Abstract
OBJECTIVE
Diabetic retinopathy is a sight-threatening microvascular complication of diabetes with a complex multifactorial pathogenesis. A systematic meta-analysis was undertaken to collectively assess genetic studies and determine which previously investigated polymorphisms are associated with diabetic retinopathy.
RESEARCH DESIGN AND METHODS
All studies investigating the association of genetic variants with the development of diabetic retinopathy were identified in PubMed and ISI Web of Knowledge. Crude odds ratios (ORs) and 95% CIs were calculated for single nucleotide polymorphisms and microsatellite markers previously investigated in at least two published studies.
RESULTS
Twenty genes and 34 variants have previously been studied in multiple cohorts. The aldose reductase (AKR1B1) gene was found to have the largest number of polymorphisms significantly associated with diabetic retinopathy. The z−2 micro satellite was found to confer risk (OR 2.33 [95% CI 1.49–3.64], P = 2 × 10−4) in type 1 and type 2 diabetes and z+2 to confer protection (0.58 [0.36–0.93], P = 0.02) against diabetic retinopathy in type 2 diabetes regardless of ethnicity. The T allele of the AKR1B1 promoter rs759853 variant is also significantly protective against diabetic retinopathy in type 1 diabetes (0.5 [0.35–0.71], P = 1.00 × 10−4), regardless of ethnicity. These associations were also found in the white population alone (P < 0.05). Polymorphisms in NOS3, VEGF, ITGA2, and ICAM1 are also associated with diabetic retinopathy after meta-analysis.
CONCLUSIONS
Variations within the AKR1B1 gene are highly significantly associated with diabetic retinopathy development irrespective of ethnicity. Identification of genetic risk factors in diabetic retinopathy will assist in further understanding of this complex and debilitating diabetes complication.
Diabetic retinopathy is a sight-threatening microvascular complication of diabetes. Global population–based data indicate that it is the fifth most common cause of blindness in the world, accounting for ∼4.8% of global blindness (1). Diabetic retinopathy is a leading cause of blindness in industrialized countries (1), and its significance is likely to increase with increasing frequency of diabetes (2). Diabetic retinopathy is defined by the presence of retinal microvascular lesions. Early retinal changes include microaneurysms, hemorrhages, hard exudates, cotton wool spots, intraretinal microvascular abnormalities, and venous beading. These clinical features occur in nonproliferative diabetic retinopathy (NPDR). Growth of abnormal new blood vessels that frequently lead to preretinal and vitreous hemorrhage are the principal hallmarks of proliferative diabetic retinopathy (PDR). Visual impairment in diabetic retinopathy occurs secondary to preretinal or vitreous hemorrhage and diabetic maculopathy (either macular edema or macular ischemia) (3).
The prevalence of diabetic retinopathy is higher in type 1 diabetic patients, with sight-threatening retinopathy reported to be up to 2.5 times more common than in those with type 2 diabetes (4). Conversely, the incidence of macular edema has been reported to be up to two times higher among those with type 2 diabetes (5).
The pathogenesis of diabetes is believed to be multifactorial, with genetic risk factors playing a fundamental role. Recent genome-wide association studies have identified several genetic loci involved in the pathogenesis of both type 1 diabetes and type 2 diabetes (6,7). Pathogenesis of diabetic retinopathy is complex, also having a multifactorial biochemical pathogenesis, primarily because of altered glucose metabolism (8). Glycemic control (9,10) and the duration of diabetes (11,12) have been identified through large longitudinal prospective studies as being major risk factors for the development of diabetic retionopathy. However, genetic factors are also likely to account for the susceptibility to this blinding disease as well as the differences in diabetic retinopathy incidence between individuals with type 1 and type 2 diabetes. It has become evident through familial aggregation studies that susceptibility to complications of diabetes such as diabetic retinopathy also have a heritable component independent of glycemic control and duration of diabetes. The Diabetes Control and Complications Trial revealed a familial tendency for severe diabetic retinopathy in type 1 and type 2 diabetes (13). These findings were also replicated in Mexican Americans (14,15) and other familial risk studies (16,17), independent of associated environmental risk factors. There have been numerous studies investigating candidate genes for diabetic retinopathy susceptibility in subjects of various ethnicities.
The aim of this systematic meta-analysis was to analyze all published studies that investigated the association between genetic risk factors and the development of diabetic retinopathy and met specified inclusion criteria. We sought to determine which of the previously investigated genetic variants are significantly associated with the development of diabetic retinopathy in type 1 or type 2 diabetes and to examine the strength of these associations.
RESEARCH DESIGN AND METHODS
Literature search and data collection.
A systematic literature search was performed to identify all studies published between January 1990 and August 2008 that investigated the association of genetic variants with the development of any form of diabetic retinopathy. The PubMed database (National Center for Biotechnology Information; NCBI), ISI Web of Knowledge (version 4.5), and the Cochrane Library were explored independently by two authors using the following keyword strings: “genetic” AND “diabetic retinopathy” and “gene” AND “diabetic retinopathy.” All retrieved publications were reviewed and compared and studies written in English that contained sufficient case (subjects with diabetes and diabetic retinopathy) and control (subjects with diabetes but without the complication of diabetic retinopathy) subjects' genotype information, such as allele or genotype frequency, were included. The reference list of each relevant publication was also examined to identify additional studies appropriate for inclusion in the meta-analysis. Polymorphisms were included in the meta-analysis if a minimum of two studies had assessed association with diabetic retinopathy development.
Data analysis.
Data were entered into a database and statistical analyses performed using RevMan software (version 4.2; Oxford, U.K.) and SPSS (version 16.0; SPSS, Chicago, IL). Data extraction and data entry was performed independently by two of the authors (S.A. and A.W.H.) and compared. Independent review and resolution by a senior investigator (K.P.B. and J.E.C.) was sought if disagreement between the reviewers occurred.
Ensemble (http://www.ensembl.org/index.html) and dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) were used to locate rs# identifiers for the genetic variants. When an rs# identifier could not be located, the most commonly used name for that specific variant or single nucleotide polymorphism (SNP) was selected. All rs# identifiers were confirmed independently by a senior investigator (K.P.B.).
Analyses were performed for all cases with any form of diabetic retinopathy compared with all diabetic subjects without diabetic retinopathy (control subjects). Subanalysis was subsequently undertaken for NPDR and PDR where possible. Major and minor allele frequencies were calculated from the available genetic data of all reported variants in the included studies and odds ratios (ORs) and 95% CIs were calculated. The allelic association of microsatellite markers was also investigated, whereby the risk conferred by each allele was compared against all other alleles as well as against specific alleles. The Der Simonian and Laird random-effects model was used (18,19). This model utilizes weights that incorporate both within-study and between-study variance. Heterogeneity between studies was calculated as the inverse variance estimate. To minimize genetic heterogeneity, subanalysis was performed for all cohorts of white origin and for datasets in which heterogeneity remained; outlying studies were then removed in a stepwise fashion until homogeneity was achieved (20). For the purpose of this study, white ancestry was defined as being of European descent. Studies including subjects of white and nonwhite ancestry have had their cohorts divided and analyzed individually. Funnel plots were constructed in Revman and Egger's test (21) applied in Stata (version 10.1; Stata, College Station, TX) to investigate for publication bias. A P value of <0.05 was considered statistically significant in all analyses, except for publication bias (Egger's test), where a P value of <0.1 (22) was considered as statistically significant. No adjustments were made for multiple testing.
