Abstract
DNA-damage-induced SOS mutations arise when Escherichia coli DNA polymerase (pol) V, activated by a RecA nucleoprotein filament (RecA*), catalyses translesion DNA synthesis. Here we address two longstanding enigmatic aspects of SOS mutagenesis, the molecular composition of mutagenically active pol V and the role of RecA*. We show that RecA* transfers a single RecA–ATP stoichiometrically from its DNA 3′-end to free pol V (UmuD′2C) to form an active mutasome (pol V Mut) with the composition UmuD′2C–RecA–ATP. Pol V Mut catalyses TLS in the absence of RecA* and deactivates rapidly upon dissociation from DNA. Deactivation occurs more slowly in the absence of DNA synthesis, while retaining RecA–ATP in the complex. Reactivation of pol V Mut is triggered by replacement of RecA–ATP from RecA*. Thus, the principal role of RecA* in SOS mutagenesis is to transfer RecA–ATP to pol V, and thus generate active mutasomal complex for translesion synthesis.
Pol V is a low-fidelity DNA polymerase1,2 induced as part of the SOS regulon in E. coli in response to DNA damage3. The replicative polymerase, pol III, typically stalls when it encounters a DNA template lesion, arresting movement of the replication fork. One pathway that enables restoration of fork movement involves pol V, which replaces pol III on the sliding β-clamp4–6 and catalyses translesion DNA synthesis (TLS). Pol V copies numerous types of lesions7, but in a mutagenic manner8,9. After TLS, pol III resumes normal replication.
The pol V complex consists of UmuD′2C10,11. Both in vitro1,2,12,13 and in vivo14–16, pol V activity requires the assembly of an active RecA filament on single-stranded (ss) DNA, termed RecA*17. The biological functions of RecA* in strand exchange during homologous recombination and in mediating cleavage of the repressor protein LexA and UmuD during the SOS response are well understood17. In contrast, the biochemical role of RecA* in pol-V-dependent mutagenic TLS remains poorly characterized.
Proposals for the role of RecA* in TLS have evolved from positioning UmuD′2C on primer/template (p/t) DNA proximal to a lesion18–20, to a dynamic interaction involving displacement of RecA* filaments on the template by an advancing pol V4, to a model in which RecA* need not be located in cis on the template strand being copied, but can instead assemble on a separate ssDNA strand to transactivate pol V for TLS21. What had not been contemplated in previous models, and what previous experimental designs were unable to detect, is whether a RecA* filament might function remotely from the site of translesion synthesis. In this unexplored scenario, RecA* would transfer a RecA monomer and ATP from the 3′-tip of RecA* to free UmuD′2C, thereby converting pol V (UmuD′2C) to an activated form that can be used elsewhere for TLS.
Formation of activated pol V Mut
In earlier studies, either free RecA protein or RecA filament was added to pol V and p/t DNA substrate. Here, we first form RecA* by incubating RecA with biotinylated ssDNA bound to a streptavidin-agarose resin matrix in the presence of ATPγS (adenosine 5′[γ-thio] triphosphate; see Methods). Pol V (UmuD′2C) is then incubated with RecA* in the absence of p/t DNA, forming an activated pol V species that can be isolated, pol V Mut (Supplementary Figs 1 and 2). Only once RecA* is removed by centrifugation is p/t DNA added, allowing DNA synthesis in the absence of RecA* (Fig. 1a, b). In these studies, the p/t template is a hairpin with a 3-nucleotide overhang21 instead of, for example, an oligonucleotide annealed to a ssDNA circle; this prevents formation of activated RecA* on the p/t DNA itself because the hairpin lacks free ssDNA on which RecA* can assemble (the 3-nucleotide overhang is too short to permit this). Throughout, we use the properties of two key RecA mutant proteins, highly active RecA E38KΔC17 (also known as RecA730ΔC17; ref. 22), and mutagenically inactive RecA F117S (also known as RecA1730; refs 15, 23).
Figure 1. DNA synthesis by pol V Mut or pol V transactivated by RecA*.
