Abstract
Microsatellite sequences are ubiquitous in the human genome and are important regulators of genome function. Here, we examine the mutational mechanisms governing the stability of highly abundant mono-, di-, and tetranucleotide microsatellites. Microsatellite mutation rate estimates from pedigree analyses and experimental models range from a low of ∼10-6 to a high of ∼10-2 mutations per locus per generation. The vast majority of observed mutational variation can be attributed to features intrinsic to the allele itself, including motif size, length, and sequence composition. A greater than linear relationship between motif length and mutagenesis has been observed in several model systems. Motif sequence differences contribute up to 10-fold to variation observed in human cell mutation rates. The major mechanism of microsatellite mutagenesis is strand slippage during DNA synthesis. DNA polymerases produce errors within microsatellites at a frequency that is 10- to 100-fold higher than the frequency of frameshifts in coding sequences. Motif sequence significantly affects both polymerase error rate and specificity, resulting in strand biases within complementary microsatellites. Importantly, polymerase errors within microsatellites include base substitutions, deletions and complex mutations, all of which produced interrupted alleles from pure microsatellites. Postreplication mismatch repair efficiency is affected microsatellite motif size and sequence, also contributing to the observed variation in microsatellite mutagenesis. Inhibition of DNA synthesis within common microsatellites is highly sequence-dependent, and is positively correlated with the production of errors. DNA secondary structure within common microsatellites can account for some DNA polymerase pause sites, and may be an important factor influencing mutational specificity.
Keywords: DNA Polymerase fidelity, slipped strand mispairing, interruptions, indel mutations, microsatellite instability
Introduction
Microsatellite sequences are tandem repeats of short (1-6 base pair) DNA motifs that are ubiquitous in eukaryotic genomes. Approximately 3% of the human genome contains microsatellite DNA sequences, which are present on every chromosome at an average density of ∼14,000 basepairs/Mbp [1]. Detailed examinations of repeat loci present within the human genome have revealed that mononucleotide repeats, predominantly poly [A/T] tracts, are the most abundant class of microsatellite [1,2]. Among dinucleotides, the [GT/CA] and [AT/TA] motifs are approximately equal in abundance; among tetranucleotides, those that are A/T-rich are predominant. In contrast, trinucleotide alleles are approximately three-fold less abundant than di- and tetranucleotide repeats [1]. This review will focus on the common, highly abundant mono-, di-, and tetranucleotide microsatellites, and the pathways that regulate their stability in the human genome.
The distribution of microsatellites within eukaryotic genomes is non-random [3]. The length of common microsatellite alleles is dependent upon genome location. Generally, alleles within noncoding regions are longer than those within coding regions, supporting the hypothesis that selection against frameshift mutations limits the expansion of non-triplet microsatellites in coding sequences [2,4]. Structural properties of microsatellites also may contribute to a nonrandom distribution. The relative abundance of tetranucleotides in vertebrate genomes is inversely proportional to their capacity to form stable secondary structures [5]. Genomic microsatellites display an extraordinary degree of sequence heterogeneity. The precise base composition of microsatellites ranges from pure arrays of a single repetitive sequence, to compound and complex arrays containing several types of repetitive units in tandem, to arrays that are interrupted by single base changes or insertion/deletion (indel) mutations [6].
A comparative analysis of the mouse and human genomes has shown that coding regions of genes account for less than half of the DNA regions under evolutionary selection [7]. This observation is consistent with the concept that the genome contains many features, in addition to protein-coding sequences, that are biologically functional. Microsatellites located in promoter regions, UTRs, and introns can be important regulators of several aspects of gene expression, such as transcription rate, RNA stability, splicing efficiency, and RNA-protein interactions [8,9]. Because microsatellite sequences are highly polymorphic and contribute to gene regulation, changes in microsatellites may provide a large pool of heritable, phenotypic variants for subsequent biologic selection [10]. Indeed, microsatellite sequences have been described as advantageous mutators in evolution, illustrating the potential positive effect of phenotypic changes due to microsatellite variation [11]. Finally, microsatellites located within intergenic regions might also have functional roles. For example, microsatellites can alter chromatin organization and are associated with recombination hotspots (reviewed in [2]).
Allele-length polymorphisms at common mono-, di- and tetranucleotide microsatellites are implicated as genetic risk factors in several diseases [5]. A well known example of this is [GT]n and [T]n allele length changes that affect CFTR gene expression via altered splicing efficiency, which consequently affects cystic fibrosis disease status [12,13]. Similarly, allele-length changes occurring at microsatellite loci associated with cancer-related genes may be a significant source of genetic variation during neoplastic progression. For example, the length of a polymorphic [CA]n allele is inversely correlated with transcription of the EGFR gene [14], and interethnic differences in [CA]n allele lengths are associated with varying EGFR levels in breast cancer patients [15]. The length of a pure [GGAA]n allele in specific promoters directly affects binding of the EWS/FL1 oncogenic transcription factor, thereby modulating target gene expression during oncogenesis [16]. Because the full impact of microsatellite changes on genome function has yet to be elucidated, it is of utmost importance to gain knowledge about the biochemical mechanisms governing the stability of microsatellite sequences.
Intrinsic Features Dictating Microsatellite Mutagenesis
Genomic Estimates of Mutation Rates and Mutability
Many microsatellite loci are highly polymorphic in human populations, hence their widespread use as markers in population genetics, forensics and oncology. Direct observations of allele length changes between parents and offspring have been used to estimate the rates of microsatellite mutation in humans (reviewed in [17,18]). Mutation rates at di- and tetranucleotide repeat loci have been estimated to be 10-2 to 10-4 per locus per gamete per generation [18]. As a point of comparison, quantitative measurements of somatic mutations arising in the hprt gene in normal human cells range from 0.4 to 1.6 × 10-6 in newborns, and 3.1 to 9.5 × 10-6 in adults [19]. Unfortunately, the lower end for microsatellite mutation rates cannot be confidently analyzed by the direct observation approach, as loci with mutation rates in the range of 10-5 or lower would require very large population samples, because the absence of any observed allele length changes is uninformative. The wide 100-fold variation among loci can be attributed to the sequence composition and varying lengths of the microsatellite sequences analyzed [18,20]. Several of the most highly mutable loci are actually compound microsatellites comprised of two or more repeated motifs [20-22], and the possibility exists that each motif is capable of influencing the mutability of the adjacent motif. Estimated mutation rates appear to increase exponentially as the number of repeat units within an allele increases [20,23], although a caveat to this conclusion is that loci of different sequences were used to make the comparisons.
