF binding to the DMR with a reciprocal increase in binding by BORIS, an effect that seems to involve binding of BORIS to unmethylated DNA.
Thus, these results suggest a regulatory mechanism whereby BORIS preferentially binds to methylated cis-acting CTCF DNAbinding sites because it is prevented from binding to unmethylated CTCF DNA-binding sites by the physical occupancy of CTCF. In isolation, BORIS preferentially binds to methylated DNA-binding sites but also binds readily to unmethylated DNA-binding sites. In contrast, when both CTCF and BORIS are present within the cell, which is the case in tumor (HCT116) and immortalized (A911P/A911M) cell lines, BORIS can bind to unmethylated CTCF DNA-binding sites only when CTCF is displaced. This explains why BORIS preferentially binds to the methylated paternal H19 DMR in the hybrid mouse cells.
In conclusion, these results strongly suggest that DNA methylation may be at least one mechanism governing the specificity of insulator DNA-binding proteins in the regulation of target gene expression. Because CTCF seems to be ubiquitously expressed in all cells, we would expect that the main targets of BORIS in tumor cells would be the induction of genes that contain methylated promoter regions. However, the empirical fact that CTCF only binds to the unmethylated DMR suggests that the DNA binding of BORIS and CTCF in tumor cells is quite complicated, and although the DNA-binding domains are highly similar, there might be key difference that leads to different binding properties. In addition, different NH2 and COOH termini of these two proteins might interact with different protein complexes leading to dissimilar DNA-binding behavior. The answers to these questions should help to better understand the binding dynamic under different physiologic conditions leading to better understanding of the role of BORIS and CTCF in normal and malignant physiology.
Supplementary Material
Acknowledgments
Grant support: CA65145 (A.P. Feinberg) and the Intramural Research Program of the NIH, the National Cancer Institute, and the Center for Cancer Research.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
We thank Melissa Stauffer, Ph.D. (Scientific Editing Solutions) for editorial assistance.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
References
- 1.Bell AC, Felsenfeld G. Methylation of a CTCFdependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–5. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 2.Takai D, Gonzales FA, Tsai YC, Thayer MJ, Jones PA. Large scale mapping of methylcytosines in CTCFbinding sites in the human H19 promoter and aberrant hypomethylation in human bladder cancer. Hum Mol Genet. 2001;10:2619–26. doi: 10.1093/hmg/10.23.2619. [DOI] [PubMed] [Google Scholar]
- 3.Bjornsson HT, Cui H, Gius D, Fallin MD, Feinberg AP. The new field of epigenomics: implications for cancer and other common disease research. Cold Spring Harb Symp Quant Biol. 2004;69:447–56. doi: 10.1101/sqb.2004.69.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurukuti S, Tiwari VK, Tavoosidana G, et al. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A. 2006;103:10684–9. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanduri C, Pant V, Loukinov D, et al. Functional association of CTCF with the insulator upstreamof the H19 gene is parent of origin-specific and methylationsensitive. Curr Biol. 2000;10:853–6. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 6.Klenova EM, Morse HC, III, Ohlsson R, Lobanenkov VV. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin Cancer Biol. 2002;12:399–414. doi: 10.1016/s1044-579x(02)00060-3. [DOI] [PubMed] [Google Scholar]
- 7.Vatolin S, Abdullaev Z, Pack SD, et al. Conditional expression of the CTCF-paralogous transcriptional factor BORIS in normal cells results in demethylation and derepression of MAGE-A1 and reactivation of other cancer-testis genes. Cancer Res. 2005;65:7751–62. doi: 10.1158/0008-5472.CAN-05-0858. [DOI] [PubMed] [Google Scholar]
- 8.Hong JA, Kang Y, Abdullaev Z, et al. Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells. Cancer Res. 2005;65:7763–74. doi: 10.1158/0008-5472.CAN-05-0823. [DOI] [PubMed] [Google Scholar]
- 9.Kim TH, Abdullaev ZK, Smith AD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–45. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green AR, Krivinskas S, Young P, et al. Loss of expression of chromosome 16q genes DPEP1 and CTCF in lobular carcinoma in situ of the breast. Breast Cancer Res Treat. 2008;107:41–5. doi: 10.1007/s10549-008-9905-8. [DOI] [PubMed] [Google Scholar]
- 11.Gius D, Cao XM, Rauscher FJ, III, Cohen DR, Curran T, Sukhatme VP. Transcriptional activation and repression by Fos are independent functions: the C terminus represses immediate-early gene expression via CArG elements. Mol Cell Biol. 1990;10:4243–55. doi: 10.1128/mcb.10.8.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62:6442–6. [PubMed] [Google Scholar]
- 13.Rhee I, Bachman KE, Park BH, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–6. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 14.Engel N, Thorvaldsen JL, Bartolomei MS. CTCF binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus. Hum Mol Genet. 2006;15:2945–54. doi: 10.1093/hmg/ddl237. [DOI] [PubMed] [Google Scholar]
- 15.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–9. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 16.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006;23:733–42. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Jelinic P, Stehle JC, Shaw P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006;4:e355. doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.