RESULTS
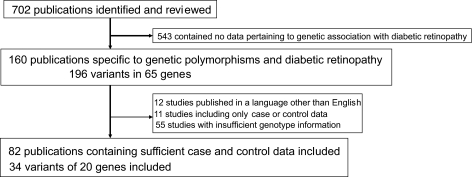
There were 702 publications identified. Of these, 160 were specific to genetic polymorphisms and the development of diabetic retinopathy (Fig. 1). Twelve of these studies were in a language other than English and were excluded, and a final total of 82 studies were suitable for inclusion as a result of having presented sufficient case and control genotype information. There were 196 polymorphisms identified, with 34 of these having adequate genotype data for inclusion in the meta-analysis. Visual inspection of the funnel plots revealed a symmetrical inverted V shape and Egger's test did not detect significant publication bias (P > 0.1) for all polymorphisms examined by more than five studies (supplementary Fig. 1, available in an online appendix at http://diabetes.diabetesjournals.org/cgi/content/full/db09-0059/DC1).
FIG. 1.
Flow chart of study selection process and included studies.
Seven polymorphisms were investigated by more than five studies and are discussed in detail (Table 1). Details of their study design are provided in supplementary Table 1. Forty-eight studies were included in the analyses for the seven polymorphisms, 20 (42%) of which included subjects of white ancestry (Table 2). Heterogeneity was eliminated in the subanalyses of studies with participants of white ancestry without the requirement of removal of outlying studies, indicating that ethnicity is a major source of heterogeneity generally.
TABLE 1.
Meta-analysis of genetic variants and risk for diabetic retinopathy in type 1 diabetes, type 2 diabetes, and both types of diabetes regardless of participant's ethnicity
| Gene | Variant | Risk allele | Comparison | Type of diabetes | Total case subjects with risk allele (%) | Total control subjects with risk allele (%) | Weight | OR (95% CI) | SE | Number of studies | χ2 | P | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACE | INS/DEL at intron 16 | 287 base pair deletion | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 651 (55) | 458 (52) | 32.76 | 1.06 (0.88–1.27) | 0.09 | 6 | 3.67 | 0.5402 | (S1–S6) |
| Type 2 | 986 (44) | 931 (43) | 67.24 | 1.01 (0.89–1.15) | 0.06 | 7 | 2.67 | 0.866 | (S3) (S7–S12) | ||||
| Total | 1,637 (47) | 1,389 (45) | 100.00 | 1.03 (0.93–1.14) | 0.05 | 13 | 6.51 | 0.6249 | |||||
| No diabetic retinopathy vs. NPDR | Type 2 | 276 (35) | 438 (36) | 100.00 | 0.91 (0.75–1.12) | 0.10 | 3 | 1.51 | 0.3786 | (S9) (S10) (S12) | |||
| No diabetic retinopathy vs. PDR | Type 1 | 387 (60) | 220 (55) | 48.63 | 1.19 (0.77–1.86) | 0.23 | 4 | 8.01 | 0.4355 | (S1) (S2) (S4) (S5) | |||
| Type 2 | 221 (41) | 438 (36) | 51.37 | 1.12 (0.89–1.42) | 0.12 | 3 | 0.21 | 0.3237 | (S9) (S10) (S12) | ||||
| Total | 608 (51) | 658 (40) | 100.00 | 1.17 (0.94–1.44) | 0.11 | 7 | 8.55 | 0.1515 | |||||
| NPDR vs. PDR | Type 1 | 120 (64) | 72 (45) | 17.02 | 2.21 (1.44–3.39) | 0.22 | 1 | 0 | 0.0003 | (S4) | |||
| Type 2 | 353 (45) | 481 (41) | 82.98 | 1.12 (0.92–1.35) | 0.1 | 6 | 4.75 | 0.2512 | (S9) (S10) (S12–S15) | ||||
| Total | 473 (49) | 553 (41) | 100.00 | 1.25 (0.95–1.65) | 0.14 | 7 | 12.77 | 0.1064 | |||||
| NOS3 | rs3138808 | 393 base pair insertion | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 21 (11) | 42 (22) | 9.91 | 0.43 (0.24–0.76) | 0.29 | 1 | 0.00 | 0.0036 | (S16) |
| Type 2 | 498 (21) | 705 (26) | 90.09 | 0.93 (0.74–1.17) | 0.12 | 7 | 13.63 | 0.5408 | (S17–S23) | ||||
| Total | 519 (21) | 747 (26) | 100.00 | 0.87 (0.68–1.11) | 0.13 | 8 | 19.38 | 0.2578 | |||||
| No diabetic retinopathy vs. NPDR | Type 1 | 21 (11) | 42 (22) | 32.47 | 0.43 (0.24–0.76) | 0.29 | 1 | 0.00 | 0.0036 | (S16) | |||
| Type 2 | 95 (20) | 116 (24) | 67.53 | 0.53 (0.12–2.46) | 0.78 | 2 | 22.10 | 0.421 | (S17) (S23) | ||||
| Total | 116 (17) | 158 (24) | 100.00 | 0.5 (0.19–1.31) | 0.49 | 3 | 22.99 | 0.157 | |||||
| No diabetic retinopathy vs. PDR | Type 2 | 43 (17) | 116 (24) | 100.00 | 0.42 (0.05–3.69) | 1.11 | 2 | 28.95 | 0.4341 | (S17) (S23) | |||
| NPDR vs. PDR | Type 2 | 43 (17) | 95 (20) | 100.00 | 0.79 (0.42–1.5) | 0.33 | 2 | 2.61 | 0.4723 | (S17) (S23) | |||
| VEGF | rs2010963 | G | No diabetic retinopathy vs. any diabetic retinopathy | Type 2 | 1,204 (55) | 1,144 (59) | 100.00 | 0.86 (0.7–1.05) | 0.10 | 7 | 14.65 | 0.1407 | (S22) (S24–S29) |
| No diabetic retinopathy vs. NPDR | Type 2 | 242 (53) | 328 (65) | 100.00 | 0.62 (0.48–0.81) | 0.14 | 3 | 1.93 | 0.0005 | (S24) (S28) (S29) | |||
| No diabetic retinopathy vs. PDR | Type 2 | 212 (56) | 643 (59) | 100.00 | 0.8 (0.6–1.06) | 0.14 | 4 | 0.92 | 0.1166 | (S24) (S26) (S28) (S29) | |||
| NPDR vs. PDR | Type 2 | 211 (56) | 242 (53) | 100.00 | 1.32 (0.99–1.76) | 0.15 | 3 | 0.81 | 0.0559 | (S24) (S28) (S29) | |||
| AKR1B1 | rs759853 | T | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 114 (29) | 149 (45) | 33.33 | 0.49 (0.36–0.68) | 0.16 | 3 | 0.02 | <0.0001 | (S30–S32) |
| Type 2 | 508 (34) | 460 (26) | 66.67 | 1.16 (0.98–1.36) | 0.08 | 5 | 3.96 | 0.0885 | (S33–S36) | ||||
| Total | 622 (33) | 609 (29) | 100.00 | 0.9 (0.66–1.22) | 0.16 | 8 | 25.19 | 0.4857 | |||||
| No diabetic retinopathy vs. NPDR | Type 1 | 8 (27) | 28 (43) | 14.13 | 0.49 (0.19–1.27) | 0.48 | 1 | 0.00 | 0.143 | (S32) | |||