Comparative genomics studies also have been used to follow human microsatellite mutations occurring over an evolutionary timescale [24,25]. A working assumption is that the microsatellites chosen for comparison are not subject to any selection bias, such as those within intergenic regions, so that any observed sequence divergence is proportional to mutation [18]. The causes of variation among microsatellite loci are uncovered computationally by deriving the mutability, or the average squared difference in the number of repeats, between orthologous microsatellite loci [24,25]. Using this approach, a direct effect of allele length on mutability was found to be greater than a linear relationship, and significant differences in mutability could be attributed to motif size [25]. A computational model that incorporates the intrinsic microsatellite properties of motif length (repeat number), motif size, and motif sequence explained the majority (∼90%) of the observed variation in mutability genome-wide [24]. In this model, mutability was inversely related to motif size and directly related to the number of repeat units. A strong effect of motif sequence on mutability of microsatellites was observed, which may be due to differences in secondary DNA structure. Within 5-Mb windows, the local genome position of a microsatellite was found to affect mutability approximately 10-fold for mononucleotide and dinucleotide alleles, but only four-fold for tetranucleotide repeats [24].
Evolutionary models of microsatellite mutation have been developed for use in estimates of genetic distances between populations (reviewed in [18]). De novo mutations arising in the germline are predominantly the gain or loss of a single unit, and a bias for the gain of repeat units within short alleles has been observed [23]. However, the rate of deletion mutation increases with allele length, such that long alleles tend to mutate to shorter alleles, thus preventing infinite growth of microsatellites [26,27]. Mutations that interrupt pure repetitive arrays, such as base substitutions and indels, also have been observed, and result in reduced levels of allelic polymorphism [18,28]. Stepwise mutation models assume that additions or deletions of one unit occur at fixed rates as a function of allele length. However, the observed distribution of microsatellite lengths within eukaryotic genomes is best described by a model that incorporates length-dependent slippage together with point mutations [29]. Thus, the observed microsatellites within the human genome likely reflect an equilibrium between expansion and deletion errors of repeat units, and point mutations within the allele that disrupt the repetitive array.
Experimental Analyses of Microsatellite Mutation
Several experimental model systems have been developed to measure microsatellite mutagenesis. In these approaches, artificial repeats of a specified length and sequence are cloned into reporter gene cassettes, either as in-frame or out-of-frame alleles. Mutations within the repeated motifs are then scored as either loss of functional activity (from in-frame alleles) or gain of activity (from out-of-frame alleles) of the downstream reporter gene after replication in cells. In wild-type E. coli, the mutation frequencies of [G/C]10, [GT/CA]10 and [TC/AG]11 alleles were estimated to range from 2 to 5 × 10-7, comparable in magnitude to the downstream reporter gene [30]. In wild-type S. cerevisiae, mutation rates of [G/C]18 and [GT/CA]16 alleles were 5 to 7 × 10−6 [31,32]. Increasing the [GT/CA] allele length resulted in an increased mutation rate [32], and the presence of a single base substitution to interrupt the allele lowered the mutation rate [33]. Integration of a [GT/CA] reporter cassette at various locations within the yeast genome resulted in a 16-fold variation in mutation rate, depending on the site of integration [34]. This magnitude of variation due to genomic features corresponds to that estimated by computational studies [24]. In non-transformed human fibroblasts, estimates of the mutation rate of an integrated [GT/CA]17 allele ranged from 1-10 × 10-7 [35]. Overall, the microsatellite mutation rates measured for pure mononucleotide and dinucleotide alleles in experimental systems are several orders of magnitude lower than those estimated by pedigree analyses of genomic loci. As discussed below, the greater length and rich sequence complexity of the genomic microsatellites analyzed in the latter studies is likely to account for this difference.
We have developed reporter cassettes containing defined, in-frame microsatellite sequences placed within the coding region of the herpes simplex virus type 1 thymidine kinase (HSV-tk) gene. Our system allows us to study microsatellite mutagenesis during DNA replication in human cells ex vivo [36,37] and by purified human DNA polymerases in vitro [38-40]. Our ex vivo system utilizes an oriP-based episomal shuttle vector system to study the relationships between DNA sequence and microsatellite mutagenesis. Replication of oriP-based vectors utilizes cellular replication proteins at both the initiation and elongation stages, and is regulated in a manner similar to chromosomal replication [41]. Importantly, this experimental approach allows us to control two critical parameters that are, at best, working assumptions in the genomic approaches described above. First, the oriP-based shuttle vectors allow us to monitor unbiased mutagenesis in human cells, as we exert no selective pressure on the target sequence during human cell replication. Shuttle vectors are introduced into lymphoblastoid cells, and independent clones are isolated to ensure that the founder cells contained only wild-type vectors. Individual plasmid-bearing clones are expanded 24-35 cell generations, during which time mutational events occur within each plasmid in the absence of genetic selection. After extracting shuttle vector DNA, HSV-tk mutant frequencies are determined for each clone by selection in E. coli. Second, the episomal nature of our vector ensures that genomic features associated with the point of vector integration do not affect HSV-tk mutagenesis, allowing strict analyses of specified microsatellite sequences and lengths. For comparison, we quantitate mutation rates in the artificial microsatellite, relative to the natural downstream HSV-tk coding sequence, which is the same in all vectors.
