Summary
Accurate chromosome segregation depends on sister kinetochores coming under tension when they make bioriented attachments to microtubules from opposite poles. The spindle checkpoint halts the cell cycle in response to defects in generating proper attachments or tension on kinetochores [1, 2], although the precise signal that triggers the checkpoint is unclear because tension and attachment are coupled [3]. The target of the checkpoint is the Cdc20 protein that initiates the anaphase promoting complex (APC)-dependent degradation of the anaphase inhibitor Pds1/securin [4]. Although the molecular details of spindle checkpoint activation are still being elucidated, phosphorylation by at least four kinases is a crucial requirement [5]. However, less is known about the mechanisms that silence the checkpoint once kinetochores biorient. Here, we show that the catalytic subunit of the budding yeast protein phosphatase I, Glc7, regulates exit from the checkpoint. Glc7 overexpression prevents spindle checkpoint activation in response to both tension and attachment defects. Although glc7 mutant cells are able to efficiently release from a non-checkpoint-mediated metaphase arrest, they are uniquely sensitive to transient spindle checkpoint activation due to a failure in spindle checkpoint exit. We therefore propose that PP1 activity silences the checkpoint by reversing key phosphorylation events.
Results and Discussion
Glc7 overexpression leads to chromosome missegregation
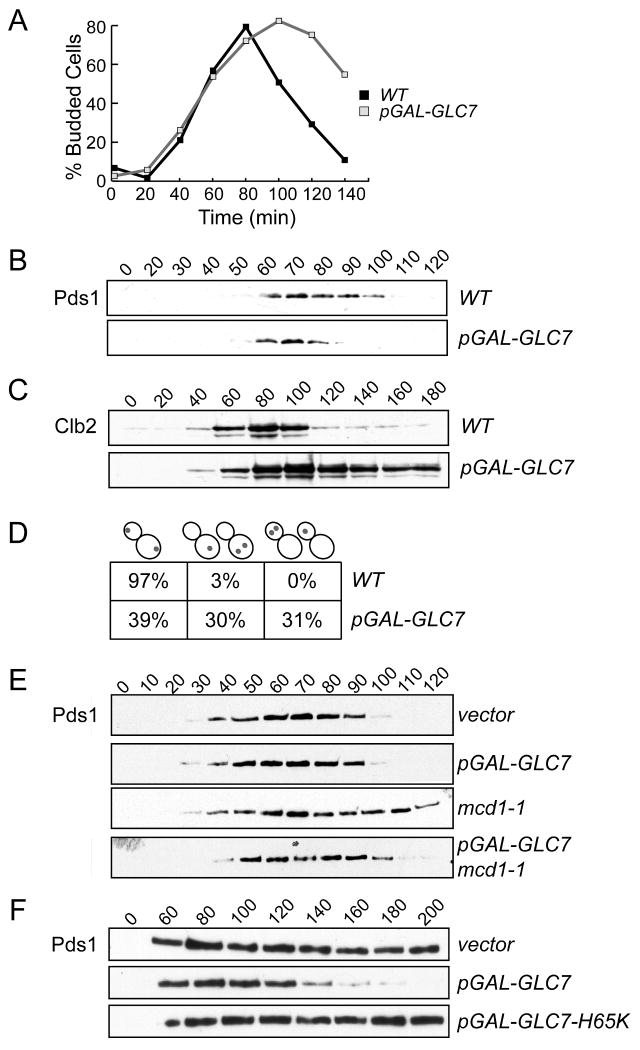
We hypothesized that exit from the spindle checkpoint would require phosphatase activity to reverse the phosphorylation that occurs upon checkpoint activation. Because the budding yeast spindle checkpoint response to defects in kinetochore tension requires Ipl1/Aurora protein kinase activity [6], we tested whether overexpression of the Glc7 protein that opposes Ipl1 function [7] would prevent cells from engaging the checkpoint. First, we analyzed the phenotype of cells overexpressing Glc7 by releasing WT and pGAL-GLC7 cells from G1 into galactose media to induce Glc7 overexpression. We monitored cell cycle progression by scoring the budding index and found that both strains began budding at similar times, but cells overexpressing Glc7 accumulated with large-buds (Fig 1A). To determine whether these cells were delayed in metaphase or anaphase, we monitored the levels of the anaphase inhibitor Pds1 and the mitotic cyclin Clb2 in a similar experiment. Although there was little difference in the kinetics of Pds1 accumulation and destruction between WT and pGAL-GLC7 cells (Fig 1B), Clb2 levels rose with similar timing in both strains but remained high in the cells that overexpressed Glc7 (Fig 1C). Clb2 is stabilized when cells do not properly position the mitotic spindle via Bub2-mediated inhibition of the mitotic exit network (for reviews, see [8, 9]), so we analyzed Clb2 levels in bub2 pGAL-GLC7 cells. We found that Clb2 remained high in these cells (Fig S1), indicating that Glc7 overexpression prevents mitotic exit in a manner that is downstream of Bub2 activity and the spindle position checkpoint. Although the role of Glc7 in mitotic exit is not clear, future research may reveal a previously unknown function for PP1 in regulating mitotic exit.