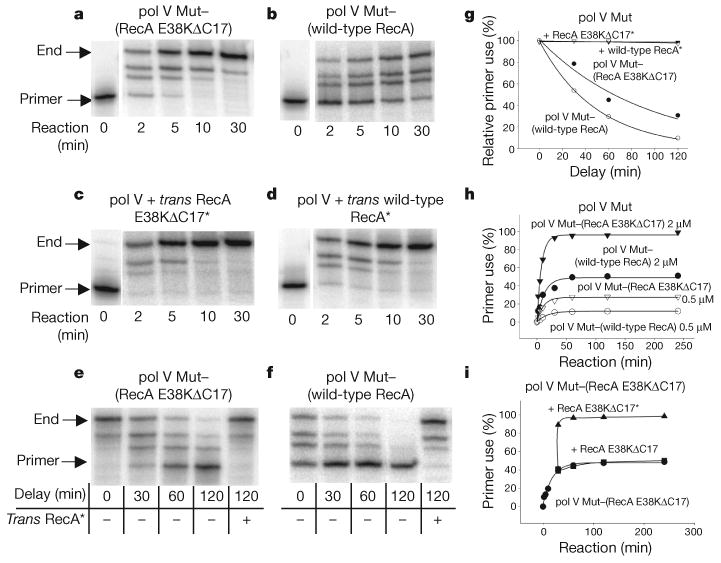
The p/t DNA is a hairpin containing a 3-nucleotide template overhang. a, b, DNA synthesis by pol V Mut–(RecA E38KΔC17) (a) or pol V Mut–(wild-type RecA) (b) in the absence of RecA*. c, d, DNA synthesis by pol V transactivated by RecA E38KΔC17* (c) or wild-type RecA* (d). e, f, DNA synthesis by pol V Mut–(RecA E38KΔC17) (e) or pol V Mut–(wild-type RecA) (f) undergoing deactivation as a function of time. g, Deactivation of pol V Mut–(RecA E38KΔC17) and pol V Mut–(wild-type RecA) measured by quantifying DNA synthesis obtained from data in panels e and f, respectively. Complete reactivation of deactivated pol V Mut is observed by addition of trans RecA* at each delay time. h, pol V Mut performs one round of DNA synthesis and cannot reinitiate synthesis on separate p/t DNA substrate. The concentration of p/t DNA is 1 μM. We estimate that the fraction of active polymerase in the reaction is 50% for pol V Mut–(RecA E38KΔC17) and 25% for pol V Mut–(wild-type RecA). i, Addition of trans RecA E38KΔC17* enables pol V Mut–(RecA E38KΔC17) to reinitiate DNA synthesis on a separate p/t DNA substrate. Cycling also occurs using pol V Mut–(wild-type RecA) in conjunction with trans wild-type RecA* (data not shown). Trans RecA* is at 1 μM when present.
Removal of the RecA*-resin by centrifugation allows activated pol V Mut to remain in solution (Supplementary Fig. 2). Filaments containing RecA E38KΔC17 activate pol V with approximately twofold higher efficiency (Fig. 1a) than wild-type RecA* (Fig. 1b). Pol V avidly copies p/t DNA when RecA* is present in trans, as reported previously21 (Fig. 1c, d). Thus, exposure of UmuD′2C to RecA* is required for pol V activity. Most importantly, pol V remains highly active after removal of RecA* from the reaction (Fig. 1a, b).
To investigate how long pol V Mut remains active in the absence of RecA*, we incubated the enzyme in standard reaction buffer at 37 °C for various times before adding the p/t DNA and dNTP substrates to initiate DNA synthesis. Polymerase activity decreases as the delay time increases (Fig. 1e, f). The decrease in activity is approximately exponential, and is similar for pol V Mut activated with either RecA E38KΔC17* or wild-type RecA* (Fig. 1g). The loss in activity is reversible; addition of RecA* in trans at each time point restores pol V Mut to a fully activated state (Fig. 1g). ATP hydrolysis has no role in pol V activation or in the activity decay, because synthesis by pol V Mut and the loss of activity during the time delay is identical when RecA* is assembled with ATP, ATPγS or AMP-PNP (β-γ-imidoadenosine 5′-triphosphate; Supplementary Figs 3 and 4).