| Type 2 | 175 (43) | 147 (39) | 85.87 | 1.17 (0.88–1.55) | 0.15 | 2 | 0.09 | 0.2945 | (S33) | ||||
| Total | 183 (42) | 175 (39) | 100.00 | 1.02 (0.7–1.5) | 0.20 | 3 | 3.00 | 0.9099 | |||||
| No diabetic retinopathy vs. PDR | Type 1 | 7 (25) | 28 (43) | 12.07 | 0.45 (0.17–1.21) | 0.50 | 1 | 0.00 | 0.1144 | (S32) | |||
| Type 2 | 123 (35) | 147 (39) | 87.93 | 0.84 (0.6–1.19) | 0.18 | 2 | 1.19 | 0.3285 | (S33) | ||||
| Total | 130 (34) | 175 (39) | 100.00 | 0.79 (0.55–1.13) | 0.18 | 3 | 2.51 | 0.1949 | |||||
| NPDR vs. PDR | Type 1 | 7 (25) | 8 (27) | 6.65 | 0.92 (0.28–2.98) | 0.60 | 1 | 0.00 | 0.8848 | (S32) | |||
| Type 2 | 123 (35) | 175 (43) | 93.35 | 0.76 (0.47–1.2) | 0.24 | 2 | 1.95 | 0.2364 | (S33) | ||||
| Total | 130 (34) | 183 (42) | 100.00 | 0.73 (0.54–0.99) | 0.16 | 3 | 2.12 | 0.0428 | |||||
| AKR1B1 | (CA)n dinucleotide repeat | z | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 344 (36) | 313 (32) | 39.83 | 1.09 (0.89–1.35) | 0.11 | 6 | 5.24 | 0.3945 | (S30) (S32) (S37–S40) |
| Type 2 | 436 (29) | 612 (25) | 60.17 | 1.04 (0.70–1.57) | 0.21 | 9 | 46.91 | 0.8330 | (S36) (S41–S48) | ||||
| Total | 780 (32) | 925 (27) | 100.00 | 1.05 (0.81–1.35) | 0.13 | 15 | 52.88 | 0.7264 | |||||
| z | No diabetic retinopathy vs. NPDR | Type 1 | 25 (41) | 20 (31) | 28.59 | 1.55 (0.75–3.23) | 0.37 | 1 | 0.00 | 0.2366 | (S32) | ||
| Type 2 | 97 (27) | 81 (37) | 71.41 | 0.65 (0.45–0.94) | 0.19 | 2 | 0.03 | 0.0215 | (S48) (S42) | ||||
| Total | 122 (29) | 101 (36) | 100.00 | 0.83 (0.49–1.42) | 0.27 | 3 | 4.38 | 0.4937 | |||||
| z | No diabetic retinopathy vs. PDR | Type 1 | 132 (43) | 147 (37) | 37.17 | 1.29 (0.95–1.75) | 0.15 | 2 | 0.30 | 0.0979 | (S39) (S32) | ||
| Type 2 | 179 (31) | 171 (38) | 62.83 | 0.77 (0.59–1.00) | 0.13 | 4 | 1.63 | 0.0482 | (S48) (S41–S43) | ||||
| Total | 311 (35) | 318 (37) | 100.00 | 0.94 (0.72–1.24) | 0.14 | 6 | 8.40 | 0.6738 | |||||
| z | NPDR vs. PDR | Type 1 | 25 (41) | 20 (31) | 28.59 | 1.55 (0.75–3.23) | 0.37 | 1 | 0.00 | 0.2366 | (S32) | ||
| Type 2 | 97 (27) | 81 (37) | 71.41 | 0.65 (0.45–0.94) | 0.19 | 2 | 0.03 | 0.0215 | (S48) (S42) | ||||
| Total | 122 (29) | 101 (36) | 100.00 | 0.83 (0.49–1.42) | 0.27 | 3 | 4.38 | 0.4937 | |||||
| Z−2 | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 362 (37) | 261 (27) | 41.98 | 1.95 (1.04–3.66) | 0.32 | 6 | 40.66 | 0.0367 | (S30) (S32) (S37–S40) | ||
| Type 2 | 385 (26) | 171 (7) | 58.02 | 2.64 (1.39–5.01) | 0.33 | 9 | 69.93 | 0.0029 | (S36) (S41–S48) | ||||
| Total | 747 (30) | 432 (13) | 100.00 | 2.33 (1.49–3.64) | 0.23 | 15 | 120.27 | 0.0002 | |||||
| Z−2 | No diabetic retinopathy vs. NPDR | Type 1 | 22 (36) | 23 (35) | 22.71 | 1.03 (0.50–2.13) | 0.37 | 1 | 0.00 | 0.9400 | (S32) | ||
| Type 2 | 137 (38) | 55 (25) | 77.29 | 1.87 (1.29–2.71) | 0.19 | 2 | 0.20 | 0.0010 | (S48) (S42) | ||||
| Total | 159 (38) | 78 (28) | 100.00 | 1.64 (1.14–2.35) | 0.18 | 3 | 2.26 | 0.0075 | |||||
| Z−2 | No diabetic retinopathy vs. PDR | Type 1 | 41 (24) | 35 (23) | 24.74 | 1.18 (0.69–2.01) | 0.27 | 2 | 0.31 | 0.5363 | (S32) (S40) | ||
| Type 2 | 173 (30) | 85 (19) | 75.26 | 1.64 (1.21–2.22) | 0.16 | 4 | 2.50 | 0.0016 | (S48) (S41–S43) | ||||
| Total | 214 (29) | 120 (20) | 100.00 | 1.51 (1.16–1.97) | 0.14 | 6 | 3.89 | 0.0023 | |||||
| Z−2 | NPDR vs. PDR | Type 1 | 19 (18) | 12 (14) | 18.52 | 1.39 (0.64–3.04) | 0.40 | 1 | 0.00 | 0.4076 | (S40) | ||
| Type 2 | 137 (38) | 55 (25) | 81.48 | 1.87 (1.29–2.71) | 0.19 | 2 | 0.20 | 0.0010 | (S48) (S42) | ||||
| Total | 156 (34) | 67 (22) | 100.00 | 1.77 (1.26–2.48) | 0.17 | 3 | 0.64 | 0.0009 | |||||
| z+2 | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 153 (16) | 224 (23) | 40.68 | 0.58 (0.36–0.93) | 0.24 | 6 | 18.16 | 0.0243 | (S30) (S32) (S37–S40) | ||
| Type 2 | 331 (22) | 521 (21) | 59.32 | 0.97 (0.57–1.65) | 0.27 | 9 | 64.22 | 0.9038 | (S36) (S41–S48) | ||||
| Total | 484 (20) | 745 (22) | 100.00 | 0.79 (0.53–1.17) | 0.20 | 15 | 104.32 | 0.2388 | |||||
| z+2 | No diabetic retinopathy vs. NPDR | Type 1 | 7 (12) | 13 (20) | 22.07 | 0.52 (0.19–1.40) | 0.51 | 1 | 0.00 | 0.1955 | (S32) | ||
| Type 2 | 34 (10) | 29 (14) | 77.93 | 0.71 (0.42–1.20) | 0.27 | 2 | 0.07 | 0.2001 | (S48) (S42) | ||||
| Total | 41 (10) | 42 (15) | 100.00 | 0.66 (0.41–1.05) | 0.24 | 3 | 0.36 | 0.0820 | |||||
| z+2 | No diabetic retinopathy vs. PDR | Type 1 | 48 (16) | 70 (18) | 38.34 | 0.84 (0.50–1.40) | 0.26 | 2 | 1.26 | 0.4971 | (S39) (S32) | ||
| Type 2 | 82 (14) | 98 (22) | 61.66 | 0.62 (0.39–0.99) | 0.24 | 4 | 4.71 | 0.0464 | (S41–S43) (S48) | ||||
| Total | 130 (15) | 168 (20) | 100.00 | 0.71 (0.51–0.98) | 0.17 | 6 | 6.92 | 0.0383 | |||||
| z+2 | NPDR vs. PDR | Type 1 | 7 (12) | 13 (20) | 22.07 | 0.52 (0.19–1.40) | 0.51 | 1 | 0.00 | 0.1955 | (S32) | ||
| Type 2 | 34 (10) | 29 (14) | 77.93 | 0.71 (0.42–1.20) | 0.27 | 2 | 0.07 | 0.2001 | (S48) (S42) | ||||
| Total | 41 (10) | 42 (15) | 100.00 | 0.66 (0.41–1.05) | 0.24 | 3 | 0.36 | 0.0820 |
All variants have been examined by a minimum of five studies. Subanalyses have been performed for NPDR and PDR if data were available. References are preceded with S as they correlate with the online supplementary appendix.
