Abstract
OBJECTIVE
The generation of mature cell types during pancreatic development depends on the expression of many regulatory and signaling proteins. In this study, we tested the hypothesis that the transcriptional regulator Islet-1 (Isl-1), whose expression is first detected in the mesenchyme and epithelium of the developing pancreas and is later restricted to mature islet cells, is involved in the terminal differentiation of islet cells and maintenance of islet mass.
RESEARCH DESIGN AND METHODS
To investigate the role of Isl-1 in the pancreatic epithelium during the secondary transition, Isl-1 was conditionally and specifically deleted from embryonic day 13.5 onward using Cre/LoxP technology.
RESULTS
Isl-1–deficient endocrine precursors failed to mature into functional islet cells. The postnatal expansion of endocrine cell mass was impaired, and consequently Isl-1 deficient mice were diabetic. In addition, MafA, a potent regulator of the Insulin gene and β-cell function, was identified as a direct transcriptional target of Isl-1.
CONCLUSIONS
These results demonstrate the requirement for Isl-1 in the maturation, proliferation, and survival of the second wave of hormone-producing islet cells.
The vertebrate pancreas is crucial in maintaining nutritional homeostasis. The pancreas is composed of an exocrine compartment, consisting of acinar and ductal cells that secrete and transport digestive enzymes into the duodenum, and an endocrine compartment, consisting of the islets of Langerhans that produce hormones for regulating glucose metabolism. Each islet comprises five cell types: α, β, δ, ε, and PP cells expressing the hormones glucagon, insulin, somatostain, ghrelin, and pancreatic polypeptide, respectively (1–4).
Development of the endocrine pancreas occurs in two phases during mouse embryogenesis (5). The primary transition at embryonic day (E)9.5 is marked by the epithelial outgrowth of foregut endoderm into the surrounding splanchnic mesoderm forming the dorsal and ventral pancreatic buds. During this time, clusters of first-wave glucagon+ and/or insulin+ cells appear by budding from the pancreatic epithelium. However, the cells lack key proteins associated with functional α- and β-cells and are not believed to populate the adult islet (3,5). A distinct second wave of hormone+ cells delaminate from the pancreatic epithelium around E13.5 to E15.5; these cells proliferate and mature into the islet cells (6,7). Final organization of the islet structure is completed soon after birth (6).
Loss- and gain-of-function studies in mice have revealed the importance of transcription factors in endocrine pancreas development. Although some transcription factors are required in multiple stages of endocrine cell development, others are required at specific stages of the formation of islet cell types (8). For example, neurogenin 3 (Ngn3) is a central regulator of endocrine cell specification whose expression is essential for all endocrine cell development (9,10). However, many transcription factors are essential for early embryonic survival; therefore, tissue-specific gene deletion strategies in mice are needed to uncover their function in late development. For instance, Foxa2 null mice die shortly after gastrulation because of abnormal development of node and notochord (11,12), and specific roles in endocrine cell differentiation and islet cell function were only revealed using tissue-specific gene ablation approaches (13–16). Similarly, the embryonic and postnatal roles of the panendocrine transcription factor Pax6 were identified only when both Pax6 null and conditional mice were investigated (17,18). In contrast to Pax6, MafB, though expressed in developing α- and β-cells, is required for cell maturation (e.g., hormone gene expression) but not cell specification (19,20). On the other hand, MafA is uniquely expressed in insulin+ cells of the secondary transition in rodents; however, no obvious developmental defects were observed in MafA null mice, likely because of compensation by MafB (21–23).
Like Pax6, the panendocrine cell transcription factor, Islet-1 (Isl-1) is expressed during development in the central nervous system, cardiac mesoderm, and pancreas and is critical for the differentiation of these organs (24–27). Unfortunately, Isl-1 null embryos die by E10.5 because of defective heart formation; thus, it remains unclear as to whether Isl-1 is involved in the establishment of pancreatic endocrine cells during the secondary transition (24–26). However, Isl-1 null mice exhibit impaired genesis of the embryonic dorsal pancreatic bud and lack glucagon+, insulin+, or somatostatin+ cells in vivo or after culturing mutant pancreatic explants in vitro (24).
To overcome the early lethality of Isl-1 null animals we have derived mice specifically lacking this factor in the pancreatic epithelium to examine its function in islet cell development during the secondary transition. We demonstrate that loss of Isl-1 from the pancreatic epithelium at E13.5 leads to a severe reduction in hormone-expressing cells and the eventual loss of islet mass. We show that Isl-1 controls the proliferation and survival of endocrine cells in the postnatal pancreas. Furthermore, reduced insulin gene transcription and β-cell function appear to at least partially result from a specific defect in MafA gene transcription, which we identify as a direct Isl-1 target. These results demonstrate the importance of Isl-1 in the production of endocrine hormones and the maintenance of endocrine cell mass during and after the secondary transition.
RESEARCH DESIGN AND METHODS
Animals and breeding strategy.
The derivation of the Isl-1LoxP and Pdx1-Cre transgenic line has been reported previously (28,29). All mice were kept on a mixed outbred CD1 background. Pdx1-Cre;Isl-1L/+ and Isl-1L/L mice were mated to generate Pdx1-Cre;Isl-1L/L mutant mice. Littermate Isl-1L/L, Isl-1L/+, and Pdx1-Cre;Isl-1L/+ were indistinguishable from mixed CD1 Isl-1+/+ controls in our assays. Animal experiments were approved by the Children's Hospital of Philadelphia's institutional animal care and use committee.
Glucose tolerance tests and analytical procedures.
Overnight fasted animals were injected intraperitoneally with 2 g glucose (Sigma) per kilogram of body weight. Blood glucose values were monitored at 0, 15, 30, 60, 90, and 120 min after injection using an automatic glucometer (One Touch Ultra; LifeScan). To prepare plasma, blood was collected in heparinized tubes (BD Microtainer), spun, and stored at −80°C until assayed. Plasma insulin levels were measured using a Luminex kit (Linco). Total pancreatic insulin and glucagon content were assessed by radioimmune assay of acid-ethanol extracts at the University of Pennsylvania Diabetes Center.
Immunofluorescence/immunohistochemistry.