Figure 1.
Glc7 overexpression leads to chromosome missegregation and bypasses the spindle checkpoint. A and B. WT and pGAL-GLC7 strains (SBY2076 and SBY2077) were arrested in G1 and released into galactose media. The percentage of budded cells was scored every 20 min (A) and Pds1 levels (B) were analyzed by immunoblotting. C. Cells containing Clb2-myc with GLC7 (SBY3116) or pGAL-GLC7 (SBY3113) were treated as in (A). D. WT (SBY214) and pGAL-GLC7 (SBY2973) cells with GFP-marked ChrIV were treated as in (A) and scored by microscopy for sister chromatid segregation at anaphase. Cells were categorized based on sister segregation to opposite poles (left), the mother cell (middle), or the daughter cell (right). E. WT and mcd1-1 strains containing a control (WT, SBY1650 and mcd1-1, SBY1849) or pGAL-GLC7 vector (WT, SBY1293 and mcd1-1, SBY1848) were released from G1 into galactose media at 37 degrees and Pds1 levels were monitored at the indicated time points (min). F. WT cells containing a vector control (SBY8259), pGAL-GLC7 (SBY8260) or catalytically inactive pGAL-GLC7-H65K (SBY8261) were released from G1 into nocodazole at 30 degrees and lysates were immoblotted for Pds1 at the indicated time points.
Because cells overexpressing Glc7 entered anaphase with normal timing, we tested whether chromosomes segregated properly. WT and pGAL-GLC7 cells containing fluorescently marked ChrIV [10] were released from G1 into galactose and analyzed for segregation at anaphase (Fig 1D). Glc7 overexpression caused ∼60% of the cells to segregate ChrIV to the same pole instead of opposite poles. This phenotype is similar to ipl1-321 mutant cells that segregate sisters to the same pole due to a defect in kinetochore biorientation [11], although ipl1-321 mutant cells show a bias in segregation toward the bud that is not observed when Glc7 is overexpressed [12].
Glc7 overexpression bypasses the tension and attachment checkpoints
Our finding that Glc7 overexpression causes chromosome missegregation without stabilizing Pds1 suggested that it might prevent spindle checkpoint activation. Ipl1 activity is required for the checkpoint when kinetochores are not under tension [6], so we tested whether Glc7 overexpression also bypasses the tension checkpoint. We created a tension defect using a temperature sensitive mutation in the Mcd1/Scc1 protein that joins sister chromatids [13, 14]. Although kinetochores can bind to microtubules in these cells, the spindle checkpoint is activated because tension cannot be generated on unlinked sister chromatids [15, 16]. We arrested WT and mcd1-1 cells containing pGAL-GLC7 or a control vector in G1 and then released them into galactose media to induce Glc7 at the non-permissive temperature. Spindle checkpoint activation was monitored by analyzing Pds1. As expected, Pds1 levels cycled similarly in wild-type cells regardless of whether Glc7 was overexpressed (Fig 1E). However, although Pds1 was stabilized in the mcd1-1 mutant, it was destroyed when Glc7 was overexpressed in mcd1-1 cells. Therefore, high levels of the Glc7 phosphatase prevent spindle checkpoint activation in response to tension defects, consistent with a role in reversing Ipl1 phosphorylation.
In budding yeast, mutants in the IPL1 gene can engage the spindle checkpoint as long as there are unattached kinetochores [6, 17]. We therefore tested whether Glc7 overexpression could bypass the spindle checkpoint in response to unattached kinetochores that were created by the addition of the microtubule depolymerizing drug nocodazole. Wild-type cells with or without pGAL-GLC7 were arrested in G1 and then released into galactose media containing nocodazole. As a control, we also analyzed cells containing a catalytically dead pGAL-GLC7-H65K mutant [18]. We confirmed that microtubules were efficiently depolymerized in the strains by analyzing GFP-Tub1 by microscopy (data not shown). However, although Pds1 was stabilized in control cells containing the vector or catalytically inactive Glc7-H65K, it cycled in cells overexpressing wildtype Glc7 (Fig 1F). Therefore, in contrast to ipl1 mutants, Glc7 overexpression acts similarly to spindle checkpoint mutants that prevent checkpoint arrest in the presence of tension or attachment defects. These data are consistent with the observation that Ipl1/Aurora kinase activity is required for the spindle checkpoint in response to attachment defects in some other organisms [19-21]. One possibility is that Ipl1/Aurora is required for all spindle checkpoint responses in budding yeast, but its role in the response to attachment defects has not been detected because it is necessary to use conditional mutants that retain residual function [22]. An alternative, and not mutually exclusive possibility, is that Glc7 overexpression reverses the phosphorylation of kinases in addition to Ipl1/Aurora, consistent with the requirement for multiple kinases in spindle checkpoint activation [5].