TABLE 2.
Meta-analysis of genetic variants and risk for diabetic retinopathy in patients with type 1 diabetes, type 2 diabetes, and both types of diabetes, for studies of white populations
| Gene | Variant | Risk allele | Comparison | Type of diabetes | Total case subjects with risk allele (%) | Total control subjects with risk allele (%) | Weight | OR (95% CI) | SE | Number of studies | χ2 | P | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACE | INS/DEL at intron 16 | 287 base pair deletion | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 651 (55) | 458 (52) | 50.735 | 1.06 (0.88–1.27) | 0.09 | 6 | 3.67 | 0.5402 | (S1–S6) |
| Type 2 | 504 (53) | 501 (54) | 49.265 | 0.99 (0.83–1.19) | 0.09 | 4 | 1.43 | 0.9182 | (S7) (S8) (S10) (S3) | ||||
| Total | 1,155 (54) | 959 (53) | 100 | 1.02 (0.9–1.16) | 0.07 | 10 | 5.35 | 0.7157 | |||||
| No diabetic retinopathy vs. PDR | Type 1 | 387 (60) | 220 (55) | 76.8488 | 1.19 (0.77–1.86) | 0.23 | 4 | 8.01 | 0.4355 | (S1) (S2) (S4) (S5) | |||
| Type 2 | 75 (53) | 78 (49) | 23.1512 | 1.14 (0.73–1.79) | 0.23 | 1 | 0.00 | 0.5617 | (S10) | ||||
| Total | 462 (59) | 298 (53) | 100 | 1.19 (0.86–1.66) | 0.17 | 5 | 8.11 | 0.2995 | |||||
| NPDR vs. PDR | Type 1 | 120 (64) | 72 (45) | 33.9165 | 2.21 (1.44–3.39) | 0.22 | 1 | 0.00 | 0.0003 | (S4) | |||
| Type 2 | 156 (54) | 224 (50) | 66.0835 | 1.23 (0.9–1.67) | 0.16 | 2 | 0.83 | 0.1905 | (S10) (S14) | ||||
| Total | 276 (58) | 296 (49) | 100 | 1.52 (1–2.32) | 0.22 | 3 | 5.55 | 0.0509 | |||||
| NOS3 | rs3138808 | 393 base pair insertion | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 21 (11) | 42 (22) | 47.0284 | 0.43 (0.24–0.76) | 0.29 | 1 | 0.00 | 0.0036 | (S16) |
| Type 2 | 123 (22) | 58 (21) | 52.9716 | 1.09 (0.77–1.55) | 0.18 | 1 | 0.00 | 0.6248 | (S21) | ||||
| Total | 144 (19) | 100 (21) | 100 | 0.71 (0.28–1.75) | 0.46 | 2 | 7.49 | 0.4512 | |||||
| VEGF | rs2010963 | G | No diabetic retinopathy vs. any diabetic retinopathy | Type 2 | 387 (55) | 247 (62) | 100 | 0.83 (0.65–1.07) | 0.13 | 2 | 0.43 | 0.1601 | (S27) (S28) |
| AKR1B1 | rs759853 | T | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 99 (29) | 121 (46) | 45.1355 | 0.5 (0.35–0.71) | 0.18 | 2 | 0.01 | 0.0001 | (S30) (S31) |
| Type 2 | 324 (42) | 202 (41) | 54.8645 | 1.05 (0.83–1.33) | 0.12 | 2 | 0.91 | 0.6756 | (S33) | ||||
| Total | 423 (38) | 323 (43) | 100 | 0.76 (0.5–1.16) | 0.22 | 4 | 12.83 | 0.2061 | |||||
| AKR1B1 | (CA)n dinucleotide repeat | z | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 277 (42) | 247 (39) | 75.2466 | 1.11 (0.88–1.39) | 0.117 | 3 | 0.88 | 0.3785 | (S30) (S37) (S39) |
| Type 2 | 79 (32) | 67 (42) | 24.7534 | 0.66 (0.44–1.00) | 0.21 | 1 | 0.00 | 0.05 | (S42) | ||||
| Total | 356 (39) | 314 (40) | 100 | 0.97 (0.73–1.27) | 0.14 | 4 | 5.47 | 0.8076 | |||||
| z | No diabetic retinopathy vs. PDR | Type 1 | 107 (43) | 127 (38) | 51.9056 | 1.24 (0.89–1.74) | 0.17 | 1 | 0.00 | 0.2008 | (S39) | ||
| Type 2 | 79 (32) | 67 (42) | 48.0944 | 0.66 (0.44–1.00) | 0.21 | 1 | 0.00 | 0.05 | (S42) | ||||
| Total | 186 (38) | 194 (39) | 100 | 0.92 (0.5–1.70) | 0.314 | 2 | 5.42 | 0.7872 | |||||
| z−2 | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 226 (34) | 167 (27) | 74.1019 | 1.83 (0.85–3.95) | 0.393 | 3 | 15.42 | 0.1238 | (S30) (S37) (S39) | ||
| Type 2 | 97 (40) | 43 (27) | 25.8981 | 1.78 (1.15–2.74) | 0.22 | 1 | 0.00 | 0.0091 | (S42) | ||||
| Total | 323 (36) | 210 (27) | 100 | 1.80 (1.06–3.06) | 0.271 | 4 | 15.89 | 0.0301 | |||||
| z−2 | No diabetic retinopathy vs. PDR | Type 1 | 87 (35) | 121 (36) | 52.3594 | 0.96 (0.68–1.35) | 0.175 | 1 | 0.00 | 0.8023 | (S39) | ||
| Type 2 | 97 (40) | 43 (27) | 47.6406 | 1.78 (1.15–2.74) | 0.22 | 1 | 0.00 | 0.0091 | (S42) | ||||
| Total | 184 (37) | 164 (33) | 100 | 1.29 (0.7–2.36) | 0.309 | 2 | 4.85 | 0.4166 | |||||
| z+2 | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 103 (16) | 159 (25) | 76.3483 | 0.51 (0.25–1.06) | 0.372 | 3 | 12.85 | 0.0722 | (S39) | ||
| Type 2 | 28 (12) | 24 (15) | 23.6517 | 0.73 (0.41–1.31) | 0.299 | 1 | 0.00 | 0.2958 | (S42) | ||||
| Total | 131 (15) | 183 (23) | 100 | 0.56 (0.32–0.97) | 0.282 | 4 | 13.71 | 0.039 | |||||
| z+2 | No diabetic retinopathy vs. PDR | Type 1 | 41 (17) | 57 (17) | 63.9459 | 0.97 (0.62–1.50) | 0.224 | 1 | 0.00 | 0.8814 | (S30) (S37) (S39) | ||
| Type 2 | 28 (12) | 24 (15) | 36.0541 | 0.73 (0.41–1.31) | 0.299 | 1 | 0.00 | 0.2958 | (S42) | ||||
| Total | 69 (14) | 81 (17) | 100 | 0.87 (0.62–1.24) | 0.179 | 2 | 0.56 | 0.455 |
Subanalyses were performed for NPDR and PDR if data were available. References are preceded with S as they correlate with the online supplementary appendix.