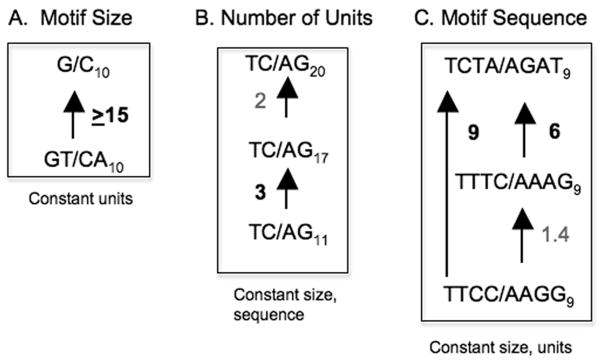
Using this system, we examined the effects of intrinsic DNA features identified by genome-wide studies on microsatellite mutagenesis, after replication in a nontumorigenic human lymphoblastoid cell line (Table 1). As a point of comparison, the median mutation rate estimated for the natural HSV-tk gene is 0.95 × 10-6 (range 0.52-1.8 × 10-6) mutations/cell generation. The median mutation rate of a [GT/CA]10 allele was not significantly higher than that of the HSV-tk gene, consistent with the above studies. Motif size (mono-, di- or tetra) was found to be inversely related to mutability in comparative genome studies; e.g., mononucleotide alleles are more mutable than di- or tetranucleotide alleles [24,25]. We have measured a 15-fold higher mutation rate for the mononucleotide [G/C]10 allele than for the corresponding dinucleotide [GT/CA]10 allele of the same number of repeat units, consistent with the computational studies (Figure 1A). However, this inverse relationship between motif size and mutagenesis was not universal. Keeping constant the total allele length, base composition, and the potential for secondary structure in the comparison, the mutation rate of a [TC/AG]17 dinucleotide (allele length =34 basepairs) was similar to that of a [TTCC/AAGG]9 tetranucleotide (allele length = 36 basepairs), suggesting that motif size is not a factor in mutagenesis of polypyrimidine/polypurine repeats. We also analyzed the effect of the number of repeat units on mutation rate within dinucleotide alleles, holding the motif size and sequence constant (Figure 1B). We observed that the median mutation rate of the [TC/AG] allele increased approximately seven-fold as the number of repeat units within the allele increased 1.8-fold, from 11 to 20 units. This result is consistent with a greater than linear relationship between the number of units and mutation rate, as has been suggested in several previous computational publications [20,24,25], as well as in yeast and E. coli studies [42,43]. Finally, we examined the effect of motif sequence on mutation rate (Figure 1C). Holding the number of repeat units constant, we observed up to a nine-fold variation in mutation rate among the three tetranucleotide alleles examined ([TTCC/AAGG]9, [TTTC/AAAG]9, and [TCTA/AGAT]9,) and a three-fold variation between two dinucleotide alleles. The highest microsatellite mutation rate observed in human cells from our studies to date is ∼5 × 10-5 for the [TCTA/AGAT]9 motif (Table 1). This value compares favorably with the reported mutation rate of a pure [AGAT/TCTA]13 allele on the Y chromosome that was estimated to be ∼6 × 10-4 (based on one observed mutational event) [21], assuming an increasing mutation rate with allele length and/or a ∼10-fold variation in mutation rate due to genome location. This comparison suggests that the discordance between the range of mutation rates estimated by pedigree and experimental analyses is actually due to sampling biases resulting from the precise motif sequences and structures of the loci included in the two types of study [25]. The extent to which a compound locus structure affects the mutability of individual microsatellite motifs can be readily tested using the oriP mutagenesis assay.
Table 1. Allelic variation in mitotic human cell microsatellite mutation rates, as determined in the oriP-tk shuttle vector assay.
| Microsatellite Allele | HSV-tk Mutation Frequencyper Cell Generation × 10-6 | |
|---|---|---|
| Median* | Range | |
| [G/C]10 | 29 (5) | 14-34 |
| [GT/CA]10 [GT/CA]16 |
≤ 1.9 (5) | ≤ 1.6-3.1 |
| 3.4 (5) | 2.1-11 | |
| [TC/AG]11 [TC/AG]17 [TC/AG]20 |
3.2 (6) | 0.59-7.5 |
| 9.8 (6) | 6.0-28 | |
| 21 (3) | 16-25 | |
| [TTCC/AAGG]9 [TTTC/AAAG]9 [TCTA/AGAT]9 |
5.6 (10) | 3.8-20 |
| 35 (9) | 12-57 | |
| 48 (6) | 36-110 | |
Number of independent clones analyzed for DNA sequences changes is indicated in parentheses.
Data are from reference [36] and K.E., unpublished data.
Figure 1. Quantitative effects of intrinsic microsatellite features on human cell mutation rates.
(A). Motif Size; (B). Number of repeat units per allele; (C). Motif Sequence. Arrows point to the microsatellite allele with the higher mutation rate. Values are the relative difference in median mutation rate. Numbers in bold indicate differences that are statistically significant (p≤0.03, Mann-Whitney test). Data are taken from Table 1.
Common microsatellite sequences have the potential for adopting several non-B form DNA conformations, including Z-DNA, H-DNA (triplex DNA) and cruciform structures. Consistent with the formation of triplex DNA, we observed S1 nuclease sensitive DNA conformations within oriP-tk cassettes containing [TC/AG]n, [TTCC/AAGG]9, and [TTTC/AAAG]9 sequences, but not within [GT/CA]10 and [TCTA/AGAT]9 sequences [36]. Secondary structure formation may influence the specificity of mutations arising within the various microsatellite motifs. We observed that the proportion of expansion mutations within a [TC/AG] allele increased as the length of the allele increased, from 39% expansions for 11 units, to 74% for 17 units and 78% for 20 units. A similar bias towards expansion mutations was observed for the [TTCC/AAGG]9 allele (71%), which is similar to the longer [TC/AG] motifs in total allele length, secondary structure, and sequence composition (50% G+C content). In contrast, microsatellite expansion and deletion mutations occurred at similar proportions (∼50% each) within both the [GT/AG]16 and [TCTA/AGAT]9 alleles. A significantly greater incidence of expansion mutations at [TC/AG]20 and [TTCC/AAGG]9 alleles, relative to a [GT/CA]10 allele, was also observed in an E. coli model [44]. These limited studies are consistent with the effect of motif sequence composition on mutability being due, in part, to the production of non-B DNA secondary structures within the repeated elements.