Glc7 is required for exit from Mps1-induced spindle checkpoint
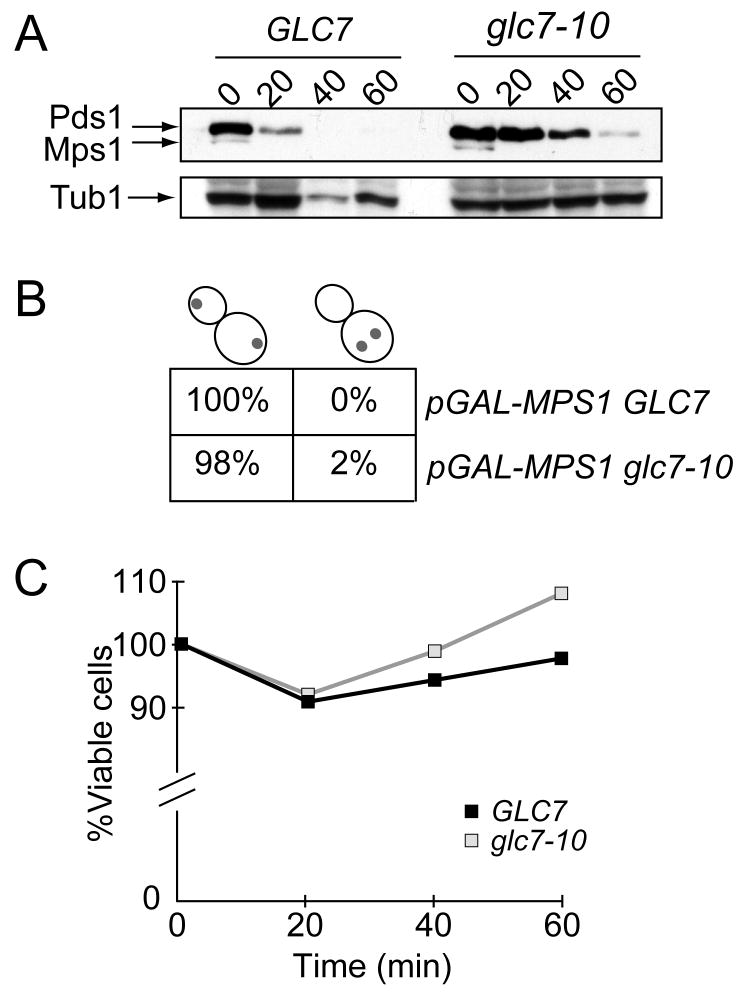
Our observation that Glc7 overexpression prevented activation of the spindle checkpoint suggested that endogenous levels of Glc7 may be required to exit from a spindle checkpoint arrest. Although it was previously reported that glc7 mutants stabilize Pds1 in a checkpoint-dependent manner [23, 24], it was not clear whether this reflects a role for Glc7 in turning off the spindle checkpoint and/or reversing Ipl1-mediated phosphorylation of kinetochore proteins to stabilize bioriented attachments [7, 23-26]. Therefore, to separate the role of Glc7 in kinetochore-microtubule attachments from a potential role in silencing the spindle checkpoint, we utilized cells overexpressing the Mps1 protein kinase. Mps1 overexpression causes constitutive activation of the spindle checkpoint in the absence of detectable spindle or kinetochore defects and it does not depend on kinetochores for checkpoint activation [27, 28]. This allowed us to assay the role of Glc7 in spindle checkpoint recovery after normal bioriented kinetochore attachments were achieved. We arrested pGAL-MPS1 GLC7 and pGAL-MPS1 glc7-10 cells in galactose media at the permissive temperature to allow cells to establish bioriented attachments and a spindle checkpoint arrest. The cells were then shifted to the non-permissive temperature to inactivate glc7-10 and released into glucose media to repress Mps1. While Mps1 and Pds1 were degraded within 20 min of release from the spindle checkpoint arrest in GLC7 cells, Pds1 degradation was delayed up to 40 minutes in glc7-10 cells despite normal kinetics of Mps1 repression (Fig 2A). To ensure that the cells had achieved normal bioriented attachments, we analyzed the segregation of a fluorescently marked chromosome and found that it segregated to opposite poles in both the GLC7 and glc7-10 cells that entered anaphase (Fig 2B). As additional confirmation that chromosome segregation was normal, we also assayed the viability of the cells as they were released from the checkpoint arrest and found no difference between the strains (Fig 2C). Therefore, cells crippled for Glc7 activity appear to be defective in exiting a spindle checkpoint arrest after they have made bioriented attachments.