Data on the remaining 27 SNPs analyzed in the current study, which have been examined by a minimum of two and maximum of five cohorts, are presented in Table 3. The rs2910964 SNP of the α2β1 integrin (ITGA2) gene (OR 1.65 [95% CI 1.26–2.15], P = 2 × 10−4) and rs13306430 of the intercellular cell adhesion molecule 1 (ICAM1) gene (0.56 [0.39–0.81], P = 1.70 × 10−3) were significantly associated with diabetic retinopathy, both being examined in type 2 diabetes only and by two studies in each case.
TABLE 3.
Pooled ORs for 27 SNPs and risk for diabetic retinopathy in type 1 diabetes, type 2 diabetes, and both types of diabetes
| Gene | Variant | Risk allele | Comparison | Type of diabetes | Total case subjects with risk allele (%) | Total control subjects with risk allele (%) | Weight | OR (95% CI) | SE | Number of studies | χ2 | P | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AGTR1 | rs5186 | C | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 98 (29) | 200 (33) | 73.99 | 0.82 (0.61–1.10) | 0.15 | 2 | 1.87 | 0.1852 | (S6; S12) |
| Type 2 | 22 (4) | 46 (5) | 34.26 | 0.73 (0.43–1.22) | 0.26 | 1 | 0.00 | 0.2251 | (S12) | ||||
| Total | 142 (14) | 246 (15) | 100.00 | 0.8 (0.62–1.03) | 0.13 | 3 | 2.05 | 0.0788 | |||||
| ITGA2 | rs2910964 | A | No diabetic retinopathy vs. any diabetic retinopathy | Type 2 | 258 (46) | 137 (34) | 100.00 | 1.65 (1.26–2.15) | 0.14 | 2 | 0.05 | 0.0002 | (S49; S50) |
| ADRB3 | rs4994 | C | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 62 (7) | 12 (10) | 48.01 | 0.73 (0.38–1.39) | 0.33 | 1 | 0.00 | 0.3320 | (S51; S52) |
| Type 2 | 49 (27) | 34 (15) | 51.99 | 2.16 (1.32–3.51) | 0.25 | 1 | 0.00 | 0.0020 | (S51; S52) | ||||
| Total | 111 (11) | 46 (13) | 100.00 | 1.28 (0.44–3.72) | 0.54 | 2 | 6.95 | 0.6516 | |||||
| No diabetic retinopathy vs. NPDR | Type 1 | 27 (7) | 12 (10) | 47.68 | 0.71 (0.35–1.44) | 0.36 | 1 | 0.00 | 0.3420 | (S51; S52) | |||
| Type 2 | 21 (22) | 34 (15) | 52.32 | 1.71 (0.94–3.14) | 0.31 | 1 | 0.00 | 0.0811 | (S51; S52) | ||||
| Total | 48 (10) | 46 (13) | 100.00 | 1.12 (0.47–2.67) | 0.44 | 2 | 3.44 | 0.7900 | |||||
| No diabetic retinopathy vs. PDR | Type 1 | 35 (7) | 12 (10) | 48.89 | 0.74 (0.37–1.47) | 0.35 | 1 | 0.00 | 0.3892 | (S51; S52) | |||
| Type 2 | 28 (31) | 34 (15) | 51.11 | 2.68 (1.51–4.75) | 0.29 | 1 | 0.00 | 0.0008 | (S51; S52) | ||||
| Total | 63 (11) | 46 (13) | 100.00 | 1.43 (0.41–5.03) | 0.64 | 2 | 7.95 | 0.5797 | |||||
| NPDR vs. PDR | Type 1 | 35 (7) | 27 (7) | 61.48 | 1.04 (0.62–1.76) | 0.27 | 1 | 0.00 | 0.8706 | (S51; S52) | |||
| Type 2 | 28 (31) | 21 (22) | 38.52 | 1.56 (0.81–3.01) | 0.34 | 1 | 0.00 | 0.1829 | (S51; S52) | ||||
| Total | 63 (11) | 48 (10) | 100.00 | 1.22 (0.81–1.83) | 0.21 | 2 | 0.89 | 0.3399 | |||||
| AGT | rs4762 | C | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 111 (55) | 189 (63) | 37.28 | 0.73 (0.51–1.05) | 0.18 | 1 | 0.00 | 0.0875 | (S6; S12) |
| Type 2 | 90 (14) | 161 (17) | 62.72 | 0.84 (0.63–1.11) | 0.14 | 1 | 0.00 | 0.2102 | (S6; S12) | ||||
| Total | 201 (24) | 350 (27) | 100.00 | 0.79 (0.64–0.99) | 0.11 | 2 | 0.35 | 0.0418 | |||||
| APOE | ϵ2/ϵ3/ϵ4 | ϵ2 | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 21 (14) | 19 (10) | 19.55 | 1.46 (0.75–2.83) | 0.34 | 1 | 0.00 | 0.2620 | (S53; S54) |
| Type 2 | 223 (10) | 51 (11) | 80.45 | 1.05 (0.76–1.45) | 0.17 | 2 | 0.05 | 0.7681 | (S53; S54) | ||||
| Total | 244 (11) | 70 (11) | 100.00 | 1.12 (0.84–1.5) | 0.15 | 3 | 0.81 | 0.4470 | |||||
| FGF2 | rs41456044 | A | No diabetic retinopathy vs. PDR | Type 2 | 31 (5) | 47 (5) | 100.00 | 1.19 (0.74–1.91) | 0.24 | 2 | 0.27 | 0.4672 | (S55; S56) |
| FGF2 | rs308395 | G | No diabetic retinopathy vs. PDR | Type 2 | 90 (14) | 131 (13) | 100.00 | 1.08 (0.81–1.45) | 0.15 | 2 | 0.29 | 0.5882 | (S55; S56) |
| NOS3 | rs1799983 | G | No diabetic retinopathy vs. any diabetic retinopathy | Type 2 | 393 (30) | 478 (24) | 100.00 | 1.11 (0.94–1.31) | 0.08 | 4 | 1.69 | 0.2279 | (S17-S20) |
| NOS3 | rs41322052 | C | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 105 (42) | 106 (42) | 25.37 | 1.01 (0.71–1.44) | 0.18 | 1 | 0.00 | 0.9525 | (S57) |
| Type 2 | 313 (27) | 334 (24) | 74.63 | 1.04 (0.75–1.45) | 0.17 | 3 | 4.33 | 0.7956 | (S17; S19) | ||||
| Total | 418 (29) | 440 (27) | 100.00 | 1.06 (0.85–1.33) | 0.11 | 4 | 4.77 | 0.5962 | |||||
| No diabetic retinopathy vs. PDR | Type 1 | 105 (42) | 106 (42) | 77.27 | 1.01 (0.71–1.44) | 0.18 | 1 | 0.00 | 0.9525 | (S57) | |||
| Type 2 | 16 (12) | 29 (14) | 22.73 | 0.83 (0.43–1.59) | 0.33 | 1 | 0.00 | 0.5686 | (S17; S19) | ||||
| Total | 121 (31) | 135 (29) | 100.00 | 0.97 (0.71–1.32) | 0.16 | 2 | 0.28 | 0.8263 | |||||
| SLC2A1 | rs841853 | XbaI- | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 227 (35) | 142 (64) | 66.08 | 0.41 (0.13–1.23) | 0.57 | 2 | 8.27 | 0.1118 | (S58; S59) |
| HLA | DR1–8 | DR1 | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 45 (14) | 40 (15) | 100.00 | 0.99 (0.62–1.56) | 0.2359 | 2 | 0.2 | 0.9506 | (S60; S61) |
| HLA | DR1–8 | DR7 | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 39 (12) | 38 (14) | 100.00 | 0.59 (0.10–3.60) | 0.9247 | 2 | 1.86 | 0.5658 | (S60; S61) |
| ICAM1 | rs13306430 | G | No diabetic retinopathy vs. any diabetic retinopathy | Type 2 | 150 (36) | 89 (50) | 100.00 | 0.56 (0.39–0.81) | 0.18 | 2 | 0.01 | 0.0017 | (S62; S63) |
| MTHFR | rs1801133 | T | No diabetic retinopathy vs. any diabetic retinopathy | Type 2 | 318 (44) | 405 (37) | 100.00 | 1.39 (0.99–1.94) | 0.17 | 4 | 8.34 | 0.0563 | (S35; S64–S66) |