Biochemical Processes Affecting Microsatellite Mutagenesis
Microsatellite mutations have been proposed to occur by slipped strand mispairing within the repetitive DNA sequences [45]. Mechanistically, two distinct pathways of slipped strand mispairing can explain the production of microsatellite mutations. During recombination, unequal crossing over between repetitive arrays located on separate DNA molecules may result in mutant products. Alternatively, DNA strand slippage may occur transiently during DNA synthesis, resulting in the addition or deletion of repeat units within the microsatellite. The characteristics of microsatellite mutation observed in genome analyses are consistent with the polymerase slippage model [2,18]. The genomic and experimental studies summarized above demonstrate that the intrinsic DNA features of motif size, length (number of units), and sequence predominantly affect microsatellite mutagenesis. Here, we describe the molecular pathways relevant to the DNA polymerase slippage model that may underlie this variation in mutagenesis among microsatellite loci. We note that microsatellite mutations do not necessarily have to arise only during replicative (S-phase) DNA synthesis, as DNA polymerases operate during most long tract DNA repair and homologous recombination pathways.
Correction of DNA Polymerase Errors
Postreplication mismatch repair (MMR), which acts during DNA synthesis to remove DNA polymerase errors, is a dominant pathway affecting microsatellite mutation rates in all organisms [42,46-48]. In a yeast model, mismatch repair was observed to efficiently remove mutations within a [GT/CA] allele up to 25 units in length, but was less efficient at repairing a longer allele (∼50 units) [32]. MMR correction efficiency is affected both by motif size and sequence composition. In E. coli, the [G/C] mononucleotide mutation frequency per cell generation was increased ∼104-fold in MMR deficient cells, compared to MMR proficient cells, while mutation frequencies for the [GT/CA] or [TC/AG] dinucleotides were increased only ∼103-fold [30]. The loss of MMR in E. coli destabilizes tetranucleotide alleles by only ∼10-fold (K.E., unpublished observations). In S. cerevisiae, loss of MMR destabilized microsatellites with a motif size of 1-8 basepairs; however, the absolute quantitative effect was again dependent upon motif size [31]. MMR was demonstrated to remove errors within [A/T]10 alleles more efficiently than within [G/C]10 mononucleotide alleles [49].
The 3′ to 5′ proofreading exonuclease activity of replicative DNA polymerases is another important biochemical pathway that limits the introduction of polymerase errors into the genome. The proofreading exonucleases of replicative polymerases δ and ε were shown to limit errors within short mononucleotide [A/T] sequences [50]. However, the magnitude of proofreading correction is much lower than that of MMR, and differs between the two polymerases. Moreover, the relationship of proofreading correction to microsatellite motif size, length and sequence has not been formally investigated. Overall, the available studies suggest that the efficiency of repairing premutational intermediates within microsatellites is affected by motif size, length and sequence, thereby contributing to the observed variation in cellular microsatellite mutagenesis.
DNA Polymerase Error Rates within Microsatellites
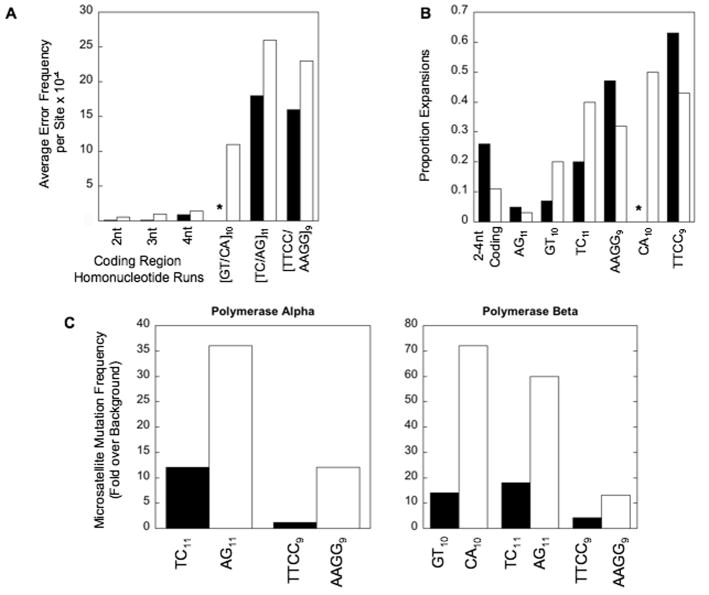
Because human polymerases differ significantly in fidelity [51] and genome function [52], the identity of the polymerase(s) synthesizing nascent microsatellite DNA is expected to impact microsatellite stability. We used the in vitro HSV-tk DNA polymerase microsatellite mutation assay [38] to examine the contribution of polymerase errors to the mutational variation caused by intrinsic microsatellite features. DNA polymerase α-primase (pol α-primase) and DNA polymerase β (pol β) produce errors within microsatellite sequences at rates that are 10- to 100-fold higher than the rate of errors produced at short repeated sequences in the downstream HSV-tk coding region (frameshifts) (Figure 2A). A predominance of one unit errors was observed within the alleles examined. The proportion of errors larger than one unit ranged from 6% to 25%, and was dependent on the motif sequence and the polymerase [38,39]. Analyses of both polα-primase and pol β-induced mutations within motifs of varying sequence suggest a greater than linear relationship between the polymerase error frequency and the number of units within the repeated sequences [38], consistent with computational models. The in vitro polymerase assay is being used to definitively determine the mathematical relationship between microsatellite allele length and mutagenesis, by altering the number of units in the target sequence in an incremental, systematic manner. Such an approach can also be used to establish the relationship between allele length and mutational specificity, as computational studies have suggested that deletion errors will dominate in long alleles.
Figure 2. Microsatellite mutagenesis by purified polymerases α-primase and β.
(A). Frameshift error frequencies at 2-4 nucleotide repeated sequences within the HSV-tk coding region are compared to artificial microsatellite sequences for pol α-primase (filled bars) and pol β (open bars). The average polymerase error frequency for complementary strands at each template sequence is graphed. Asterisk, not determined. (B). Specificity of pol α-primase (filled bars) and pol β (open bars) within the indicated frameshift and microsatellite regions. The proportion of expansion errors, relative to the total number of insertion and deletion errors, at each site is graphed. Asterisk, not determined. (C). Strand bias of errors produced by pol α-primase and pol β within microsatellite sequences. Data are from Refs. [38,39].