Figure 2.
Glc7-10 mutants are delayed in exit from a Gal-Mps1-induced spindle checkpoint arrest. A. GLC7 (SBY8289) and glc7-10 mutant (SBY8290) cells containing pGAL-MPS1 were arrested in galactose media at 23 degrees for 3 hours to activate the spindle checkpoint. The cells were then shifted to 35 degrees to inactivate Glc7-10 for 30 min and subsequently released into glucose media at 35 degrees. The levels of Pds1 (top arrow) and Mps1 (middle arrow) were monitored at the indicated time points. Tubulin is shown as a loading control. B. Segregation of a fluorescent chromosome to opposite poles (left) or the same pole (right) was scored in the cells from (A) that had entered anaphase 60 min after release into glucose media. C. The experiment described in (A) was performed on GLC7 (SBY3854) and glc7-10 mutant (SBY3856) cells that were plated for viability at 23 degrees on glucose media.
Defects in PP1 sensitize cells to increased Mps1 kinase activity
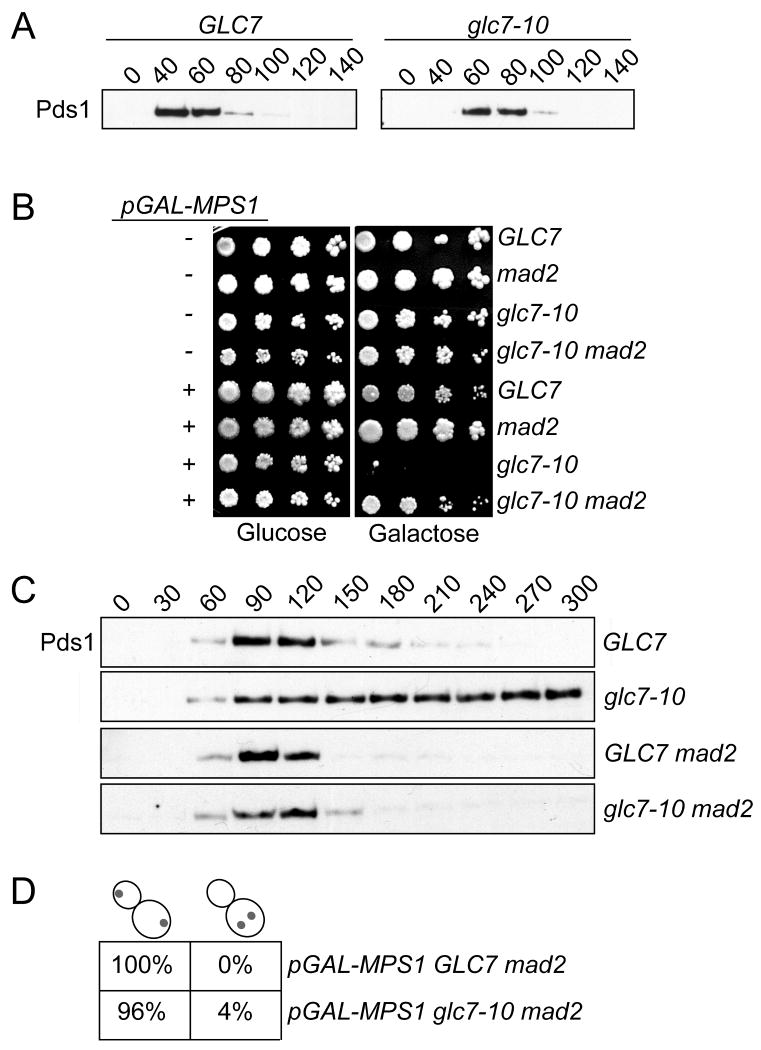
While our data suggested a role for Glc7 in recovery from checkpoint activation, we had to perform the experiment at a non-permissive temperature where glc7-10 mutants are known to make defective kinetochore-microtubule attachments [24, 29]. Although we allowed cells to establish bioriented attachments prior to shifting cells to the high temperature, we could not eliminate the possibility that the stabilization of Pds1 was due to transient activation of the checkpoint because of an undetectable kinetochore defect. Indeed, glc7-10 mutant cells activate the spindle checkpoint and stabilize the Pds1 protein at the non-permissive temperature ((Fig S2) and [24]). To address this concern, we looked for a semi-permissive temperature where glc7-10 mutant cells no longer triggered the spindle checkpoint. At 30 degrees, we found that there was a 20 min delay in both Pds1 accumulation and destruction in glc7-10 cells relative to WT (Fig 3A). When we monitored the budding index, we confirmed that glc7-10 cells take ∼20 min longer than WT cells to enter the cell cycle when released from a pheromone arrest at 30 degrees (Fig S3). When this delay in cell cycle entry is taken into account, Pds1 levels cycle similarly in WT and glc7-10 cells at 30 degrees.