Each active pol V Mut complex can promote only one round of DNA synthesis (Fig. 1h, i). A plot of primer use against time shows that, after the addition of one or more nucleotides, pol V Mut cannot reinitiate synthesis on a different p/t DNA (Fig. 1h, i). Deactivated pol V Mut is reactivated by adding RecA* in trans (Fig. 1i), but not by adding either RecA (Fig. 1i) or ssDNA alone (data not shown). After synthesis, deactivated pol V Mut dissociates into solution (Supplementary Fig. 5). Pol V Mut catalyses TLS in the absence of RecA*, but without pre-activation, pol V is inactive even on undamaged DNA (Supplementary Fig. 6).
Pol V Mut consists of UmuD′2C–RecA–ATP
Pol V Mut activated with RecA E38KΔC17* synthesizes DNA equally well with or without a nucleotide cofactor, ATP or ATPγS, added post-activation (Fig. 2a). In marked contrast, pol V Mut activated with wild-type RecA* is inactive in the absence of added ATP or ATPγS (Fig. 2b). These properties indicate that activation entails formation of a pol V–RecA complex. A denaturing gel of activated pol V Mut provides visual evidence that UmuD′2C forms a complex with RecA (Fig. 2c). A plot of RecA released from RecA* as a function of increasing pol V concentration shows a 1:1:1 ratio of RecA:UmuC:UmuD′2 over a tenfold range of pol V Mut (Fig. 2d). When resin-bound RecA* is pre-incubated in the absence of pol V and centrifuged, an insignificant amount of RecA is released from the resin into the supernatant (Fig. 2c, lane 1) compared with the amount of RecA bound to pol V Mut (Fig. 2c, lanes 2–5). Quantification of ATPγS released into the supernatant also indicates that a RecA–ATP complex is transferred from RecA* to pol V to form pol V Mut, in a 1:1:1 stoichiometry of UmuD′2C–RecA–ATP (Fig. 2c, d).
Figure 2. Activated pol V Mut complex is composed of UmuD′2C–RecA–ATP.

a, b, DNA synthesis by pol V Mut–(wild-type RecA) (b), but not pol V Mut–(RecA E38KΔC17) (a), requires the presence of ATPγS or ATP. c, d, Activated pol V Mut–(wild-type RecA) complex contains UmuC, UmuD′ and wild-type RecA, in a 1:1:1 ratio. e, Fl-wild-type RecA and ATPγS remain bound in deactivated pol V Mut–(Fl-wild-type RecA) complexes as measured by affinity chromatography. f, Reactivation of pol V Mut–(Fl-wild-type RecA) is achieved by replacing Fl-wild-type RecA and [35S]ATPγS bound to deactivated pol V Mut–(Fl-wild-type RecA) with unlabelled wild-type RecA and ATPγS from wild-type RecA*. Reactivated pol V Mut–(Fl-wild-type RecA) complexes were isolated as in panel e.
To obtain direct evidence that RecA is part of pol V Mut, we formed pol V Mut using fluorescein-labelled (Fl-) wild-type RecA (Fig. 2e); its behaviour is indistinguishable from pol V Mut activated with unlabelled wild-type RecA (Supplementary Fig. 7). We identified the components of the activated and deactivated pol V Mut species by affinity chromatography of pol V Mut–(Fl-wild-type RecA) either immediately after activation or after delay times of 1 or 2 h. After elution from the column, Fl-wild-type RecA and [γ-35S]ATP are retained in a complex with UmuC and UmuD′ irrespective of delay time (Fig. 2e). Thus, deactivation of pol V Mut is neither caused nor accompanied by a concomitant loss of RecA or ATPγS. We verified the composition of reactivated pol V Mut by incubating pol V–(Fl-wild-type RecA) complexes isolated after a 2 h delay time with increasing amounts of unlabelled wild-type RecA* (Fig. 2f). After adsorption and elution of pol V–RecA from the column, the Fl-wild-type RecA and [γ-35S]ATP are readily replaced with unlabelled wild-type RecA and ATPγS, with greater replacement occurring as the concentration of unlabelled wild-type RecA* is increased (Fig. 2f). These data confirm that UmuD′2C, RecA and ATP are present in 1:1:1 stoichiometry (Fig. 2d).