Tissues were fixed in 4% paraformaldehyde (PFA) overnight at 4°C and then embedded in either paraffin or optimal cutting temperature freezing medium. Slides (8–10 μm sections) were subjected to microwave antigen retrieval in 10 mmol/l citric acid buffer (pH6.0) and blocked with protein blocking reagent (Immunotech). Slides were incubated with primary antibodies overnight at 4°C, and appropriate secondary antibodies were added for 2 h at room temperature. The following primary antibodies were used: Glucagon (1:3,000; Linco), insulin (1:1,000; Linco), somatostatin (1:3,000; Santa Cruz), pancreatic polypeptide (1:50; Zymed), ghrelin (1:200; Santa Cruz), Isl-1 (1:50; Developmental Studies Hybridoma Bank 39.4D5 and 40.2D6), Pax6 (1:500; Covance), MafA (1:1,000; Bethyl Laboratories), and Pdx1 (1:200; Santa Cruz). Ngn3 (1:500; Developmental Studies Hybridoma Bank), Cy2, Cy3, and Cy5 conjugated secondary antibodies (1:600; Jackson ImmunoResearch). The Developmental Studies Hybridoma Bank Isl-1 monoclonal antibodies were developed under the auspices of the National Institute of Child Health and Human Development and are maintained by The University of Iowa, Department of Biology (Iowa City, IA).
RNA isolation, cDNA synthesis, and real-time PCR reactions.
Total RNA from homogenized pancreata was extracted using the RNA Easy Kit (Qiagen). RNA was reverse transcribed using 1 μg Oligo(dT) primer, Superscript II Reverse Transcriptase, and accompanying reagents (Invitrogen). Real-time PCR reactions were set up using the Brilliant SYBR Green PCR Master Mix (Stratagene). All reactions were performed in triplicate with reference dye normalization, and median cycling threshold values were used for analysis. Primer sequences are available upon request.
Measurement of β-cell mass.
Pancreata were removed, weighed, fixed in 4% PFA overnight at 4°C, and embedded in paraffin. Sections (8–10 μm) with maximum footprint were used for insulin staining. Images were taken under 4× magnification, and pancreatic tissue positive for insulin staining was measured by using IP Lab software. The β-cell mass was obtained by measuring the fraction of insulin-positive staining to total cross-sectional area and multiplying by the pancreatic weight. One section was used per pancreas with at least four control and mutant mice analyzed at each time point.
Pax6+ cell and hormone cell quantitation.
Immunostaining of Pax6 was used to detect late endocrine progenitors (hormone+ or hormone− (30); individual hormone immunostains were used to mark mature endocrine cell subtypes. Pancreatic cross-sectional area was measured using IP Lab software. At E15.5 (n = 6), two adjacent sections with the largest pancreatic footprint were selected. Pax6+ cells were counted, and the results were normalized to cross-sectional area. To measure the differentiation of endocrine cell subtypes, the number of hormone-expressing cells was expressed relative to the number of Pax6+ cells. The same technique was used to measure endocrine cell subtypes at E18.5 (n = 3), except that the average was taken of ∼10 sections taken at seven-section intervals through the block. At P4 (n = 4), Pax6+ cells were counted in four sections and normalized to pancreas cross-sectional area.
Transferase-mediated dUTP nick-end labeling and bromodeoxyuridine assays.
The rate of apoptosis was evaluated by transferase-mediated dUTP nick-end labeling (TUNEL) staining using the ApopTag Peroxidase In Situ Apoptosis Detection Kit (Chemicon). Sections were processed for the TUNEL assay, and adjacent sections were stained with an anti-Pax6 antibody. The percentage of TUNEL+ endocrine cells was generated by dividing the number of TUNEL+ cells to the number of Pax6+ cells. Insulin staining was performed on the same section as TUNEL assay to locate islets. For bromodeoxyuridine (BrdU) assays, pregnant animals (E18.5) and pups were injected with 10 μl/g of BrdU Labeling Reagent (Zymed) and killed 6 h later. BrdU staining was performed with the BrdU Staining Kit (Invitrogen). Sections were labeled with BrdU antibody, and adjacent sections were stained with an anti-Pax6 antibody. The percentage of BrdU+ endocrine cells was generated by dividing the number of BrdU+ cells into the number Pax6+ cells. Insulin staining was performed on the same section as BrdU staining to locate islets. At least 2,000 Pax6+ cells were counted for each animal for both TUNEL and BrdU assays, and the number of Pax6+ cells was used for normalization.
Electrophoretic mobility shift assays.
pCS2 rat Isl-1-Myc plasmid (gift from Dr. Pfaff) was used as a template for in vitro translation of Isl-1-Myc using Quick Coupled Transcription/Translation System rabbit reticulocyte lysate reagents (Promega). DNA binding reactions (20 μl final volume) included 1 μl of in vitro translated protein or 10 μg of Ins-1 β-cell line nuclear extract (24–26) and 400 fmol of 32P-end labeled oligo probe. The oligonucleotide sequences are as follows: MafA Isl-1 wild type, −7,822-CGTAACGTTAATGGAAGATGCTTGCTGCAG-7793, and MafA Isl-1 mutant, 5′-GCCGTAACGTGCCGGGAAGATGCT-3′, with the mutation underlined. The reactions were allowed to proceed at 30°C for 20 min in a buffer containing 10 mmol/l HEPES (pH 7.8), 75 mmol/l KCL, 2.5 mmol/l MgCl2, 0.1 mmol/l EDTA, 1 mmol/l dithiothreitol, 3% (v/v) Ficoll, 1 mg/ml BSA, and 1 μg poly(dI-dC). Competition experiments were performed using 100-fold molar excess of unlabeled wild-type or mutant oligos. Antibody supershift analyses were performed using a cocktail of four Isl-1–specific antibodies (Developmental Studies Hybridoma Bank, 39.3F7, 39.4D5, 40.2D6, and 40.3A4) preincubated with the Isl-1 protein or nuclear extract at room temperature for 20 min before adding the DNA probes. Reactions were separated on 6% nondenaturing polyacrylamide gels at 150V for 2 h in 0.5× tris-borate-EDTA buffer. Gels were dried and visualized by autoradiography.
Transient transfections: reporter assays.
Cultured βTC3 cells were transfected using Lipofectamine reagent (Invitrogen) with 1 μg of MafA region3:pTK (wild type and Isl-1 mutant) or the pTK(An) chloramphenicol acetyltransferase (CAT) vector and 1 μg of Rous sarcoma virus enhancer–driven luciferase (pRSV-Luc). Transfection reporter constructs for MafA were as described (31). Site-directed mutagenesis (Quickchange Mutagenesis kit, Stratagene) was used to create a noncomplementary transversional mutation in the Isl-1 binding site 5′-CACGGCCGTAACGTGCCGGGAAGATGCTTGCTGC-3′ (mutation is underlined). Cell lysates were prepared 48 h after transfection, and luciferase (LUC) (Promega) and CAT (32) assays were performed. CAT values were normalized to the LUC internal control. Each experiment was performed in triplicate with at least two independently isolated plasmid preparations.
Chromatin immunoprecipitation.