Figure 3.
Glc7-10 cells are sensitive to increased Mps1 activity due to spindle checkpoint activation. A. GLC7 (SBY8132) and glc7-10 (SBY2083) cells containing Pds1-myc were arrested in G1 at 23 degrees and then released to 30 degrees. Pds1 levels were monitored by immunoblot. B. Five-fold serial dilutions of the following strains were plated on glucose or galactose media at 30 degrees: WT (SBY1308), mad2 (SBY2377), glc7-10 (SBY1306), glc7-10 mad2 (SBY2400), pGAL-MPS1 (SBY3670), pGAL-MPS1 mad2 (SBY3844), pGAL-MPS1 glc7-10 (SBY3680) and pGAL-MPS1 glc7-10 mad2 (SBY3846). C. pGAL-MPS1 GLC7 (SBY3854) or pGAL-MPS1 glc7-10 (SBY3856) cells with either MAD2 (SBY3854 and SBY3856) or mad2Δ (SBY7883 and SBY7884) were released from a G1 arrest at 23 degrees into galactose media at 30 degrees. After one hour, glucose was added to repress Mps1. Pds1 levels were monitored every 30 min. D. Chromosome IV segregation to opposite poles (left) or the same pole (right) was monitored in large-budded cells at 180 min in the experiment described in (C) for pGAL-Mps1 GLC7 mad2 (SBY8137) and pGAL-MPS1 glc7-10 mad2 (SBY8136) cells.
The establishment of a temperature where the glc7-10 mutant no longer activates the spindle checkpoint due to defective kinetochores allowed us to more carefully address the requirement for Glc7 activity in exit from the checkpoint. Although high levels of Mps1 expression are lethal due to constitutive activation of the spindle checkpoint, lower levels delay the cell cycle creating a mild growth defect [28]. We reasoned that defects in a phosphatase required to exit the checkpoint would sensitize cells to low levels of Mps1 expression. We therefore analyzed the growth of cells containing an integrated pGAL-MPS1 gene in the presence and absence of the glc7-10 temperature sensitive mutation at 30 degrees. As expected, the growth of the pGAL-MPS1 strain was normal on glucose media but showed a modest defect on galactose plates (Fig 3B). The growth inhibition depended on spindle checkpoint activity because it was suppressed when the MAD2 checkpoint gene was disrupted. Strikingly, although the growth of pGAL-MPS1 glc7-10 cells was almost completely inhibited in the presence of galactose, viability was restored in the absence of MAD2. These data strongly suggest that the growth defect is related to continued activation of the spindle checkpoint.
To determine whether there is a sustained spindle checkpoint response in pGAL-MPS1 glc7-10 cells, we analyzed Pds1 levels. pGAL-MPS1 and pGAL-MPS1 glc7-10 strains were arrested in G1 in raffinose media to prevent Mps1 expression and then released into the cell cycle in the presence of galactose at 30 degrees. Glucose was added one hour after release to repress Mps1 because continued expression prevented otherwise wild-type cells from degrading Pds1 for at least 6 hours (data not shown). When Mps1 was only expressed for one hour, Pds1 levels started to decline in GLC7 cells within 150 min (Fig 3C). However, Pds1 levels remained high in glc7-10 mutants for the duration of the time course, and this was due to spindle checkpoint activation because Pds1 levels cycled normally when MAD2 was disrupted in the same strains. The sustained checkpoint response in this experiment is more dramatic than when cells overexpressing Mps1 were released at the higher temperature, suggesting that the checkpoint silencing function of the Glc7-10 protein may be more efficiently inactivated earlier in the cell cycle, or that more of the Mps1 kinase acts unopposed when cells are released from G1 instead of mitosis. Alternatively, another phosphatase may be active during the prolonged mitotic arrest in the previous experiment, therefore diminishing the requirement for Glc7.