We used laser multi-angle light scattering (MALS)24 to evaluate the molecular mass of pol V Mut–(wild-type RecA). Pol V alone was activated by incubating UmuD′2C with trans wild-type RecA* bound to resin. After centrifugation, the supernatant containing a mixture of pol V Mut–(wild-type RecA) and non-activated pol V was resolved by size-exclusion chromatography. The column effluent was passed directly on-line into a MALS detection system. Two light scattering peaks were observed (Fig. 3a, upper trace). The peak eluting at 17.3 min indicates a molecular mass of 113 kDa, in accord with the value predicted for UmuD′2C–(wild-type RecA) (110 kDa). The peak eluting at 18.4 min indicates a molecular mass of 73 kDa, in accord with UmuD′2C (72 kDa), and coinciding with the single peak exhibited by non-activated pol V (UmuD′2C; Fig. 3a, lower trace). Analysis of the two peaks shows that UmuC, UmuD′ and wild-type RecA are present in the 113 kDa peak, whereas only UmuC and UmuD′ are in the 73 kDa peak (Fig. 3b). Thus, the MALS data provide independent verification that the intact pol V Mut–(wild-type RecA) complex contains UmuD′2C bound to wild-type RecA, in accord with the affinity column binding and elution analysis of pol V Mut–(wild-type RecA) (Fig. 2e, f).
Figure 3. Determination of the molecular mass of pol V Mut–(wild-type RecA) by MALS.

a, After wild-type-RecA*-mediated transactivation of UmuD′2C and removal of wild-type RecA*, the mixture of pol V Mut–(wild-type RecA) and non-activated pol V was resolved by size-exclusion chromatography (upper trace), and the molecular mass corresponding to each peak was measured by MALS. Non-activated pol V run separately on the silica gel elutes at 18.4 min (lower trace). b, Silver-stained SDS–polyacrylamide gel showing the protein composition from the two peaks contained in panel a (upper trace).
Activation requires transfer from the 3′-proximal tip of RecA*
We have previously shown that pol V is activated only when it encounters a RecA molecule located at the 3′-proximal tip of RecA*21. We determined whether RecA is transferred to pol V from one or both RecA* filament ends by incubating pol V with RecA* having either the 3′- or 5′-proximal tip exposed (Supplementary Fig. 8a). After removing RecA* by centrifugation, the supernatant containing pol V Mut was separated by SDS–polyacrylamide gel electrophoresis (Fig. 4a). The data show that one molecule each of RecA and ATPγS is transferred to pol V from both ends of RecA* (Fig. 4a and Supplementary Fig. 8b). However, pol V becomes activated only if wild-type RecA or RecA E38KΔC17 is transferred from the 3′-proximal tip of RecA* (Fig. 4b). RecA F117S is also transferred to pol V from either the 3′- or 5′-tip, but fails to activate pol V in both cases (Fig. 4a, b).
Figure 4. Pol V activation requires transfer of a RecA molecule from the DNA 3′ end of RecA*.

a, The amount of RecA transferred to pol V from exposed 3′- or 5′-tips of RecA* are similar for wild-type RecA*, RecA E38KΔC17* and RecA F117S*. b, DNA synthesis catalysed by pol V Mut–(wild-type RecA) or pol V Mut–(RecA E38KΔC17) occurs when RecA is transferred from the 3′-RecA* tip (left gel). Pol V Mut cannot synthesize DNA when RecA transfer takes place from the 5′-RecA* tip (right gel). Mutagenically inactive RecA F117S* fails to activate pol V.
Discussion
We propose a comprehensive biochemical model depicting pol V activation, deactivation and reactivation (Fig. 5). Pol V (UmuD′2C) first migrates to a preformed RecA* filament. It is activated by the transfer of a 3′-RecA–ATP subunit from RecA* to pol V to form activated pol V Mut, that is: UmuD′2C–RecA–ATP. Pol V Mut scans the genome for sites where replication has stalled. When a suitable primer/template is located, pol V Mut binds and promotes DNA synthesis in the absence of RecA*. Pol V Mut is inactivated by one round of primer extension. Subsequent RecA*-mediated reactivation of pol V Mut occurs when the complex again encounters a RecA* filament, and replaces the ‘used’ RecA–ATP, which is still bound, but in an inactive conformation, with ‘new’ RecA–ATP (Fig. 5). The model has the unique feature that there is no need for an active RecA* filament to be present during TLS; also, although ATP is required, ATP hydrolysis is not.