βTC3 cells were cultured for 72 h at 4 × 106 cells per 10-cm dish, then isolated and cross-linked with 1% formaldehyde in DMEM for 5 min at room temperature. Protein DNA chromatin fragments were prepared by sonication and precleared with protein A-agarose/salmon sperm DNA (Millipore, Temecula, CA) for 2 h at 4°C. The precleared chromatin was then incubated overnight at 4°C with a cocktail of Isl-1 monoclonal antibodies (Developmental Studies Hybridoma Bank 39.4D5, 39.3F7, 40.3A4, 40.2D6) or as controls, normal mouse immunoglobulin (Santa Cruz Biotechnology), or no antibody. Antibody-bound chromatin complexes were precipitated with protein A-agarose beads at 4°C for 4 h. Washed complexes were eluted from the beads and cross-links reversed. Quantitative real-time PCR was performed on ∼1:20 of the Isl-1–immunoprecipitated DNA using SYBR Green PCR mater mix (Applied Biosciences) and an ABI Prism 7900 instrument. Input DNA dilutions were used to generate a standard curve and to normalize amplification of the gene of interest (ΔCt). The enrichment of target control sequences in the chromatin immunoprecipitation (ChIP) DNAs (n = 3–4) was then calculated relative to the inactive phosphoenolpyruvate carboxykinase (PEPCK) promoter set as onefold (ΔΔCt). Nonquantitative PCR reactions were performed using 1:20 of the purified immunoprecipitated DNA with Taq polymerase hotstart master mix (5 Prime, Gaithersburg, MD) and 15 pmol of primer. ChIP PCR amplicons were visualized using a 1.5% agarose gel stained with ethidium bromide in 1× tris-acetate-EDTA buffer. Each ChIP experiment was repeated at least three times using independent chromatin preparations. The sequences of the real- time and standard PCR primers are available upon request.
RESULTS
Isl-1 is efficiently and specifically deleted in the pancreatic epithelium by E13.5.
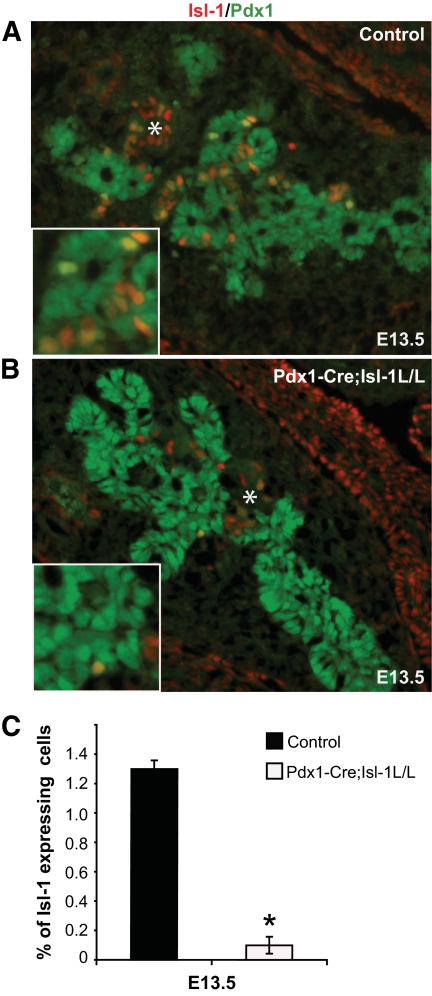
To inactivate Isl-1 specifically in the developing pancreatic epithelium, mice carrying the floxed allele of Isl-1 (29) were mated to Pdx-1–driven Cre transgenic mice (28). We anticipated that Isl-1 inactivation would occur in the pancreatic epithelium around E12.5 based on the detection of Pdx1-Cre activity in Rosa26R mice (33). However, efficient loss of Isl-1 protein in Pdx1+ pancreatic progenitors was not observed until E13.5 (Fig. 1A and B; data not shown), with the delay likely reflecting the accessibility of the Isl-1 locus to Cre and/or Cre expression levels in our strain background. By E13.5, greater than 90% of epithelial cells lacked Isl-1, as the percentage of Pdx1+Isl-1+ double-positive cells was reduced from 1.3% in control pancreata to 0.1% in the Pdx1-Cre;Isl-1L/L pancreata (Fig. 1C). The few remaining Isl-1+ cells in the Pdx1-Cre;Isl-1L/L pancreas were either unrecombined Pdx1+ epithelial cells, first-wave endocrine cells, or mesenchymal cells (24) (data not shown).
FIG. 1.
Isl-1 is efficiently deleted from the pancreatic epithelium in Pdx1-Cre;Isl-1L/L mice at E13.5. A: Immunofluorescence staining of control E13.5 pancreata shows Isl-1 (red) and Pdx1 (green) costaining in pancreatic epithelium. B: In Pdx1-Cre;Isl-1L/L pancreata, virtually no Pdx1+ cells (green) express Isl-1. The asterisk in A and B denotes Isl-1+ cells in the mesenchyme or first-wave endocrine cells that lack Pdx1 expression. The inset illustrates a higher magnification of Isl-1+/Pdx1+ (yellow) coexpressing cells and individual Pdx1+ or Isl-1+ cells. C: Quantitative analysis shows a 10-fold reduction in the proportion of Isl-1+ endocrine cells in the mutant pancreas (□) compared with controls (■). To normalize Isl-1+ cell numbers between the control and mutant, 1,000 Pdx1+ cells were counted for each pancreas, n = 3 for both groups. Data are represented as means ± SEM. *P < 0.05. (A high-quality digital representation of this figure is available in the online issue.)
Pdx1-Cre;Isl-1L/L mice exhibit severe hyperglycemia and impaired glucose tolerance.
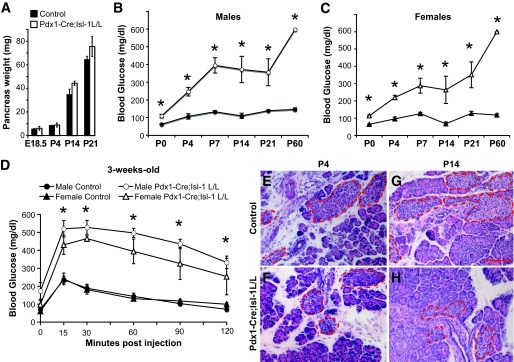
Pdx1-Cre;Isl-1L/L mice were born at the expected Mendelian ratio and did not differ from their littermate controls in size at birth (data not shown). There was also no change in total pancreas weight up to 3 weeks of age (Fig. 2A). However, both male and female mutant mice displayed elevated random-fed glucose levels as early as postnatal day (P)0 that worsened with age (Fig. 2B and C). Eventually, mutant mice died between 3 to 8 weeks after birth.
FIG. 2.