The delay in Pds1 destruction that occurs when Mps1 is expressed in glc7-10 mutants at a semi-permissive temperature strongly suggests that Glc7 is required to turn off the spindle checkpoint. Because glc7-10 mutant cells do not stabilize Pds1 without exogenous checkpoint activation at 30 degrees, these cells most likely make normal kinetochore-microtubule attachments at this temperature. To ensure that kinetochore attachments were normal, we analyzed the segregation of a pair of fluorescently marked sister chromatids. To monitor segregation in the pGAL-MPS1 GLC7 and pGAL-MPS1 glc7-10 strains, we also deleted MAD2 to prevent checkpoint activation. This experiment provides a sensitive way to determine if there are any segregation defects in glc7-10 cells at 30 degrees because the cells progress into anaphase with similar kinetics and no mitotic delay. Strikingly, greater than 96% of GLC7 mad2 and glc7-10 mad2 cells segregated chromosomes to opposite poles in anaphase after Mps1 was expressed for an hour (Fig 3D). Taken together, these data strongly argue that glc7-10 cells segregate chromosomes normally at 30 degrees, yet are defective in recovery from a transient spindle checkpoint arrest.
Glc7 is specifically required to exit from the spindle checkpoint
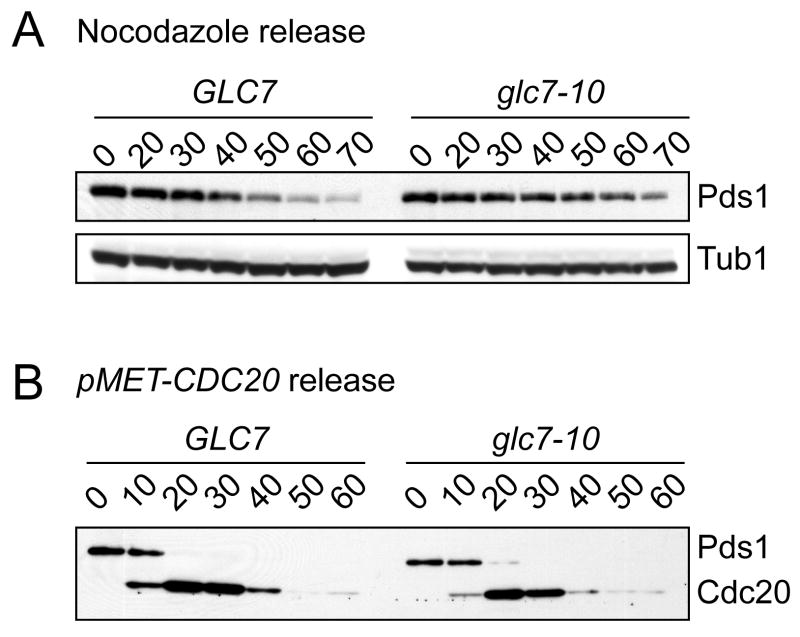
In a complimentary test for the role of Glc7 in exit from the spindle checkpoint that did not involve increasing Mps1 levels, we analyzed recovery from a nocodazole arrest. WT and glc7-10 cells were arrested in nocodazole at 23 degrees for 2 hours and then shifted to 30 degrees for 1 hour to decrease glc7-10 activity. The cells were then washed into media without nocodazole at 30 degrees and Pds1 levels were analyzed. While Pds1 levels begin to decline in GLC7 cells 40 min after release, they remained stable for up to 70 min in glc7-10 cells (Fig 4A). Therefore, Glc7 is required for exit from a nocodazole-mediated spindle checkpoint arrest, although the effect is less dramatic than exit from a Gal-Mps1 arrest.
Figure 4.
Glc7-10 cells are defective in mitotic exit specifically after spindle checkpoint activation. A. GLC7 (SBY8132) and glc7-10 (SBY2083) cells were released from a nocodazole arrest at 30 degrees and Pds1 levels were monitored at the indicated time points (min). B. pMET-HA-CDC20 cells with GLC7 (SBY4635) or glc7-10 (SBY4636) were arrested in metaphase with methionine and then released into media without methionine at 30 degrees. Pds1 and Cdc20 levels were monitored by immunoblotting at the indicated time points (min).
To determine whether the role of Glc7 is specific to reversing the spindle checkpoint, we asked whether Glc7 is required to exit from a metaphase arrest where the spindle checkpoint is not active. To do this, we depleted the APC activator Cdc20 and analyzed Pds1 levels as Cdc20 protein was restored. GLC7 and glc7-10 cells containing CDC20 under control of the methionine promoter (pMET-CDC20) were arrested in metaphase at 23 degrees by the addition of methionine for 2 hours, and then shifted to 30 degrees for 1 hour to reduce glc7-10 activity. The cells were then washed into media lacking methionine to induce Cdc20 at 30 degrees. In GLC7 cells, Pds1 was destroyed within 20 min, which corresponded to the time that Cdc20 protein started to accumulate (Fig 4B). The kinetics of Pds1 destruction and Cdc20 accumulation was similar in glc7-10 cells, indicating that glc7-10 cells recover normally from a metaphase arrest that does not involve exit from the spindle checkpoint.