Figure 5. Model depicting pol V activation, deactivation and reactivation.
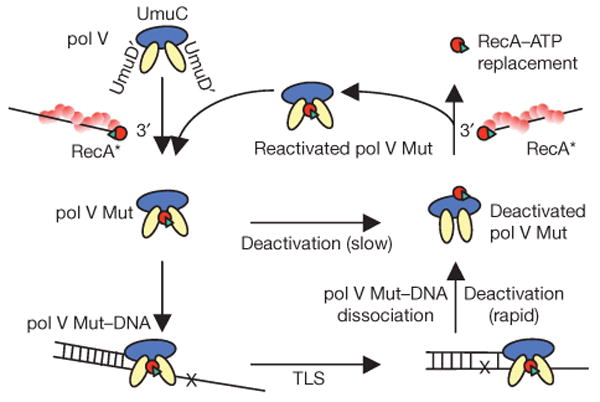
Pol V (UmuD′2C) is barely active in the absence of RecA*. A molecule of RecA (red circle) and ATP (green triangle) is transferred from the DNA 3′-end of RecA* to form a mutasomal complex (pol V Mut) containing UmuD′2C–RecA–ATP; this catalyses TLS in the absence of RecA*. After TLS, pol V Mut undergoes rapid deactivation upon dissociation from DNA, with RecA and ATP retained in the complex. When pol V Mut is activated, we surmise that RecA–ATP is bound to UmuD′2, and that dissociation from p/t DNA triggers a repositioning of RecA–ATP to bind with UmuC to deactivate pol V Mut. Reactivation of pol V Mut involves a replacement of RecA–ATP from the RecA* 3′-tip. Free pol V Mut undergoes slow deactivation, with RecA and ATP retained in the complex. A damaged DNA base is shown as a black cross in the template strand.
Pol-V-mediated TLS requires the presence of RecA* both in vivo14–16 and in vitro1,2,12,13. Since the late 1980s the most important but refractory questions have been ‘what is the molecular composition of the pol V mutasome?’ and ‘what is RecA* doing?’. The requirement for RecA* initially suggested that a RecA filament would form in cis4,13. However, RecA filaments formed in cis strongly impede pol-V-mediated DNA synthesis4. Activation in trans resolves the difficult situation of pol V replication of DNA with RecA bound to it21. However, transactivation introduces new structural and topological problems associated with a RecA filament bound to DNA in one genomic location interacting continuously with a pol V replicating in a DNA gap somewhere else. And finally, how is pol V inactivated when the SOS response is no longer needed?
The model in Fig. 5 answers these questions. The role of RecA* is both to promote the autocatalytic cleavage of UmuD to UmuD′, and to activate the resulting UmuD′2C complex by transferring a single RecA subunit to pol V from its 3′-proximal tip. Activated pol V Mut can migrate to wherever it is needed; no RecA* filament needs be near the site where TLS actually takes place. In fact, no RecA nucleoprotein filament needs to participate directly in TLS, either in cis or in trans, a fact obscured in previous studies because such filaments were always present when TLS was observed. Participation in TLS as a subunit of pol V Mut becomes the only filament-independent role documented for RecA. Rapid inactivation of pol V Mut after TLS, or slower inactivation if no suitable p/t is encountered, ensures that pol-V-catalysed error-prone DNA synthesis will cease soon after the RecA* filaments supporting the SOS response (and pol V reactivation) are gone. The mutational load associated with pol V function is thereby minimized, effectively restricting it to periods when SOS is induced.
We have shown that transfer from the 3′-tip of RecA* is an absolute requirement for pol V activation (Fig. 4b). Consistent with in vivo data23, the mutagenically inactive RecA F117S is unable to activate pol V for DNA synthesis (Fig. 4b, left gel), despite the transfer of RecA F117S from the 3′-tip of RecA F117S* (Fig. 4a, left gel). Notably, the mutation in RecA F117S is located at the surface facing 3′ on a RecA filament end20.