Pdx1-Cre;Isl-1L/L animals are severely hyperglycemic. A: There is no significant difference in total pancreatic weight between control (■) and mutant (□) animals at E18.5, P4, P14, and P21. B and C: The random-fed blood glucose level increases in aging mice. Measurement of blood glucose levels in control (○ and ▵) and mutant (● and ▲) male and female mice. D: IPGTT analysis demonstrates that 3-week-old mutant male (○) and female (▵) mice exhibit impaired glucose tolerance. ●, male controls; ▲, female controls. E–H: Hematoxylin-eosin–stained pancreatic tissues at P4 and P14 indicate that mutant mice have smaller and fewer islets. Pancreatic islets are outlined with dashed red lines. Scale bar: 100 μm, n > 3 for all groups. Data are represented as means ± SEM. *P value <0.05. (A high-quality digital representation of this figure is available in the online issue.)
We next evaluated endogenous islet β-cell function by intraperitoneal glucose tolerance tests (IPGTT) in 3-week-old male and female Pdx1-Cre;Isl-1L/L and control animals. Mutant mice had slightly increased blood glucose levels after an overnight fast that became dramatically elevated without returning to baseline after administration of exogenous glucose (Fig. 2D). Although both male and female mutant mice showed similar β-cell dysfunction, male mutant animals also exhibited growth retardation shortly after week 3, whereas females remained indistinguishable from controls (data not shown).
Pax6+ cell number is decreased postnatally in Pdx1-Cre;Isl-1L/L mice because of reduced proliferation and increased apoptosis.
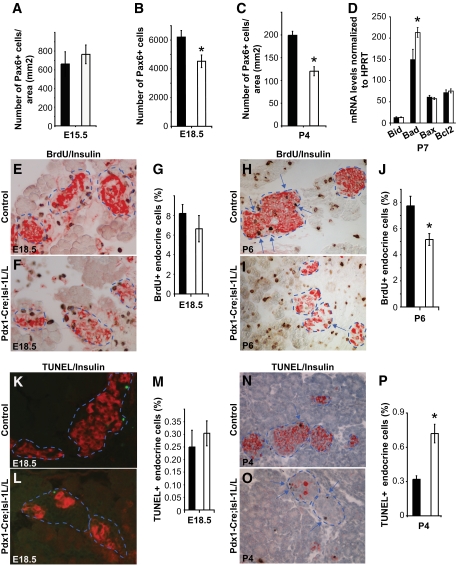
To investigate if the change in β-cell function reflected alterations in islet cell number and/or size, we compared hematoxylin-eosin–stained pancreatic sections from control and Pdx1-Cre;Isl-1L/L animals. Fewer and smaller islets were found in P4 mutant mice (Fig. 2E and F), with an even more dramatic difference by P14 (Fig. 2G and H). The number of late endocrine progenitors marked by Pax6 expression (30) was quantified at E15.5, E18.5, and P4 to determine the period of endocrine cell loss. A 27.2% reduction was observed by E18.5 (Fig. 3B), with a 39.7% decrease in Pax6+ cells by P4 (Fig. 3A–C). These data show that Isl-1 is involved in maintaining the Pax6+ endocrine progenitor population, with their loss in Pdx1-Cre;Isl-1L/L animals resulting from either a defect in cell differentiation, proliferation, and/or survival.
FIG. 3.
Postnatal Pdx1-Cre;Isl-1L/L mice show a gradual loss of islet mass because of decreased proliferation and increased apoptosis. A–C: Quantitative analysis of Pax6+ cells at E15.5, E18.5, and P4 in control (■) and mutant (□) animals. D: mRNA expression analysis of apoptotic genes at P7 in control (■) and mutant (□) total pancreata, n > 4 for both groups. Data are represented as means ± SEM. *P value <0.05. E–J: Immunostaining of E18.5 and P6 pancreatic sections for insulin (red) and BrdU (brown) reveals reduced proliferation at P6 (arrows; H–J) and not E18.5 mutants (E–G). K–P: Immunostaining for insulin (red) and TUNEL (K and L, green; N and O, brown) indicate that the rate of apoptosis is increased in P4 (arrows; N–P) but not E18.5 mutant pancreata (K–M). Islets are outlined by dashed line. Immunostaining of Pax6 was used to assess late endocrine progenitor cell number and used as a reference point to obtain percentage of BrdU+ or TUNEL+ cells (in G, J, M, and P; see research design and methods). Data are represented as means ± SEM. *P value <0.05. (A high-quality digital representation of this figure is available in the online issue.)
To determine if endocrine cell proliferation and/or survival was affected in the mutant pancreata, we next performed BrdU incorporation and TUNEL to evaluate these processes. A decrease in the rate of endocrine cell proliferation was first observed after birth in the pancreata of Pdx1-Cre;Isl-1L/L mice, with a 33.2% reduction in BrdU incorporating cells at P6 (Fig. 3E–J). Similarly, although there was not a significant change in the number of apoptotic TUNEL+ cells at E18.5, a twofold increase was found in P4 mutants (Fig. 3K–P). We next examined by real-time PCR if mRNA levels of the prosurvival and/or proapoptosis mediators Bid, Bad, Bax, and Bcl2 was changed by P7 (34). Only Bad was modestly but significantly increased (Fig. 3D). Collectively, these results suggest that a major function of Isl-1 in postnatal animals is to control the proliferation and survival of endocrine cells.
Isl-1 broadly affects endocrine hormone production.
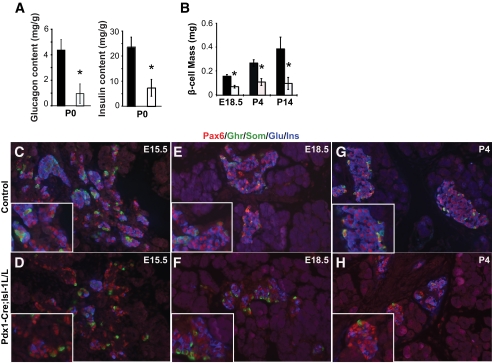
To investigate if Isl-1 is involved in endocrine hormone production, we measured the total glucagon and insulin content of control and mutant pancreata at birth. Interestingly, glucagon and insulin content were reduced in the Pdx1-Cre;Isl-1L/L pancreas by 78 and 69%, respectively (Fig. 4A). These results suggest that the reduction in total hormone content cannot be solely caused by the loss of endocrine cell mass described above (Fig. 3A–C). Rather, Isl-1 is likely to have an additional role in regulating endocrine hormone production itself. To identify the cause of this defect in mutant animals, we measured the cross-sectional area of pancreatic tissue stained for insulin as an indicator for mature β-cell mass at E18.5, P4, and P14 and found significant reductions in this population of cells in mutant animals at all stages examined (Fig. 4B).
FIG. 4.