Taken together, our data strongly suggest that the budding yeast Glc7 protein phosphatase I regulates exit from the spindle checkpoint, consistent with the need to reverse the phosphorylation required to activate the checkpoint. In S. pombe, PP1 is also required to exit the spindle checkpoint [21], so this may be a conserved mechanism for checkpoint inactivation. Although it remains possible that Glc7 also directly regulates the APC, the overexpression of Glc7 led to the stabilization of Clb2 which would not be expected if the APC were globally hyperactivated. Consistent with this, glc7 mutants showed no delay when released from a checkpoint-independent mitotic arrest.
Although PP1 is known to reverse Aurora-mediated phosphorylation [7, 25, 30-33], it is not clear whether the role for Glc7 in exiting the checkpoint is strictly related to reversing Ipl1 phosphorylation. Because Glc7 overexpression bypasses the arrest caused by unattached kinetochores but ipl1 mutants do not, we favor the possibility that Glc7 reverses the phosphorylation of additional checkpoint kinases. The sensitivity of slight increases in Mps1 dosage to decreased Glc7 activity is consistent with the possibility that Glc7 counteracts Mps1 phosphorylation. This is also supported by our finding that the defect in recovery from a checkpoint arrest induced by Mps1 overexpression is more severe than a nocodazole arrest when Glc7 is crippled at a semi-permissive temperature. Although we have not detected altered Ipl1 kinase activity in glc7 mutants [25], Glc7 may also directly regulate the activity of Mps1 and other checkpoint kinases. Taken together, our data suggest that the most likely role for Glc7 is to reverse checkpoint phosphorylation, possibly through multiple mechanisms that will need to be elucidated in the future. It will also be critical to identify and characterize the Glc7 regulatory subunit that directs it to dephosphorylate checkpoint targets to better understand the process of checkpoint silencing.
Experimental Procedures
Microbial Techniques, yeast strains and plasmids
Media and microbial techniques were essentially as described [34]. All experiments in which cells were released from a G1 arrest were carried out by α-factor arrest and release as described where pheromone was added back when cells had entered the cell cycle [6]. All sugars were used at a final concentration of 2%. Yeast strains are listed in Supplementary Table 1 and were constructed by standard genetic techniques or by PCR-based integration [35]. Specific primer sequences are available on request. Deletions and epitope tags were confirmed by PCR. Plasmids pKC991 (pSB344) and pKC1048 (pSB345) that contain pGAL-GLC7 were the generous gifts of John Cannon (U of Missouri). The catalytically dead pGAL-GLC7-H65K plasmid was generated by site directed mutagenesis of plasmid pKC1048 using primers SB1064 and SB1065 to generate pSB1589. The glc7-10 (PAY700-4) and GLC7 (PAY704-1) yeast strains that were used to generate strains for this study were the kind gifts of Michael Stark (U of Dundee).
Microscopy, Protein and immunological techniques
Analysis of budding index, sister chromatids and GFP-Tub1 in fixed cells was performed as described [11]. At least 100 cells were analyzed for all reported experiments. Protein extracts were made and immunoblotted as described [36]. 9E10 antibodies that recognize the myc tag and 12CA5 antibodies that recognize the hemagglutinin (HA) tag were obtained from Covance and used at a 1:10,000 dilution. Protein loading was confirmed in all experiments by anti-tubulin immunoblotting (data not shown).