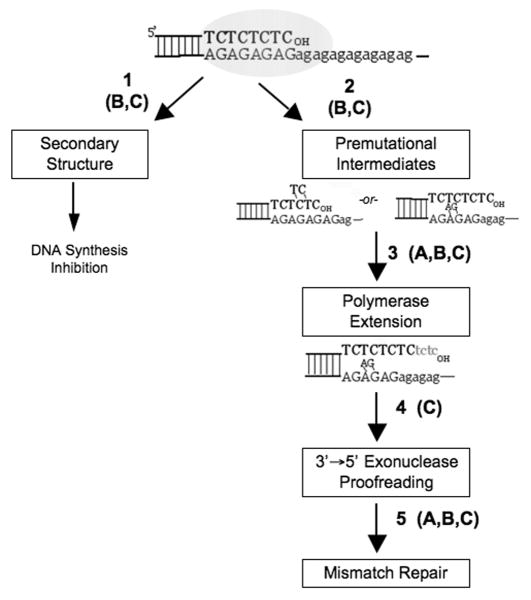
Although DNA polymerases produce strand slippage errors within microsatellite alleles, motif sequence composition directly affects the mutational outcomes (i.e., relative proportions of deletions and expansions) within each repetitive motif (Figure 2B). Moreover, a DNA strand bias exists for mammalian DNA polymerase errors within complementary microsatellite sequences (Figure 2C). The error frequencies for both pol α-primase and pol β were greater when polypurine alleles (AG and AAGG) served as the templates for DNA synthesis, relative to the complementary polypyrimidine (TC and TTCC) alleles [38,39]. This bias is specific for microsatellite sequences, as neither polymerase displayed a strand bias for template purine versus template pyrimidine repeated sequences within the coding region. However, the precise direction of the strand bias may be variable, depending on the identity of the replicating DNA polymerase and its inherent error discrimination mechanisms. Such studies are under investigation. Two models could explain mutational strand biases in microsatellite sequences. Microsatellite sequences in the starting single-stranded DNA templates may form secondary structures 5′ to the advancing DNA polymerase. DNA synthesis by purified polymerases across the base of secondary structures has been shown to result in deletion errors [53]. The physical structure of misaligned, pre-mutational intermediates is a second factor that may lead to mutational biases. For example, solution studies have shown that purine bulges adopt intrahelical positions, whereas single pyrimidine bulges adopt extrahelical structures [54]. DNA polymerases may favor certain structures for continued extension synthesis. These two factors, formation of secondary structures in the DNA template and polymerase utilization of pre-mutational intermediates, are not mutually exclusive, and both may contribute to biases in microsatellite mutagenesis (Figure 3).
Figure 3. Sources of mutational biases observed in microsatellite sequences.
The known effects of intrinsic features (Figure 1) on the efficiency of each step in the DNA slipped strand mispairing model are indicated. A, motif size; B, number of repeat units; C, motif sequence. See text for details.
DNA Polymerases Produce Interrupted Microsatellite Alleles
Intriguingly, 30%-40% of pol β errors within the [GT]10, [TC]11, and [TTCC]9 microsatellites were other than unit-length gains or losses [38]. These errors included single base substitutions within the repeated sequence, as well as single base deletions and complex base substitution and deletions, all of which produced interrupted alleles from pure microsatellites. We used a specialized mutagenesis assay allowing improved detection of interruption mutations to examine polymerase errors within a mononucleotide allele [40]. For human DNA polymerase κ (pol κ), 78% of errors produced within a [T]11 microsatellite sequences were the insertion of a single dGMP or dCMP residue to create an interrupted allele. In contrast, pol β produced primarily the expected unit-based deletions within the same microsatellite sequence. The mutational specificity difference between the polymerases is specific to the microsatellite region, as the two polymerases produced a similar proportion and type of frameshift errors within the HSV-tk coding region. These results demonstrate that DNA polymerases in vitro can directly create interrupted alleles from a pure microsatellite. In yeast, base interruptions of microsatellites result in increased genetic stability, relative to pure alleles [33]. We suggested that DNA synthesis by DNA pol κ will promote microsatellite stability, because the major type of error it produces (interruptions) within the microsatellite are protective mechanistically. The extent to which the frequency of interruptions is affected by motif sequence and the identity of the polymerase is under investigation.
The in vitro data for DNA polymerase errors within common microsatellites support the model of a microsatellite life-cycle [28]. Our direct experimental data using mammalian polymerases show that microsatellite-specific strand slippage errors occur at a high rate and produce expansion and deletion mutations (Figure 2). In the genome, microsatellites reach an equilibrium of allele lengths due to the accumulation of base substitution and short indel mutations that interrupt pure arrays. We have observed directly the degeneration of common microsatellite alleles through polymerase base substitution and indel errors that create shorter, interrupted alleles [38,40].
DNA Secondary Structures and Common Microsatellite Mutagenesis
Repetitive microsatellite DNA comprises a significant component of genome replication that must occur faithfully each cell cycle. Thus, elucidating the replication dynamics through common microsatellite sequences and the key polymerases involved is crucial for understanding genome stability. Experimentally, specific polymerase-nucleic acid interactions can be analyzed by quantitative primer extension studies, which measure how the enzymes travel along a specific substrate. DNA polymerase pausing patterns, or sites of increased DNA synthesis termination, are unique to each enzyme and template sequence. Pause sites can be caused by several factors, including primary DNA sequence and secondary structure.
Using this approach, we observed that the degree of pol α-primase DNA synthesis termination within mono-, di- and tetranucleotide microsatellites is highly dependent upon the sequence composition of the microsatellite allele, and pausing patterns are distinct for each microsatellite examined [39,40]. In order to determine whether polymerase pausing may be a causative factor in microsatellite mutagenesis, we compared pause sites and error rates [39]. A greater degree of polα-primase termination was observed within the polypurine [AG]11 and [AAGG]9 alleles than within the polypyrimidine [TC]11 and [TTCC]9 sequences. polα-primase mutational analyses also revealed strand biases for both the [TC/AG]11 and the [TTCC/AAGG]9 pairs (Figure 2C). In each case, polymerase error frequencies were increased when the purine sequence served as the template strand, relative to when the pyrimidine sequence served as the template strand. Therefore, we conclude that a positive correlation exists between overall synthesis termination and polymerase error frequency, for the polymerases and microsatellites examined.