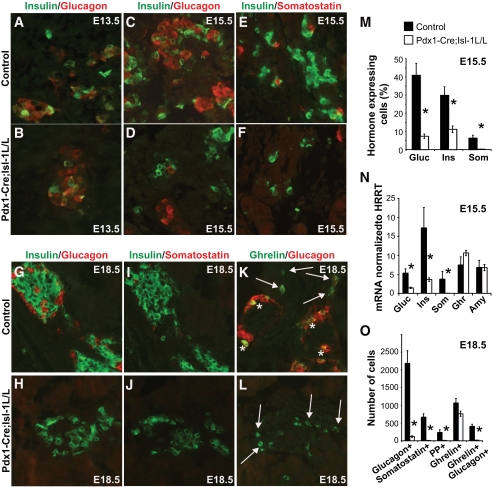
Loss of Isl-1 in the pancreatic epithelium leads to reduced endocrine hormone production. A: Total pancreatic glucagon and insulin content was reduced in P0 mutant animals by 78 and 69%, respectively. B: Morphometric analysis of insulin+ β-cell mass in E18.5, P4, and P14 pancreata, n = 4 for all groups. A and B: *P value <0.05, controls (■) and mutants (□). C–H: Immunostaining with an antibody cocktail of α-Pax6 (red), α-ghrelin (green), α-somatostatin (green), α-insulin (blue), and α-glucagon (blue) indicates that hormone expression is reduced by E15.5 in Pax6+ endocrine cells (C and D). Loss of hormone genes continues and the decline in the islet mass begins at E18.5 (E and F) and continues into postnatal stages P4 (G and H). (A high-quality digital representation of this figure is available in the online issue.)
To distinguish between changes in endocrine cell number (both mature and immature) and defects in the differentiation of islet cell subtypes, we immunostained pancreatic tissues with Pax6 (30) in conjunction with hormone cocktails (glucagon, insulin, somatostatin, and ghrelin) at E15.5, E18.5, and P4. Unlike control pancreata, in which the majority of the Pax6+ endocrine cells also coexpressed hormones, most of the Pax6+ endocrine cells in mutant pancreata lacked hormone expression at all stages examined (Fig. 4C–H). These data suggest that Isl-1 regulates the maturation of pancreatic endocrine cells.
To determine when the defects in endocrine cell differentiation were first detectable, we performed immunofluorescence analysis at several embryonic stages (E13.5, E15.5, and E18.5). E13.5 was the earliest time point at which a reduction in glucagon- and insulin-expressing cells was evident (Fig. 5A and B) coinciding with the onset of Isl-1 deletion in our model (Fig. 1). The number of glucagon+, insulin+, and somatostatin+ cells at E15.5 was reduced in mutant mice by 83, 63, and 99%, respectively (Fig. 5C–F and M). A similar pattern was obtained upon analysis of hormone mRNA expression by real-time PCR (Fig. 5N). Notably, the few hormone-expressing cells remaining in the mutant pancreas were not Isl-1+ (data not shown). The number of pancreatic polypeptide+ cells was decreased in E18.5 mutant pancreata (Fig. 5O), whereas islet ghrelin+ (ε)- and exocrine amylase+-expressing cell numbers were unchanged (Fig. 5K, L, and O and data not shown). These data strongly suggest that Isl-1 is required to regulate the formation of respective hormone-producing cell types.
FIG. 5.
Decreased hormone expression is first apparent in E13.5 Pdx1-Cre;Isl-1L/L animals. A–D and G–H: Immunostaining of insulin (green) and glucagon (red) indicates that both are affected by E13.5. E, F, and I–J: Somatostatin-expressing cells (red) are lost by E15.5 in the mutant pancreata. K and L: Immunostaining of ghrelin (green) and glucagon (red) indicates that the number of ghrelin-expressing cells remained unchanged in mutant E18.5 embryos. The ghrelin+/glucagon+ observed in control pancreas (yellow, asterisks) are absent in the mutants, whereas ghrelin+/glucagon− cells (arrows) are present in both. M: Quantitative analysis of glucagon-, insulin-, and somatostatin-expressing cells at E15.5. Immunostaining of Pax6 was used to assess late endocrine progenitor cell number and used as a reference point to obtain percentage hormone-expressing cells (see research design and methods). N: Real-time PCR analysis was used to measure total pancreas (n = 5) mRNA levels at E15.5. Amy: Amylase, HPRT: hypoxanthine-guanine phosphoribosyl transferase. O: Quantitative analysis of glucagon-, somatostatin-, PP-, ghrelin-, and ghrelin/glucagon-expressing cells at E18.5. Number of hormone-expressing cells was obtained from seven sections per pancreas (see research design and methods). Data are represented as means ± SEM. *P value <0.05. (A high-quality digital representation of this figure is available in the online issue.)
MafA expression is reduced in the E15.5 Pdx1-Cre;Isl-1L/L pancreata.
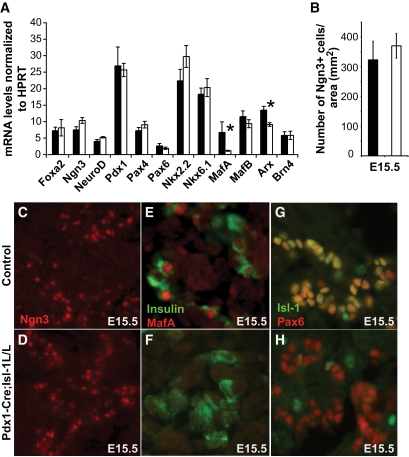
Many islet-enriched transcription factors are essential regulators of pancreatic development and/or hormone gene expression (35). Next, we examined if Isl-1 controls expression of these key regulators. A dramatic reduction in MafA mRNA levels was observed in the E15.5 Pdx1-Cre;Isl-1L/L pancreata, while the expression of other transcription factors including Ngn3 was unaffected (Fig. 6A). Quantitative analysis of Ngn3+ cells from control and mutant pancreata further confirmed Pdx1-Cre;Isl-1L/L animals have normal numbers of early endocrine progenitors (Fig. 6B–D). In addition, decreased MafA expression in β-cells was evident by immunostaining (Fig. 6E and F). Notably, the known regulators of MafA transcription (MafB, Pax6, Pdx1, Foxa2, and Nkx2.2) (19,31) were unaffected in the mutant animals (Fig. 6A, G, and H and data not shown). In summary, these results suggest that Isl-1 acts upstream of MafA in β-cells.
FIG. 6.
MafA expression is specifically reduced in Pdx1-Cre;Isl-1L/L animals. A: Real-time PCR analysis of mRNA levels of islet-enriched transcription factors in control (■) and mutant (□) pancreata indicates a significant reduction of MafA mRNA by E15.5 (n = 5). *P value <0.05. B–D: The number of Ngn3+ endocrine progenitors (red) was not changed. E and F: MafA (red) is lost from insulin+ cells in the E15.5 mutant pancreas. G and H: Pax6+ cells were unchanged in the E15.5 mutant pancreata. (A high-quality digital representation of this figure is available in the online issue.)