| NPY | rs16139 | C | No diabetic retinopathy vs. any diabetic retinopathy | Type 2 | 26 (9) | 16 (5) | 100.00 | 2.62 (0.9–7.61) | 0.54 | 2 | 1.38 | 0.0759 | (S67; S68) |
| PAI-1 | rs1799768 | 5G | No diabetic retinopathy vs. any diabetic retinopathy | Type 2 | 355 (44) | 252 (45) | 100.00 | 1.06 (0.85–1.32) | 0.11 | 3 | 0.00 | 0.6017 | (S10; S35; S69) |
| PON1 | rs662 | G | No diabetic retinopathy vs. any diabetic retinopathy | Type 2 | 310 (54) | 175 (48) | 100.00 | 0.99 (0.44–2.27) | 0.42 | 2 | 8.48 | 0.9896 | (S69; S70) |
| PON2 | rs7493 | G | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 192 (57) | 191 (49) | 54.02 | 1.37 (1.03–1.84) | 0.15 | 1 | 0.00 | 0.0322 | (S70; S71) |
| Type 2 | 144 (72) | 144 (78) | 45.98 | 0.72 (0.46–1.15) | 0.23 | 1 | 0.00 | 0.1685 | (S70; S71) | ||||
| Total | 336 (62) | 335 (58) | 100.00 | 1.02 (0.55–1.91) | 0.32 | 2 | 5.33 | 0.9422 | |||||
| PPARG | rs1801282 | G | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 33 (13) | 108 (14) | 29.35 | 0.85 (0.56–1.29) | 0.21 | 1 | 0.00 | 0.4413 | (S72-S75) |
| Type 2 | 124 (14) | 273 (16) | 70.65 | 0.83 (0.62–1.11) | 0.15 | 4 | 3.33 | 0.2135 | (S72-S75) | ||||
| Total | 157 (14) | 381 (16) | 100.00 | 0.83 (0.67–1.05) | 0.12 | 5 | 3.34 | 0.1173 | |||||
| AGER | rs1800624 | A | No diabetic retinopathy vs. any diabetic retinopathy | Type 2 | 361 (33) | 112 (18) | 100.00 | 1.89 (0.53–6.75) | 0.65 | 3 | 46.09 | 0.3271 | (S76-S78) |
| No diabetic retinopathy vs. NPDR | Type 2 | 217 (37) | 60 (12) | 100.00 | 2.94 (0.46–18.76) | 0.95 | 2 | 26.55 | 0.2540 | (S76; S78) | |||
| No diabetic retinopathy vs. PDR | Type 2 | 67 (26) | 93 (20) | 100.00 | 0.93 (0.62–1.38) | 0.20 | 2 | 0.88 | 0.7055 | ||||
| AGER | rs1800625 | C | No diabetic retinopathy vs. any diabetic retinopathy | Type 2 | 125 (13) | 94 (15) | 100.00 | 0.94 (0.7–1.27) | 0.15 | 3 | 0.32 | 0.7035 | (S76; S78) |
| No diabetic retinopathy vs. PDR | Type 2 | 35 (14) | 76 (15) | 100.00 | 0.99 (0.64–1.55) | 0.23 | 2 | 0.99 | 0.9803 | (S76; S78) | |||
| VDR | rs10735810 | C | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 207 (40) | 86 (34) | 57.59 | 1.27 (0.93–1.74) | 0.16 | 1 | 0.00 | 0.1282 | (S79) |
| Type 2 | 85 (50) | 169 (47) | 42.41 | 1.15 (0.8–1.66) | 0.19 | 1 | 0.00 | 0.4415 | (S80) | ||||
| Total | 292 (42) | 255 (42) | 100.00 | 1.22 (0.96–1.55) | 0.12 | 2 | 0.17 | 0.0978 | |||||
| VEGF | rs25648 | T | No diabetic retinopathy vs. any diabetic retinopathy | Type 2 | 86 (16) | 56 (14) | 100.00 | 1.48 (0.42–5.19) | 0.64 | 2 | 9.06 | 0.5363 | (S22; S24) |
| VEGF | rs1570360 | A | No diabetic retinopathy vs. any diabetic retinopathy | Type 2 | 78 (12) | 81 (13) | 100.00 | 0.93 (0.67–1.3) | 0.17 | 2 | 0.22 | 0.6740 | (S24; S25) |
| VEGF | rs3095039 | T | No diabetic retinopathy vs. any diabetic retinopathy | Type 2 | 86 (16) | 52 (13) | 100.00 | 1.1 (0.47–2.62) | 0.44 | 2 | 4.47 | 0.8230 | (S24; S29) |
| No diabetic retinopathy vs. NPDR | Type 2 | 45 (14) | 52 (13) | 100.00 | 0.97 (0.33–2.85) | 0.55 | 2 | 5.03 | 0.9560 | (S24; S29) | |||
| No diabetic retinopathy vs. PDR | Type 2 | 41 (18) | 52 (13) | 100.00 | 1.44 (0.88–2.36) | 0.25 | 2 | 1.13 | 0.1491 | (S24; S29) | |||
| NPDR vs. PDR | Type 2 | 41 (18) | 45 (14) | 100.00 | 1.22 (0.76–1.96) | 0.24 | 2 | 1.01 | 0.4146 | (S24; S29) | |||
| VEGF | rs35569394 | -2549DEL | No diabetic retinopathy vs. any diabetic retinopathy | Type 1 | 73 (58) | 68 (52) | 46.01 | 1.25 (0.77–2.04) | 0.25 | 1 | 0.00 | 0.3724 | (S81) |
| Type 2 | 252 (65) | 81 (45) | 53.99 | 2.28 (1.59–3.26) | 0.18 | 1 | 0.00 | 1.0 × 10.5 | (S82) | ||||
| Total | 325 (63) | 149 (48) | 100.00 | 1.73 (0.96–3.11) | 0.30 | 2 | 3.77 | 0.0677 | |||||
| VEGF | rs699947 | A | No diabetic retinopathy vs. any diabetic retinopathy | Type 2 | 204 (29) | 280 (29) | 100.00 | 1.01 (0.56–1.83) | 0.30 | 2 | 7.61 | 0.9741 | (S25; S26) |
All SNPs have been examined by a minimum of two and maximum of five studies. Subanalyses have been performed for NPDR and PDR if data were available. References are preceded with S as they correlate with the online supplementary appendix.
ACE.
The gene encoding ACE is located on chromosome 17q23. Six studies examined the insertion/deletion (INS/DEL) polymorphism in intron 16 of the ACE gene in patients with type 1 diabetes, and seven studies in patients with type 2 diabetes were included in the meta-analysis. The 287 base pair deletion was treated as the risk variant, and there was no statistically significant association with this polymorphism and the development of any form of diabetic retinopathy (Table 1).
Ten studies of subjects with white origin were subanalyzed for all diabetic retinopathy comparisons. The 287 base pair deletion was not found to be significantly associated with any diabetic retinopathy or its subtypes in type 1 or type 2 diabetes (Table 2).
Aldose reductase (AKR1B1).
The aldo-keto reductase family 1 member B1 (AKR1B1) gene (also known as ALR) is located on chromosome 7q35. Associations of two AKR1B1 SNPs with diabetic retinopathy have been reported in the literature: the promoter SNP rs759853 and the (CA)n microsatellite polymorphism located 5′ of the AKR1B1 gene. Six studies have examined the association between the AKR1B1 (CA)n microsatellite with diabetic retinopathy in type 1 and nine studies in type 2 diabetes (Table 1). The three most commonly investigated AKR1B1 variants (z, z+2, and z−2) in the literature were included for analysis.