Several studies have shown that formation of DNA secondary structures plays an important role in mutagenesis (reviewed in [55,56]). Strong DNA pol α-primase and pol κ pause sites occurring within a [TC]20 allele are caused by triplex DNA formed between the nascent DNA primer-template duplex and the downstream single-stranded DNA template [39,40]. However, no polymerase pausing was observed for a shorter [TC]11 allele, suggesting that microsatellites must exceed a threshold length before the formation of an intramolecular triplex is inhibitory to DNA synthesis. The sequence-specific trend towards expansions within certain microsatellite alleles that was observed in human cells (see above) is not consistent with computational studies that have suggested the rate of deletion mutation increases with allele length [26,27]. Perhaps, the allele length that must be reached before deletions exceed expansions is greater than 20 units. Alternatively, the biochemical analyses of polymerase pausing suggest a model wherein the formation of an intramolecular triplex structure within long [TC/AG]17 and [TC/AG]20 alleles overrides the inherent tendency for deletion errors within long alleles. In such a model, DNA polymerase inhibition due to occlusion of the nascent 3′OH within a triple-stranded structure would be resolved by retrograde slippage of the primer strand.
We also observed prominent, polar DNA polymerase pauses within a short poly(dT) microsatellite sequence during DNA synthesis by both pol κ and polα-primase [40]. This pause site is sequence-dependent, and could not be explained by intramolecular triplex DNA formation. Instead, this inhibition was proposed to result from structural changes to, or DNA bending of, the duplex primer-template stem, thus disrupting polymerase-DNA interactions critical for catalysis. The degree to which the observed sequence-specific pausing by DNA polymerases within common microsatellite sequences impacts the rate of replication fork movement is unknown. Strong strand biases for replicative DNA polymerase termination may lead to non-uniform rates of leading and lagging strand DNA synthesis. Quantitatively, the greatest intensity of DNA polymerase pausing that we have measured to date is within poly(dT) sequences, the most abundant microsatellite in the human genome. Thus, primary DNA sequence and accompanying DNA structural changes may be a factor contributing to nonrandom replication fork movements observed in vivo [57]. The available biochemical studies demonstrate that DNA secondary structure can account for at least some of the DNA polymerase pause sites observed within common microsatellites. Importantly, the inhibition of DNA synthesis caused by secondary structure formation within common microsatellites appears to be polymerase-independent, and may be a factor contributing to differences in mutational specificity that has been observed among various microsatellite motifs.
Summary
Mutation rates of common microsatellites found in the human genome are extremely variable. The combined mutation rate estimates from analyses of parent-child allele transmissions and experimental determinations in nontumorigenic human cells range from a low of ∼10-6 to a high of ∼10-2 mutations per locus per generation. The location of an allele within the genome accounts for only a factor of ∼10 in this variation. The vast majority of the observed variation in mutagenesis is due to features intrinsic to the repeated DNA sequence, including motif size, length (number of units), and sequence composition. In addition, allele structure (e.g., pure, compound, or interrupted) may play a key role in determining microsatellite stability. Several steps in the molecular pathway relevant to the DNA polymerase slippage model may underlie the observed variation in cellular mutagenesis among microsatellite loci (Figure 3). Common microsatellites can adopt non-B form DNA secondary structures in a sequence and length-dependent manner (Fig. 3, Step 1). Available DNA polymerase studies have shown that enzymatic pausing within microsatellites is correlated with both the absolute frequency and the specificity of errors within microsatellite sequences. The mechanistic relationship between secondary structure formation and mutagenesis within common mono-, di- and tetranucleotide alleles will need to be established by further experimental testing.
The DNA strand slippage model predicts that the microsatellite mutation frequency will increase as a function of the number of repeat units, due to stabilization of pre-mutational intermediates by surrounding correct basepairs. The precise mathematical relationship between the number of repeat units and the mutation rate has yet to be rigorously established, but available publications suggest a greater than linear relationship between mutagenesis and length, once a threshold length is exceded. The rate of formation of premutational strand slippage intermediates and/or their stability is also expected to be dependent upon the precise motif sequence (Fig. 3, Step 2). Once formed, premutational intermediates must be used as a substrate for continued DNA synthesis by a polymerase in order for a mutation to arise. Polymerase error frequencies for misalignment mutations within a microsatellite reflect the efficiency of such extension synthesis. Pronounced effects of motif composition on DNA polymerase error rates and DNA strand biases in polymerase error rates are apparent (Figure 2), which may reflect polymerase discrimination at this step (Fig. 3, Step 3). The effect of motif size on polymerase extension synthesis has not been directly examined, but the size of the motif unit will directly affect the structure of the bulged intermediate. Simplistically, one base bulges within mononucleotides may be less disruptive to the duplex primer-stem than two base or four base bulges within di- and tetranucleotides, respectively. Regarding the effect of motif length on DNA polymerase errors, increasing the length of the repetitive allele will increase the physical distance between the misaligned (bulged) nucleotides and the 3′OH of the nascent strand, thereby increasing the efficiency of continued DNA synthesis from the pre-mutational intermediate. However, this component of microsatellite error frequency is relevant only over a finite distance dictated by the polymerase-DNA binding interaction, which will be unique to each enzyme.
Once a polymerase error has been incorporated into the nascent DNA strand, the 3′ to 5′ proofreading exonuclease activity and MMR proteins can act to remove errors before a mutation is fixed in the genome (Fig. 3, Steps 4 and 5). Available experimental studies have shown that the efficiency of both steps varies according to the sequence of the microsatellite motif, and that the efficiency of MMR is influenced by motif size and allele length. We point out that several of the above five steps may be involved in mutational biases to differing extents, depending on the specific microsatellite allele. Elucidating the molecular mechanisms underlying microsatellite variation is necessary for future research models which seek to assess an individual's disease risk based on cis-acting genome sequence features and trans-acting proteins that promote genome stability.
Acknowledgments
We are grateful to the Jake Gittlen Cancer Research Foundation for their continued support of our research over the past 15 years. Our sincere thanks to the anonymous reviewers of this manuscript, whose thoughtful comments and criticisms helped to improve the article.
This work was supported, in part, by NIH Grant RO1 CA100060 (to K.A. E.).