Isl-1 binds to and activates MafA transcription through an evolutionarily conserved enhancer.
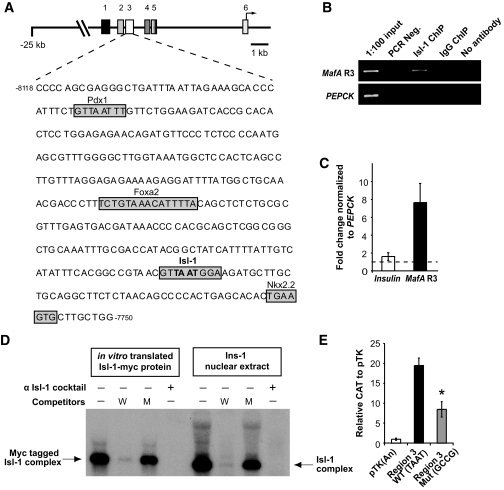
MafA was originally identified in the β-cell as a transactivator of the C1 element in the insulin enhancer (36). Because mRNA and protein levels of previously known MafA regulators were not affected in Pdx1-Cre;Isl-1L/L mice (Fig. 6A and data not shown), we hypothesized that Isl-1 directly controls MafA transcription. The MafA 5′ promoter has six evolutionarily conserved regions, of which “region 3” is not only the longest and most well conserved (Fig. 7A) but uniquely capable of driving β-cell line–specific expression in vitro (31).
FIG. 7.
Isl-1 binds and activates through the MafA region 3 control domain in β-cells. A: Diagram of the conserved sequence domains in the mouse MafA promoter, with region 3 sequence below. A putative Isl-1 binding site was identified in region 3, with other boxes illustrating known islet-enriched transcription factor binding sites (31). B and C: ChIP analysis demonstrates Isl-1 binding to region 3 of the MafA gene in βTC3 cells. Input chromatin, Isl-1 ChIP, IgG ChIP, and no antibody ChIP DNA were analyzed by standard PCR with region 3 and control (PEPCK) primers. C: Real-time PCR analysis of ChIP DNA shows a sevenfold enrichment for MafA region 3 (black bar) as compared with the inactive PEPCK promoter set as onefold (dashed line). No significant enrichment for the insulin enhancer region was observed in these experiments (white bar), although Isl-1 has been reported to bind to sequences in this region in gel shift experiments in vitro (44). D: The labeled region 3 MafA probe spanning base pairs −7,822 to −7,793 was incubated in gel shift binding assays with in vitro translated Myc-tagged Isl-1 protein or Ins-1 β-cell extract. The specificity of protein DNA binding was determined by competition with a 100-fold molar excess of unlabeled wild-type or TAAT mutant (M) competitors. Additionally, Isl-1 binding to the region 3 probe was blocked with Isl-1 antibodies; a faint antibody super-shifted complex was detected after longer exposure (data not shown). E: The Isl-1 binding site is required for region 3 activity. Wild-type (TAAT; black bar) and Isl-1 binding site mutant (GCCG; gray bar), region 3–driven CAT reporters, and CAT reporter alone (white bar) were transfected into βTC3 cells (n = 4). Data are represented as means ± SEM. *P < 0.05.
ChIP analysis was performed in mouse βTC3 cells to determine if Isl-1 occupied region 3 of the MafA gene. A significant and quantitative enrichment of region 3 was found compared with PEPCK control sequences by standard or real-time PCR only after α–Isl-1 immunoprecipitation (Fig. 7B and C). Conversely, insulin enhancer sequences were not enriched in Isl-1 ChIP DNA (Fig. 7C).
Next, we performed electrophoretic mobility shift assays with either in vitro translated Myc-tagged Isl-1 or Ins-1 β-cell line nuclear extract to analyze binding to the Isl-1-like site in a MafA region 3 base pair −7,816 to −7,808 spanning probe (Fig. 7A). Isl-1 indeed bound to this site because the level of the protein DNA complex was reduced by addition of excess wild-type and not mutant competitors (Fig. 7D). Additionally, antibodies against Isl-1 blocked binding to the MafA probe (Fig. 7D). Mutation of the Isl-1 binding site in region 3 also reduced reporter activity in βTC-3 cells (Fig. 7E). Collectively, these in vitro and endogenous binding data strongly suggest that Isl-1 is a direct regulator of MafA transcription in β-cells.
DISCUSSION
Ahlgren et al. (24) had shown previously through tissue recombination studies using Isl-1 null embryos that Isl-1 expression in the mesenchyme is essential for proper differentiation of the exocrine pancreas, while the epithelial expression of Isl-1 is required for early endocrine pancreas development). We have identified novel roles for Isl-1 later in the pancreatic epithelium. Our findings establish Isl-1 as transcriptional regulator required in the formation, proliferation, and survival of islet α-, β-, δ-, and PP cells after the secondary transition and in postnatal lives.
A striking phenotype of Pdx1-Cre;Isl-1L/L animals is the dramatic loss of glucagon, insulin, somatostatin, and pancreatic polypeptide expression in developing cells, with a severe reduction in islet cell mass after birth. Lack of Isl-1 in the developing endocrine pancreas has a global impact on islet cells, with the exception of ghrelin expression. We showed that the decrease in α, β, δ, and PP production was not simply the result of a reduction in the Ngn3+ endocrine progenitor population but was because of the incomplete endocrine cell maturation. Although it is not clear from this study why ghrelin expression was not affected in the Pdx1-Cre;Isl-1L/L animals, it is possible that ε-cells represent an immature endocrine population that does not require Isl-1 action. This notion is supported by the fact that ghrelin-expressing cells are present only during fetal development and in young animals but not in the mature pancreas (1).
Although Isl-1 was identified more than a decade ago, its direct downstream targets in the islet remain largely uncharacterized. We identified MafA as a novel transcriptional target of Isl-1. Notably, MafA mRNA expression was compromised very early and highly selectively upon Isl-1 inactivation in vivo. Furthermore, Isl-1 binding was detected within the principal MafA transcriptional control region in vitro and by ChIP in β-cell lines. Finally, the luciferase construct containing mutations in the Isl-1 response element exhibited lower activity than the wild-type region 3 reporter. However, Isl-1 was unable to independently activate transfected MafA promoter-driven reporters in non–β-cells, as, for example, the relative activity level of Isl-1 to the vector control with the MafA region 3 reporter equaled 0.76 (±0.10)-fold. Similarly, Pdx-1, E47, and NeuroD1/BETA2, widely accepted insulin gene activators, are also unable to activate insulin-driven reporters in non–β-cell lines on their own (37–39). Moreover, Isl-1 is incapable of stimulating a variety of different promoter-driven reporters under circumstances wherein other LIM-homeodomain proteins could potentiate (i.e., with E47) (37). The coactivators that are necessary for mediating Isl-1 activation in β-cells or for region 3 specifically have not yet been identified. The loss of only MafA in β-cells cannot explain the pancreatic phenotype observed in our Pdx1-Cre;Isl-1L/L animals. For example, although insulin transcription is reduced in MafA−/− mice, their total insulin content remains normal. Again, unlike Pdx1-Cre;Isl-1L/L mice, the MafA mutant has no developmental phenotype but displays compromised β-cell activity soon after birth because of defects in insulin secretion (40).