In the model (Fig. 5), we show RecA bound to UmuD′2 when pol V Mut is either activated or reactivated, and to UmuC when pol V Mut is deactivated. This assignment is based on our previous identification of two distinct modes of RecA binding to pol V25. RecA and pol V form a stably bound complex (dissociation constant Kd ≈ 250 nM) through the UmuC subunit in the absence of DNA; we have called this ‘mode 1’ binding25. RecA also binds to pol V, with a similar apparent Kd, through its UmuD′2 subunit, requiring DNA and ATP, but not ATP hydrolysis, referred to as ‘mode 2’ binding25. Therefore, we speculate that RecA binds proximally to UmuD′2 during DNA synthesis, and that dissociation from p/t DNA triggers a rapid conformational rearrangement, repositioning RecA to bind with UmuC. When a newly activated pol V Mut is free in solution, the presumed conformational rearrangement shifting RecA binding from UmuD′2 to UmuC occurs much more slowly (Fig. 1g). Subsequent reactivation of pol V Mut (Fig. 1e–g) could occur by a RecA*-mediated replacement of RecA–ATP with the deactivated form of pol V Mut (Fig. 2f).
A decade before the discovery of pol V, the term “mutasome” was coined by Echols to define the protein components that assemble at a lesion in the presence of a stalled replication complex to mediate TLS19. We now propose that the mutasome envisioned by Echols is a ‘stand alone’ activated multiprotein complex, pol V Mut, composed of UmuD′2C–RecA–ATP.
Methods Summary
Pol V was activated by incubation with RecA* immobilized on streptavidin-coated resin. RecA* was removed by centrifugation, leaving pol V Mut in the supernatant. The polymerase activity of pol V or pol V Mut was measured as the rate of deoxynucleotide addition to the 3′-OH end of a hairpin p/t DNA containing a 32P label at its 5′-end. The sequences of template and biotinylated oligomers are shown in Supplementary Fig. 9. Gel band intensities were quantified by phosphorimaging with IMAGEQUANT software (Molecular Dynamics), and primer utilization was calculated from the integrated gel band intensities of extended p/t DNA divided by the total DNA intensity. Deactivated and reactivated His–pol V Mut were bound to a nickel-affinity column and washed twice with reaction buffer. Bound pol V Mut was eluted from the column by washing with buffer containing imidazole. The amount of Fl-RecA and [γ-35S]ATP remaining bound to pol V Mut were visualized by SDS–PAGE or by urea PAGE, respectively. For MALS analysis, pol V Mut was subjected to size-exclusion chromatography using a silica-based column (KW-802.5, Shodex). Chromatography was performed at 0.5ml min−1 and the column effluent was passed directly on-line into a MALS detection system (Dawn Heleos, Wyatt Technology) to determine the molecular masses of eluted species; fractions were collected manually after passing through the light scattering detector. Data analysis was performed with ASTRA software.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants to M.F.G. (ES12259; R37GM21422) and M.M.C. (GM32335), and funds from the NICHD/NIH Intramural Research Program to K.K. and R.W. The authors thank R. Britt for preparation of the RecA E38KΔC17 protein used in this study. MALS data were collected using the USC NanoBiophysics Core Facility, with the aid and cooperation of N. Chelyapov. We thank J. Bertram for his help in performing the MALS experiment and analysing the data. We thank J. Petruska for his comments.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions Q.J. performed the experiments and experimental analyses; K.K. performed the construction and purification of His-tagged pol V; M.M.C. provided the purified RecA E38KΔC17 protein; Q.J. and M.F.G. designed the experiments with input from M.M.C. and R.W.; M.F.G., Q.J., M.M.C. and R.W. wrote the manuscript.