Supplementary Material
Acknowledgments
We are very grateful to Damien D'Amours, John Cannon, Michael Stark and Dave Toczyski for generously sharing reagents. We thank Kevin Hardwick for discussing data prior to publication and helpful comments on the manuscript. We also thank the Biggins lab and Chitra Kotwaliwale for critical reading of the manuscript. This work was supported by a Paul Allen Foundation fellowship to B. A. P. and a National Institutes of Health Grant to S. B. (R01-GM64386). S. B. is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- 2.Rieder CL, Schultz A, Cole R, Sluder G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J Cell Biol. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinsky BA, Biggins S. The Spindle Checkpoint: Tension vs. Attachment. Trends Cell Biol. 2005;15(9):486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 5.Kang J, Yu H. Kinase signaling in the spindle checkpoint. J Biol Chem. 2009 doi: 10.1074/jbc.R900005200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Francisco L, Chan CS. Regulation of yeast chromosome segregation by Ipl1 protein kinase and type 1 protein phosphatase. Cell Mol Biol Res. 1994;40:207–213. [PubMed] [Google Scholar]
- 8.Fraschini R, Venturetti M, Chiroli E, Piatti S. The spindle position checkpoint: how to deal with spindle misalignment during asymmetric cell division in budding yeast. Biochem Soc Trans. 2008;36:416–420. doi: 10.1042/BST0360416. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan M, Morgan DO. Finishing mitosis, one step at a time. Nat Rev Mol Cell Biol. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- 10.Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- 11.Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 13.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 15.Stern BM, Murray AW. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Current Biology. 2001;11:1462–1467. doi: 10.1016/s0960-9822(01)00451-1. [DOI] [PubMed] [Google Scholar]
- 16.Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Zhang Z, Brew K, Lee EY. Mutational analysis of the catalytic subunit of muscle protein phosphatase-1. Biochemistry. 1996;35:6276–6282. doi: 10.1021/bi952954l. [DOI] [PubMed] [Google Scholar]
- 19.Kallio MJ, McCleland ML, Stukenberg PT, Gorbsky GJ. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr Biol. 2002;12:900–905. doi: 10.1016/s0960-9822(02)00887-4. [DOI] [PubMed] [Google Scholar]
- 20.Petersen J, Hagan IM. S. pombe aurora kinase/survivin is required for chromosome condensation and the spindle checkpoint attachment response. Curr Biol. 2003;13:590–597. doi: 10.1016/s0960-9822(03)00205-7. [DOI] [PubMed] [Google Scholar]
- 21.Vanoosthuyse V, Hardwick KG. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Current Biology. 2009 doi: 10.1016/j.cub.2009.05.060. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotwaliwale CV, Frei SB, Stern BM, Biggins S. A pathway containing the Ipl1/aurora protein kinase and the spindle midzone protein Ase1 regulates yeast spindle assembly. Dev Cell. 2007;13:433–445. doi: 10.1016/j.devcel.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloecher A, Tatchell K. Defects in Saccharomyces cerevisiae protein phosphatase type I activate the spindle/kinetochore checkpoint. Genes Dev. 1999;13:517–522. doi: 10.1101/gad.13.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sassoon I, Severin FF, Andrews PD, Taba MR, Kaplan KB, Ashford AJ, Stark MJ, Sorger PK, Hyman AA. Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev. 1999;13:545–555. doi: 10.1101/gad.13.5.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinsky BA, Kotwaliwale CV, Tatsutani SY, Breed CA, Biggins S. Glc7/Protein Phosphatase-1 Regulatory Subunits can oppose the Ipl1/Aurora Protein Kinase by redistributing Glc7. Mol Cell Biol. 2006;26:2648–2680. doi: 10.1128/MCB.26.7.2648-2660.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR, Chan CS, Drubin DG, Barnes G. Phospho-Regulation of Kinetochore-Microtubule Attachments by the Aurora Kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 27.Poddar A, Stukenberg PT, Burke DJ. Two complexes of spindle checkpoint proteins containing Cdc20 and Mad2 assemble during mitosis independently of the kinetochore in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:867–878. doi: 10.1128/EC.4.5.867-878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 29.Andrews PD, Stark MJ. Type 1 protein phosphatase is required for maintenance of cell wall integrity, morphogenesis and cell cycle progression in Saccharomyces cerevisiae. J Cell Sci. 2000;113(Pt 3):507–520. doi: 10.1242/jcs.113.3.507. [DOI] [PubMed] [Google Scholar]
- 30.Emanuele MJ, Lan W, Jwa M, Miller SA, Chan CS, Stukenberg PT. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J Cell Biol. 2008;181:241–254. doi: 10.1083/jcb.200710019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murnion ME, Adams RR, Callister DM, Allis CD, Earnshaw WC, Swedlow JR. Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J Biol Chem. 2001;276:26656–26665. doi: 10.1074/jbc.M102288200. [DOI] [PubMed] [Google Scholar]
- 32.Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 33.Bharucha JP, Larson JR, Gao L, Daves LK, Tatchell K. Ypi1, a positive regulator of nuclear protein phosphatase type 1 activity in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:1032–1045. doi: 10.1091/mbc.E07-05-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rose MD, Winston F, Heiter P. Methods in yeast genetics. Cold Spring Harbor, N. Y.: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 35.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 36.Minshull J, Straight A, Rudner A, Dernburg A, Belmont A, Murray AW. Protein Phosphatase 2A Regulates MPF Activity and Sister Chromatid Cohesion in Budding Yeast. Curr Biol. 1996;6:1609–1620. doi: 10.1016/s0960-9822(02)70784-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.