There was a significant association with the z−2 allele and the development of any diabetic retinopathy (OR 2.33 [95% CI 1.49–3.64], P = 2 × 10−4). Subanalyses revealed a significant association between the z−2 allele and diabetic retinopathy in patients with type 2 diabetes (2.64 [1.39–5.01], P = 2.9 × 10−3), with weaker but statistically significant association also being found for patients with type 1 diabetes (1.95 [1.04–3.66], P = 0.04). A significant association was also found in the NPDR and PDR subgroups.
No statistically significant association was found between the z allele and the development of diabetic retinopathy overall (OR 1.05 [95% CI 0.81–1.35], P = 0.73). However, in the subanalysis for type of diabetic retinopathy, the z allele was significantly protective against NPDR development in type 2 diabetes (0.65 [0.45–0.94], P = 0.02). A significant difference between NPDR and PDR development for the presence of z allele was found also in type 2 diabetes (0.65 [0.45–0.94], P = 0.02). Similarly, the z+2 allele was found to be significantly protective against the development of diabetic retinopathy (0.58 [0.36–0.93], P = 0.02).
Only four studies examining the AKR1B1 (CA)n microsatellite have included subjects of white ancestry (Table 2), with the majority of studies including subjects of only Asian ancestry. In the analyses of studies of white origin, only the z−2 (OR 1.80 [95% CI 1.06–3.06], P = 0.03) and z+2 (0.56 [0.32–0.97], P = 0.04) polymorphisms remained significantly associated with diabetic retinopathy, with the z−2 allele conferring risk and z+2 conferring protection against diabetic retinopathy.
Three studies examined the association of a second AKR1B1 polymorphism (promoter SNP rs759853) with diabetic retinopathy in type 1 diabetes and five studies in type 2 diabetes (Table 1). The T allele was considered the risk variant. Interestingly, analyses revealed protection against diabetic retinopathy with the T allele in type 1 diabetes (OR 0.49 [95% CI 0.36–0.68], P < 1.00 × 10−4). There was no statistically significant association between diabetic retinopathy and rs759853 in type 2 diabetes. However, a borderline association between NPDR and PDR was found (0.73 [0.54–0.99], P = 0.04).
Four studies investigating the AKR1B1 rs759853 included subjects of white ancestry. A significant protection of the T allele against diabetic retinopathy in type 1 diabetes (OR 0.5 [95% CI 0.35–0.71], P = 1.00 × 10−4) remained. Insufficient studies were available for subanalysis of diabetic retinopathy subtypes.
Vascular endothelial growth factor.
The vascular endothelial growth factor (VEGF) gene is located on chromosome 6p12. Six VEGF polymorphisms were included in this meta-analysis, with the rs2010963 polymorphism being the most frequently studied. No studies examining this polymorphism in type 1 diabetes and diabetic retinopathy were located. Seven studies examining this polymorphism in type 2 diabetes and diabetic retinopathy were included in the analyses (Table 1). The G allele has been considered as the risk variant. A significant association between patients with no diabetic retinopathy and those with NPDR (OR 0.62 [95% CI 0.48–0.81], P = 5.0 × 10−4) was identified, yet no significant differences were found between NPDR and PDR development. Meta-analysis revealed no significant association between the VEGF polymorphisms: rs25648, rs1570360, rs3095039, rs35569394, or rs699947 and any type of diabetic retinopathy (Table 3).
Only two studies of participants were available for inclusion in the subanalysis for white ancestry (Table 2). No statistically significant association of the VEGF rs2010963 polymorphism with diabetic retinopathy was found (OR 0.83 [95% CI 0.65–1.07], P = 0.16) and insufficient studies were available for diabetic retinopathy subtype analyses.
Endothelial nitric oxide synthase.
The endothelial nitric oxide synthase (NOS3) gene is located on chromosome 7q35–36. Three NOS3 SNPs (rs1799983, rs41322052, and rs3138808) met the inclusion criteria for meta-analysis. The rs3138808 variant has been the most commonly examined NOS3 diabetic retinopathy polymorphism, and the 393 base pair insertion has been classified as the risk variant. One study examining this polymorphism and diabetic retinopathy in type 1 diabetes and seven studies in type 2 diabetes were included in the analysis (Table 1). There was no statistically significant association between rs3138808 and any form of diabetic retinopathy. Additionally, no significant association between any form of diabetic retinopathy and the rs1799983 (OR 1.11 [95% CI 0.94–1.31], P = 0.23) or rs41322052 (1.06 [0.85–1.33], P = 0.60) were identified (Table 3).
Only two studies were available to be included in the subanalysis for subjects of white ancestry (Table 2). No statistically significant association was found with the NOS3 rs3138808 polymorphism and diabetic retinopathy development (OR 0.71 [95% CI 0.28–1.75], P = 0.45) and insufficient studies were available for diabetic retinopathy subtype analysis.
DISCUSSION
Diabetic retinopathy remains one of the leading causes of blindness in the developed world (1). There is strong evidence for a genetic component in the development of diabetic retinopathy, independent of other established risk factors (13–17). Despite a large number of candidate gene studies on diabetic retinopathy, the genetic evidence thus far for an association with the development of or severity of diabetic retinopathy has been conflicting. This is largely accounted for by varying participant ethnicity, study design, differences in retinopathy grading scales, and in particular suboptimal power.
The current meta-analysis comprehensively assesses the risk of diabetic retinopathy in relation to every published candidate gene meeting inclusion criteria. Eighty-two studies examining 20 genes and 34 SNPs were analyzed. Pathways involved in the pathogenesis of diabetic retinopathy relevant to the genes with the most studied polymorphisms in this meta-analysis are described in detail below.
Renin-angiotensin system.
ACE is involved in the conversion of angiotensin I to angiotensin II (ATII). ATII mediates its hemodynamic effects through signaling via angiotensin type 1 (AT1) and type 2 (AT2) receptors. Vascular remodeling and proliferation occurs mainly via the AT1 receptor (23). All components of the renin-angiotensin system (RAS) have been shown to be expressed in the retina (24). The physiologic effects of ATII in the eye include the regulation of intraocular blood flow and pressure, promotion of capillary growth, enhancing vascular permeability, increasing oxidative stress, and the regulation of cell growth via the expression of various growth factors including VEGF, insulin-like growth factor, and platelet-derived growth factor (23).
Clinical evidence also supports the role for the RAS system in diabetic retinopathy pathogenesis. The EURODIAB Controlled Trial of Lisinopril in Insulin-Dependent Diabetes (EUCLID) trial provided evidence for lisinopril, an ACE inhibitor, decreasing the progression of diabetic retinopathy by 50% (25). In addition, animal studies have shown that ACE inhibitors and AT1 receptor blockers can prevent retinal neovascularization (26).
The ACE INS/DEL polymorphism in intron 16 had the largest number of subjects to be genotyped for any polymorphism and the largest number of studies to include participants of white ancestry. Three-quarters of the studies included participants of white ancestry. There was no statistically significant association of the INS/DEL polymorphism of the ACE gene with diabetic retinopathy or diabetic retinopathy subtypes in type 1, type 2, or combined diabetes. This is consistent with the findings of the majority of the included studies examining this polymorphism in this meta-analysis. Two other meta-analyses have examined the association of the INS/DEL of the ACE gene with diabetic retinopathy development. Fujisawa et al. (27) examined 12 studies, including type 1 and type 2 diabetic subjects. Wiwanitkit (28) examined four studies and included type 2 diabetic subjects only. Both analyses also found no statistically significant association with this polymorphism and the development of diabetic retinopathy.