Isl-1 has been shown to regulate preproglucagon gene expression in vitro (41) and presumably directly influences preproglucagon mRNA levels in Pdx1-Cre;Isl-1L/L mice. A small but significant decrease in the mRNA level of the α-cell–enriched Arx transcription factor was also observed in mutant animals. Although much is known about how Arx regulates α-cell specification (35,42,43), little is known about its significance to α-cell maturation and function. It is also remains to be determined how Isl-1 controls somatostatin and pancreatic polypeptide gene expression in islet δ- and PP cell, respectively.
Unlike first wave Isl-1+ cells that are postmitotic (24), subsets of Isl-1+ cells after the secondary transition have the ability to proliferate (data not shown). Although it is unknown whether proliferation and/or survival of early endocrine cells was affected in the Isl-1 null pancreata (24), our data demonstrated a crucial role of Isl-1 in regulating endocrine cell growth and survival in young animals. Growth and maintenance of β-cell mass is crucial to prevent or delay loss of β-cells in diabetic patients. We propose that there are at least three transcriptional mechanisms utilized by Isl-1 in islet β-cell development and function: 1) direct regulation of targets bound by Isl-1 alone; 2) targets bound by both Isl-1 and MafA (or other factors); and perhaps to a lesser extent 3) indirect regulation of targets, bound for example by MafA alone. The former two are likely involved in regulating genes involved in cell proliferation and survival, processes unaffected in MafA null mice. In this regard, Bad gene expression was downregulated in Pdx1-Cre;Isl-1L/L mice. However, it is unclear if this is a direct or indirect response since we were unable to detect binding of Isl-1 to the proximal Bad promoter in ChIP assays. Combined genome-wide transcription factor binding and expression profiling studies will need to be performed on factors like Isl-1 to determine how islet cell differentiation is regulated in mammalian cells.
Severe hyperglycemia observed in the mutant animals is likely because of both the loss of islet mass and decreased hormone expression by the remaining islet cells. The decrease in islet cell number results from decreased cell proliferation and increased cell death, changes that became significant soon after birth. Isl-1 has important functions in the differentiation of hormone-producing cells beyond what was shown previously, but the exact mechanism of how Isl-1 regulates its target genes is largely unknown. In addition, future studies in which Isl-1 is deleted conditionally in mature β-cells will determine if this factor controls β-cell function as well as β-cell survival and proliferation in adult animals.
Acknowledgments
This work was supported by National Institutes of Health Grants DK078606, DK019525, and JDRF 2-2007-703 to C.L.M. C.S.H. was supported by National Institutes of Health Grants DK007061 and DK083160 and R.S. by DK078606.
No potential conflicts of interest relevant to this article were reported.
We thank Klaus Kaestner, Joshua Friedman, and Rana Gupta for careful reading of the manuscript. We thank Gary Swain and the members of the Morphology Core in the Center for Molecular Studies in Digestive and Liver Disease (P30-DK050306) as well as the Radioimmunoassay and Biomarkers Core of the Penn Diabetes Center (P30-DK19525) for sample processing. We are also grateful for the Isl-1 expression vector obtained from Sam Pfaff and the Pdx1-Cre mouse from Pedro Herrera. We are also thankful to Jeffrey Raum, Min Guo, and Brian McKenna for reagents and assistance with the ChIP and EMSA experiments.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Heller RS, Jenny M, Collombat P, Mansouri A, Tomasetto C, Madsen OD, Mellitzer G, Gradwohl G, Serup P: Genetic determinants of pancreatic epsilon-cell development. Dev Biol 2005; 286: 217– 224 [DOI] [PubMed] [Google Scholar]
- 2.Prado CL, Pugh-Bernard AE, Elghazi L, Sosa-Pineda B, Sussel L: Ghrelin cells replace insulin-producing β-cells in two mouse models of pancreas development. Proc Natl Acad Sci U S A 2004; 101: 2924– 2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slack JM: Developmental biology of the pancreas. Development 1995; 121: 1569– 1580 [DOI] [PubMed] [Google Scholar]
- 4.Wierup N, Svensson H, Mulder H, Sundler F: The ghrelin cell: a novel developmentally regulated islet cell in the human pancreas. Regul Pept 2002; 107: 63– 69 [DOI] [PubMed] [Google Scholar]
- 5.Pictet RL, Clark WR, Williams RH, Rutter WJ: An ultrastructural analysis of the developing embryonic pancreas. Dev Biol 1972; 29: 436– 467 [DOI] [PubMed] [Google Scholar]
- 6.Habener JF, Kemp DM, Thomas MK: Minireview: transcriptional regulation in pancreatic development. Endocrinology 2005; 146: 1025– 1034 [DOI] [PubMed] [Google Scholar]
- 7.Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV: PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 1996; 122: 983– 995 [DOI] [PubMed] [Google Scholar]
- 8.Murtaugh LC: Pancreas and β-cell development: from the actual to the possible. Development 2007; 134: 427– 438 [DOI] [PubMed] [Google Scholar]
- 9.Gradwohl G, Dierich A, LeMeur M, Guillemot F: neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A 2000; 97: 1607– 1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee CS, Perreault N, Brestelli JE, Kaestner KH: Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev 2002; 16: 1488– 1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ang SL, Rossant J: HNF-3 β is essential for node and notochord formation in mouse development. Cell 1994; 78: 561– 574 [DOI] [PubMed] [Google Scholar]
- 12.Weinstein DC, Ruiz i Altaba A, Chen WS, Hoodless P, Prezioso VR, Jessell TM, Darnell JE, Jr.: The winged-helix transcription factor HNF-3 β is required for notochord development in the mouse embryo. Cell 1994; 78: 575– 588 [DOI] [PubMed] [Google Scholar]
- 13.Gao N, White P, Doliba N, Golson ML, Matschinsky FM, Kaestner KH: Foxa2 controls vesicle docking and insulin secretion in mature β-cells. Cell Metab 2007; 6: 267– 279 [DOI] [PubMed] [Google Scholar]
- 14.Lantz KA, Vatamaniuk MZ, Brestelli JE, Friedman JR, Matschinsky FM, Kaestner KH: Foxa2 regulates multiple pathways of insulin secretion. J Clin Invest 2004; 114: 512– 520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee CS, Sund NJ, Behr R, Herrera PL, Kaestner KH: Foxa2 is required for the differentiation of pancreatic alpha-cells. Dev Biol 2005; 278: 484– 495 [DOI] [PubMed] [Google Scholar]