Abbreviations used
- HSV-tk
herpes simplex virus type 1 thymidine kinase
- Indel
insertion/deletion
- pol α-primase
calf thymus DNA polymerase α-primase
- pol β
recombinant rat DNA polymerase β
- pol κ
human DNA polymerase κ
- MMR
postreplication mismatch repair
References
- 1.Subramanian S, Mishra RK, Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li YC, Korol AB, Fahima T, Beiles A, Nevo E. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol. 2002;11:2453–2465. doi: 10.1046/j.1365-294x.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- 3.Katti MV, Ranjekar PK, Gupta VS. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Molecular Biology and Evolution. 2001;18:1161–1167. doi: 10.1093/oxfordjournals.molbev.a003903. [DOI] [PubMed] [Google Scholar]
- 4.Metzgar D, Bytof J, Wills C. Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res. 2000;10:72–80. [PMC free article] [PubMed] [Google Scholar]
- 5.Bacolla A, Larson JE, Collins JR, et al. Abundance and length of simple repeats in vertebrate genomes are determined by their structural properties. Genome Res. 2008;18:1545–1553. doi: 10.1101/gr.078303.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers GK, MacAvoy ES. Microsatellites: consensus and controvery. Comparative Biochemistry and Physiology Part B. 2000;126:455–476. doi: 10.1016/s0305-0491(00)00233-9. [DOI] [PubMed] [Google Scholar]
- 7.Waterson RH, Consortium MGS. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 8.Li YC, Korol AB, Fahima T, Nevo E. Microsatellites within genes: structure, function, and evolution. Mol Biol Evol. 2004;21:991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- 9.Hui J, Hung LH, Heiner M, et al. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 2005;24(11):1988–1998. doi: 10.1038/sj.emboj.7600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rockman MV, Wray GA. Abundant raw material for cis-regulatory evolution in humans. Molecular Biology and Evolution. 2002;19:1991–2004. doi: 10.1093/oxfordjournals.molbev.a004023. [DOI] [PubMed] [Google Scholar]
- 11.Kashi Y, King DG. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006;22:253–259. doi: 10.1016/j.tig.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Chu CS, Trapnell BC, Curristin S, Cutting GR, Crystal RG. Genetic basis of variable exon 9 skipping in cystic fibrosis transmembrane conductance regulator mRNA. Nat Genet. 1993;3:151–156. doi: 10.1038/ng0293-151. [DOI] [PubMed] [Google Scholar]
- 13.Cuppens H, Lin W, Jaspers M, et al. Polyvariant mutant cystic fibrosis transmembrane conductance regulator genes. The polymorphic (Tg)m locus explains the partial penetrance of the T5 polymorphism as a disease mutation. J Clin Invest. 1998;101:487–496. doi: 10.1172/JCI639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gebhardt F, Zanker KS, Brandt B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. Journal of Biological Chemistry. 1999;274:13176–13180. doi: 10.1074/jbc.274.19.13176. [DOI] [PubMed] [Google Scholar]
- 15.Buerger H, Packeisen J, Boecker A, et al. Allelic length of a CA dinucleotide repeat in the egfr gene correlates with the frequency of amplifications of this sequence--first results of an inter-ethnic breast cancer study. J Pathol. 2004;203:545–550. doi: 10.1002/path.1542. [DOI] [PubMed] [Google Scholar]
- 16.Gangwal K, Sankar S, Hollenhorst PC, et al. Microsatellites as EWS/FLI response elements in Ewing's sarcoma. Proc Natl Acad Sci U S A. 2008;105:10149–10154. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikitina TV, Nazarenko SA. Human microsatellites: mutation and evolution. Russian Journal of Genetics. 2004;40:1064–1079. [PubMed] [Google Scholar]
- 18.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5(6):435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 19.Albertini RJ, Nicklas JA, O'Neill JP, Robison SH. In vivo somatic mutations in humans: measurement and analysis. Annu Rev Genet. 1990;24:305–326. doi: 10.1146/annurev.ge.24.120190.001513. [DOI] [PubMed] [Google Scholar]
- 20.Brinkmann B, Klintschar M, Neuhuber F, Huhne J, Rolf B. Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am J Hum Genet. 1998;62:1408–1415. doi: 10.1086/301869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dupuy BM, Stenersen M, Egeland T, Olaisen B. Y-chromosomal microsatellite mutation rates: differences in mutation rate between and within loci. Hum Mutat. 2004;23:117–124. doi: 10.1002/humu.10294. [DOI] [PubMed] [Google Scholar]
- 22.Kayser M, Roewer L, Hedman M, et al. Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am J Hum Genet. 2000;66:1580–1588. doi: 10.1086/302905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai Y, Sun F. The relationship between microsatellite slippage mutation rate and the number of repeat units. Mol Biol Evol. 2003;20:2123–2131. doi: 10.1093/molbev/msg228. [DOI] [PubMed] [Google Scholar]
- 24.Kelkar YD, Tyekucheva S, Chiaromonte F, Makova KD. The genome-wide determinants of human and chimpanzee microsatellite evolution. Genome Res. 2008;18:30–38. doi: 10.1101/gr.7113408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webster MT, Smith NG, Ellegren H. Microsatellite evolution inferred from human-chimpanzee genomic sequence alignments. Proc Natl Acad Sci U S A. 2002;99:8748–8753. doi: 10.1073/pnas.122067599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellegren H. Heterogeneous mutation processes in human microsatellite DNA sequences. Nature Genetics. 2000;24:400–402. doi: 10.1038/74249. [DOI] [PubMed] [Google Scholar]
- 27.Xu X, Peng M, Fang Z, Xu X. The direction of microsatellite mutations is dependent upon allele length. Nature Genetics. 2000;24:396–399. doi: 10.1038/74238. [DOI] [PubMed] [Google Scholar]
- 28.Buschiazzo E, Gemmell NJ. The rise, fall and renaissance of microsatellites in eukaryotic genomes. Bioessays. 2006;28:1040–1050. doi: 10.1002/bies.20470. [DOI] [PubMed] [Google Scholar]