References
- 1.Tang MJ, et al. UmuD′2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J Biol Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 3.Friedberg EC, et al. DNA Repair and Mutagenesis. 2nd. ASM Press; 2006. pp. 463–555. [Google Scholar]
- 4.Pham P, Bertram JG, O'Donnell M, Woodgate R, Goodman MF. A model for SOS-lesion-targeted mutations in Escherichia coli. Nature. 2001;409:366–370. doi: 10.1038/35053116. [DOI] [PubMed] [Google Scholar]
- 5.Becherel OJ, Fuchs RP, Wagner J. Pivotal role of the β-clamp in translesion DNA synthesis and mutagenesis in E. coli cells. DNA Repair. 2002;1:703–708. doi: 10.1016/s1568-7864(02)00106-4. [DOI] [PubMed] [Google Scholar]
- 6.Lenne-Samuel N, Wagner J, Etienne H, Fuchs RP. The processivity factor β controls DNA polymerase IV traffic during spontaneous mutagenesis and translesion synthesis in vivo. EMBO Rep. 2002;3:45–49. doi: 10.1093/embo-reports/kvf007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang M, et al. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- 8.Kato T, Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet. 1977;156:121–131. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- 9.Steinborn G. Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. I. Isolation of uvm mutants and their phenotypical characterization in DNA repair and mutagenesis. Mol Gen Genet. 1978;165:87–93. doi: 10.1007/BF00270380. [DOI] [PubMed] [Google Scholar]
- 10.Bruck I, Woodgate R, McEntee K, Goodman MF. Purification of a soluble UmuD′C complex from Escherichia coli: cooperative binding of UmuD′C to single-stranded DNA. J Biol Chem. 1996;271:10767–10774. doi: 10.1074/jbc.271.18.10767. [DOI] [PubMed] [Google Scholar]
- 11.Woodgate R, Rajagopalan M, Lu C, Echols H. UmuC mutagenesis protein of Escherichia coli: purification and interaction with UmuD and UmuD′. Proc Natl Acad Sci USA. 1989;86:7301–7305. doi: 10.1073/pnas.86.19.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang M, et al. Biochemical basis of SOS-induced mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD′2C mutagenic complex and RecA protein. Proc Natl Acad Sci USA. 1998;95:9755–9760. doi: 10.1073/pnas.95.17.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujii S, Gasser V, Fuchs RP. The biochemical requirements of DNA polymerase V-mediated translesion synthesis revisited. J Mol Biol. 2004;341:405–417. doi: 10.1016/j.jmb.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 14.Nohmi T, Battista JR, Dodson LA, Walker GC. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci USA. 1988;85:1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutreix M, et al. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriol. 1989;171:2415–2423. doi: 10.1128/jb.171.5.2415-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sweasy JB, Witkin EM, Sinha N, Roegner-Maniscalco V. RecA protein of Escherichia coli has a third essential role in SOS mutator activity. J Bacteriol. 1990;172:3030–3036. doi: 10.1128/jb.172.6.3030-3036.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bridges BA, Woodgate R. Mutagenic repair in Escherichia coli. X. The umuC gene product may be required for replication past pyrimidine dimers but not for the coding error in UV-mutagenesis. Mol Gen Genet. 1984;196:364–366. doi: 10.1007/BF00328073. [DOI] [PubMed] [Google Scholar]
- 19.Echols H, Goodman MF. Mutation induced by DNA damage: a many protein affair. Mutat Res. 1990;236:301–311. doi: 10.1016/0921-8777(90)90013-u. [DOI] [PubMed] [Google Scholar]
- 20.Sommer S, Boudsocq F, Devoret R, Bailone A. Specific RecA amino acid changes affect RecA–UmuD′C interaction. Mol Microbiol. 1998;28:281–291. doi: 10.1046/j.1365-2958.1998.00803.x. [DOI] [PubMed] [Google Scholar]
- 21.Schlacher K, Cox MM, Woodgate R, Goodman MF. RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature. 2006;442:883–887. doi: 10.1038/nature05042. [DOI] [PubMed] [Google Scholar]
- 22.Eggler AL, Lusetti SL, Cox MM. The C terminus of the Escherichia coli RecA protein modulates the DNA binding competition with single-stranded DNA-binding protein. J Biol Chem. 2003;278:16389–16396. doi: 10.1074/jbc.M212920200. [DOI] [PubMed] [Google Scholar]
- 23.Dutreix M, Burnett B, Bailone A, Radding CM, Devoret R. A partially deficient mutant, recA1730, that fails to form normal nucleoprotein filaments. Mol Gen Genet. 1992;232:489–497. doi: 10.1007/BF00266254. [DOI] [PubMed] [Google Scholar]
- 24.Wyatt PJ. Light scattering and the absolute characterization of macromolecules. Anal Chim Acta. 1993;272:1–40. [Google Scholar]
- 25.Schlacher K, et al. DNA polymerase V and RecA protein, a minimal mutasome. Mol Cell. 2005;17:561–572. doi: 10.1016/j.molcel.2005.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