- 16.Sund NJ, Vatamaniuk MZ, Casey M, Ang SL, Magnuson MA, Stoffers DA, Matschinsky FM, Kaestner KH: Tissue-specific deletion of Foxa2 in pancreatic β-cells results in hyperinsulinemic hypoglycemia. Genes Dev 2001; 15: 1706– 1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashery-Padan R, Zhou X, Marquardt T, Herrera P, Toube L, Berry A, Gruss P: Conditional inactivation of Pax6 in the pancreas causes early onset of diabetes. Dev Biol 2004; 269: 479– 488 [DOI] [PubMed] [Google Scholar]
- 18.St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P: Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature 1997; 387: 406– 409 [DOI] [PubMed] [Google Scholar]
- 19.Artner I, Blanchi B, Raum JC, Guo M, Kaneko T, Cordes S, Sieweke M, Stein R: MafB is required for islet β-cell maturation. Proc Natl Acad Sci U S A 2007; 104: 3853– 3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Artner I, Le Lay J, Hang Y, Elghazi L, Schisler JC, Henderson E, Sosa-Pineda B, Stein R: MafB: an activator of the glucagon gene expressed in developing islet alpha- and β-cells. Diabetes 2006; 55: 297– 304 [DOI] [PubMed] [Google Scholar]
- 21.Matsuoka TA, Artner I, Henderson E, Means A, Sander M, Stein R: The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc Natl Acad Sci U S A 2004; 101: 2930– 2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang C, Moriguchi T, Kajihara M, Esaki R, Harada A, Shimohata H, Oishi H, Hamada M, Morito N, Hasegawa K, Kudo T, Engel JD, Yamamoto M, Takahashi S: MafA is a key regulator of glucose-stimulated insulin secretion. Mol Cell Biol 2005; 25: 4969– 4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishimura W, Kondo T, Salameh T, Khattabi IE, Dodge R, Bonner-Weir S, Sharma A: A switch from MafB to MafA expression accompanies differentiation to pancreatic β-cells. Dev Biol 2006; 293: 526– 539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahlgren U, Pfaff SL, Jessell TM, Edlund T, Edlund H: Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature 1997; 385: 257– 260 [DOI] [PubMed] [Google Scholar]
- 25.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S: Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell 2003; 5: 877– 889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM: Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell 1996; 84: 309– 320 [DOI] [PubMed] [Google Scholar]
- 27.Thor S, Ericson J, Brannstrom T, Edlund T: The homeodomain LIM protein Isl-1 is expressed in subsets of neurons and endocrine cells in the adult rat. Neuron 1991; 7: 881– 889 [DOI] [PubMed] [Google Scholar]
- 28.Herrera PL: Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development 2000; 127: 2317– 2322 [DOI] [PubMed] [Google Scholar]
- 29.Sun Y, Dykes IM, Liang X, Eng SR, Evans SM, Turner EE: A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci 2008; 11: 1283– 1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou QLA, Rajagopal J, Anderson WJ, Gray PA, Melton DA: A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell 2007; 13: 103– 114 [DOI] [PubMed] [Google Scholar]
- 31.Raum JC, Gerrish K, Artner I, Henderson E, Guo M, Sussel L, Schisler JC, Newgard CB, Stein R: FoxA2, Nkx2.2, and PDX-1 regulate islet β-cell-specific mafA expression through conserved sequences located between base pairs −8118 and −7750 upstream from the transcription start site. Mol Cell Biol 2006; 26: 5735– 5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nordeen SK, Green PP, 3rd, Fowlkes DM: A rapid, sensitive and inexpensive assay for chloramphenicol acetyltransferase. DNA 1987; 6: 173– 178 [DOI] [PubMed] [Google Scholar]
- 33.Heiser PW, Lau J, Taketo MM, Herrera PL, Hebrok M: Stabilization of β-catenin impacts pancreas growth. Development 2006; 133: 2023– 2032 [DOI] [PubMed] [Google Scholar]
- 34.Pawlowski J, Kraft AS: Bax-induced apoptotic cell death. Proc Natl Acad Sci U S A 2000; 97: 529– 531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collombat P, Hecksher-Sorensen J, Serup P, Mansouri A: Specifying pancreatic endocrine cell fates. Mechanisms of Development 2006; 123: 501– 512 [DOI] [PubMed] [Google Scholar]
- 36.Matsuoka TA, Zhao L, Artner I, Jarrett HW, Friedman D, Means A, Stein R: Members of the large Maf transcription family regulate insulin gene transcription in islet β-cells. Mol Cell Biol 2003; 23: 6049– 6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson JD, Zhang W, Rudnick A, Rutter WJ, German MS: Transcriptional synergy between LIM-homeodomain proteins and basic helix-loop-helix proteins: the LIM2 domain determines specificity. Mol Cell Biol 1997; 17: 3488– 3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohneda K, Mirmira RG, Wang J, Johnson JD, German MS: The homeodomain of PDX-1 mediates multiple protein-protein interactions in the formation of a transcriptional activation complex on the insulin promoter. Mol Cell Biol 2000; 20: 900– 911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiu Y, Guo M, Huang S, Stein R: Insulin gene transcription is mediated by interactions between the p300 coactivator and PDX-1, BETA2, and E47. Mol Cell Biol 2002; 22: 412– 420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Brun T, Kataoka K, Sharma AJ, Wollheim CB: MafA controls genes implicated in insulin biosynthesis and secretion. Diabetologia 2006; 50: 348– 358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang M, Drucker DJ: The LIM domain homeobox gene isl-1 is a positive regulator of islet cell-specific proglucagon gene transcription. J Biol Chem 1995; 270: 12646– 12652 [DOI] [PubMed] [Google Scholar]
- 42.Collombat P, Hecksher-Sorensen J, Broccoli V, Krull J, Ponte I, Mundiger T, Smith J, Gruss P, Serup P, Mansouri A: The simultaneous loss of Arx and Pax4 genes promotes a somatostatin-producing cell fate specification at the expense of the alpha- and β-cell lineages in the mouse endocrine pancreas. Development 2005; 132: 2969– 2980 [DOI] [PubMed] [Google Scholar]
- 43.Collombat P, Mansouri A, Hecksher-Sorensen J, Serup P, Krull J, Gradwohl G, Gruss P: Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev 2003; 17: 2591– 2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karlsson O, Edlund T, Moss JB, Rutter WJ, Walker MD: A mutational analysis of the insulin gene transcription control region: expression in β-cells is dependent on two related sequences within the enhancer. Proc Natl Acad Sci U S A 1987; 84: 8819– 8823 [DOI] [PMC free article] [PubMed] [Google Scholar]