- 29.Kruglyak S, Durrett RT, Schug MD, Aquadro CF. Equilibrium distributions of microsatellite repeat length resulting from a balance between slippage events and point mutations. Proceedings of the National Academy of Sciences USA. 1998;95:10774–10778. doi: 10.1073/pnas.95.18.10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacob KD, Eckert KA. Escherichia coli DNA polymerase IV contributes to spontaneous mutagenesis at coding sequences but not microsatellite alleles. Mutat Res. 2007;619:93–103. doi: 10.1016/j.mrfmmm.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sia EA, Kokoska RJ, Dominska M, Greenwell P, Petes TD. Microsatellite instability in yeast: dependence on repeat unit size and mismatch repair genes. Molecular and Cellular Biology. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wierdl M, Dominska M, Petes TD. Microsatellite instability in yeast: dependence on the length of the microsatellite. Genetics. 1997;146:769–779. doi: 10.1093/genetics/146.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petes TD, Greenwell PW, Dominska M. Stabilization of microsatellite sequences by variant repeats in the yeast Saccharomyces cerevisiae. Genetics. 1997;146:491–498. doi: 10.1093/genetics/146.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hawk JD, Stefanovic L, Boyer JC, Petes TD, Farber RA. Variation in efficiency of DNA mismatch repair at different sites in the yeast genome. Proc Natl Acad Sci U S A. 2005;102:8639–8643. doi: 10.1073/pnas.0503415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roques CN, Boyer JC, Farber RA. Microsatellite mutation rates are equivalent in normal and telomerase-immortalized human fibroblasts. Cancer Res. 2001;61:8405–8407. [PubMed] [Google Scholar]
- 36.Eckert KA, Yan G, Hile SE. Mutation Rate and Specificity Analysis of Tetranucleotide Microsatellite DNA Alleles in Somatic Human Cells. Molecular Carcinogenesis. 2002;34:140–150. doi: 10.1002/mc.10058. [DOI] [PubMed] [Google Scholar]
- 37.Hile SE, Yan G, Eckert KA. Somatic mutation rates and specificities at TC/AG and GT/CA microsatellite sequences in nontumorigenic human lymphoblastoid cells. Cancer Research. 2000;60:1698–1703. [PubMed] [Google Scholar]
- 38.Eckert KA, Mowery A, Hile SE. Misalignment-mediated DNA polymerase beta mutations: comparison of microsatellite and frame-shift error rates using a forward mutation assay. Biochemistry. 2002;41:10490–10498. doi: 10.1021/bi025918c. [DOI] [PubMed] [Google Scholar]
- 39.Hile SE, Eckert KA. Positive correlation between DNA polymerase alpha-primase pausing and mutagenesis within polypyrimidine/polypurine microsatellite sequences. Journal of Molecular Biology. 2004;335:745–759. doi: 10.1016/j.jmb.2003.10.075. [DOI] [PubMed] [Google Scholar]
- 40.Hile SE, Eckert KA. DNA polymerase kappa produces interrupted mutations and displays polar pausing within mononucleotide microsatellite sequences. Nucleic Acids Res. 2008;36:688–696. doi: 10.1093/nar/gkm1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugden B. In the beginning: a viral origin exploits the cell. Trends in Biochemical Sciences. 2002;27:1–3. doi: 10.1016/s0968-0004(01)02032-1. [DOI] [PubMed] [Google Scholar]
- 42.Levinson G, Gutman G. High frequencies of short frameshifts in poly-CA/TG tandem repeats borne by bacteriophage M13 in Escherichia coli K12. Nucleic Acids Research. 1987;15:5323–5338. doi: 10.1093/nar/15.13.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weirdl M, Dominska M, Petes TD. Microsatellite instability in yeast: dependence on the length of the microsatellite. Genetics. 1997;146:769–779. doi: 10.1093/genetics/146.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eckert KA, Yan G. Mutational analyses of dinucleotide and tetranucleotide microsatellites in Escherichia coli: influence of sequence on expansion mutagenesis. Nucleoic Acids Research. 2000;28:2831–2838. doi: 10.1093/nar/28.14.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levinson G, Gutman GA. Slipped strand mispairing: a major mechanism for DNA sequence evolution. Molecular Biology Evolution. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 46.Strand M, Prolla TA, Liskay RM, Petes TD. Destabilization of tracts of simple repetitve DNA in yeast by mutations affecting DNA mismatch repair. Nature (Lond) 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 47.Narayanan L, Fritzell JA, Baker SM, Liskay RM, Glazer PM. Elevated levels of mutation in multiple tissues of mice deficient in the DNA mismatch repair gene PMS2. Proc Natl Acad Sci U S A. 1997;94:3122–3127. doi: 10.1073/pnas.94.7.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vilkki S, Tsao JL, Loukola A, et al. Extensive somatic microsatellite mutations in normal human tissue. Cancer Res. 2001;61:4541–4544. [PubMed] [Google Scholar]
- 49.Gragg H, Harfe BD, Jinks-Robertson S. Base composition of mononucleotide runs affects DNA polymerase slippage and removal of frameshift intermediates by mismatch repair in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:8756–8762. doi: 10.1128/MCB.22.24.8756-8762.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tran HT, Keen JD, Kricker M, Resnick MA, Gordenin DA. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Molecular and Cellular Biology. 1997;17:2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kunkel TA, Bebenek K. DNA replication fidelity. Annual Review of Biochemistry. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 52.Sweasy JB, Lauper JM, Eckert KA. DNA polymerases and human diseases. Radiat Res. 2006;166:693–714. doi: 10.1667/RR0706.1. [DOI] [PubMed] [Google Scholar]
- 53.Ripley LS. Frameshift mutation: determinants of specificity. Annual Review of Genetics. 1990;24:189–213. doi: 10.1146/annurev.ge.24.120190.001201. [DOI] [PubMed] [Google Scholar]
- 54.Joshua-Tor L, Frolow F, Appella E, Hope H, Rabinovich D, Sussman JL. Three-dimensional structures of bulge-containing DNA fragments. J Mol Biol. 1992;225:397–431. doi: 10.1016/0022-2836(92)90929-e. [DOI] [PubMed] [Google Scholar]
- 55.Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability. Mutat Res. 2006;598:103–119. doi: 10.1016/j.mrfmmm.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 56.Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends Biochem Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Raghuraman MK, Winzeler EA, Collingwood D, et al. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]