Polyol pathway.
Aldose reductase (ALR) is a rate-limiting enzyme of the polyol pathway, which catalyzes NADPH-dependent reduction of glucose to sorbitol. This pathway leads to the intracellular accumulation of sorbitol and is primarily active under hyperglycemic conditions (29). Several mechanisms have been proposed to explain the pathogenesis of diabetic microvascular complications, including the induction of osmotic stress and the activation of protein kinase C as well as pathogenic vascular and hemodynamic alterations (8). ALR has been identified in human pericytes, which exhibit an active polyol pathway (30). Animal studies have shown that cultured mural cells from the retinal capillaries of adult rhesus monkeys undergo cellular degeneration after exposure to high glucose levels, with ALR being isolated from these mural cells (31). The sorbitol pathway is also biologically plausible as it is involved in the selective degeneration of human mural cells in NPDR (32). Induced hyperglycemia in dogs with galactosemia has similarly shown retinal vascular changes including microaneurysm formation, degeneration of retinal pericytes, retinal hemorrhages, and nonperfused or acellular vessels (33).
The AKR1B1 gene had the largest number of studies examining the relationship of its polymorphisms to diabetic retinopathy development, regardless of ethnicity. The z−2 microsatellite showed the most significant association with diabetic retinopathy, especially in type 2 diabetes, conferring risk also in PDR and NPDR subtypes. The z+2 and z microsatellite conferred protection against overall diabetic retinopathy and NPDR, respectively, and both were protective against PDR when compared with NPDR in type 2 diabetes. The majority of studies included in this meta-analysis have individually reported a risk for diabetic retinopathy with the z−2 microsatellite; however, the z microsatellite was not found to be statistically significant by most studies, with only a fifth of the studies individually reporting z+2 to be protective against diabetic retinopathy. The vast majority of studies have included participants of non-white ancestry; however, those of white ancestry found the z−2 microsatellite to confer risk and z+2 to confer protection against overall diabetic retinopathy in combined diabetes.
The T allele of the AKR1B1 promoter SNP rs759853 conferred protection against diabetic retinopathy in type 1 diabetes of any ancestry and also of white ancestry alone. This protection against diabetic retinopathy in type 1 diabetes has also been found in the individual studies examining this polymorphism.
VEGF.
VEGF is a multifunctional cytokine that promotes angiogenesis and is a potent mediator of microvascular permeability. Diabetic microvascular changes in the retina lead to hypoxia, a stimulator of VEGF production (34). VEGF has been found to have a significant role in the development of diabetic retinopathy by inducing hyperpermeability of retinal vessels, breakdown of the blood–retinal barrier, and neovascularization in PDR (35–37). Complications such as retinal edema and blinding vitreous hemorrhage arise as a result of the abnormal barrier function of vessels and the growth of new vessels, which are fragile and prone to rupture.
VEGF protein expression has been shown to be influenced by genetic variation in the VEGF gene (38). VEGF levels in the vitreous of patients with PDR are significantly elevated when compared with the vitreous of diabetic eyes without PDR and control subjects without diabetes (39,40). VEGF inhibition has been shown to result in a marked reduction in retinal neovascularization (41) and prevention of the blood–retinal barrier breakdown (42), further supporting its role in diabetic retinopathy development.
The VEGF gene had the largest number of individual SNPs examined in relation to diabetic retinopathy. Of these six polymorphisms, the G variant of the rs2010963 polymorphism was found to significantly protect against the development of NPDR in type 2 diabetes but not with overall diabetic retinopathy development. In keeping with the findings of this meta-analysis, the majority of studies reported no statistically significant association of this polymorphism with any diabetic retinopathy development in type 2 diabetes, regardless of ethnicity.
NO pathway.
Endothelial NO synthase (eNOS) is an enzyme produced by endothelial cells. NO derived from eNOS is a key endogenous vasodilator (43). It is also believed to be important in the promotion of angiogenesis and regulation of VEGF expression (44). A low concentration of eNOS is believed to be necessary to maintain endothelial function (45) with experimental deficiency of eNOS shown to significantly decrease retinal neovascularization in a mouse model (46). Aqueous NO levels have also been found to play an important role in the progression of diabetic retinopathy, with levels significantly higher in active PDR (47). Similarly, NO levels have been found to be significantly elevated in PDR vitreous when compared with nondiabetic subjects (48), making the eNOS gene (NOS3) a biologically plausible candidate for susceptibility to diabetic retinopathy development.
The NOS3 rs3138808 polymorphism was the most studied polymorphism, and the majority of studies included participants of nonwhite ancestry. No statistically significant association was found with diabetic retinopathy development regardless of ethnicity and for whites alone, consistent with the findings of the majority of the included studies. All other NOS3 polymorphisms did not show statistically significant associations with the development of diabetic retinopathy.
Data on the remaining 27 SNPs examined by a minimum of two and maximum of five studies revealed the rs2910964 SNP of the ITGA2 gene and rs13306430 of the ICAM1 gene to be significantly associated with diabetic retinopathy. Both of these variants were examined in type 2 diabetes only and by the minimum allowable two studies each. Unless further replicative studies are undertaken, the importance of these genes in diabetic retinopathy may remain unclear. Zintzaras et al. (49) performed a meta-analysis of five studies examining the link between the C677T polymorphism of the methylenetetrahydrofolate reductase (MTHFR) gene and development of diabetic retinopathy in type 2 diabetes. Unlike our results, a borderline association between C677T transition and the risk of development diabetic retinopathy (OR 1.39 [95% CI 1.05–1.83], P = 0.08) was reported.
Because of the publication deadline for included studies, some polymorphisms, such as those recently reported in the erythropoietin gene (50), were unable to be included in the meta-analysis as further studies have not as yet been reported. The definition of diabetic retinopathy requires the minimum of microaneurysm presence in a diabetic individual (34); however, several definitions and criteria exist for the subclassification of diabetic retinopathy and preferences have been variable among included studies, with many studies defining diabetic retinopathy without the use of standardized scales. Additionally, confounding factors such as glycemic control have not been adjusted for in calculation of the ORs of polymorphisms included in this meta-analysis, as this information provided by included studies have often been incomplete and nonstandardized definitions were used. Therefore, these are accepted as limitations of this meta-analysis, with some being implicit in the meta-analysis concept when dealing with a large number of studies with different design and reporting style.
In conclusion, this meta-analysis has found that sequence variation within the AKR1B1 gene are the most significantly associated with diabetic retinopathy development among those genes qualifying for inclusion. This result supports more detailed research into the role of the AKR1B1 gene in diabetic retinopathy to determine which variants are causally associated and their mechanism of action. Genetic research on diabetic retinopathy susceptibility has the potential for a direct positive impact on patient management by ultimately altering screening regimes in an individualized manner for a disease that is of major and increasing public health concern. Although much work has focused on dissecting the genetics of diabetes itself, considerably less has been conducted into the molecular mechanisms leading to its specific complications, including diabetic retinopathy and its subtypes. Although NPDR and PDR are commonly subanalyzed in genetic studies, very few studies have reported on genetic associations with the common subtype of macular edema, that is a frequent cause of visual disability in diabetic subjects. This is an area that requires further exploration. Future genetic studies should also include genome-wide association studies to identify diabetic retinopathy susceptibility loci that have not been previously considered as candidates to assist with a better understanding of the pathogenesis of this debilitating sight-threatening diabetes complication.
Supplementary Material
Acknowledgments
This research was supported by a grant from the Ophthalmic Research Institute of Australia. K.P.B. is a Peter Doherty Fellow of the National Health and Medical Research Council of Australia (NHMRC) and J.E.C. is an NHMRC Practitioner Fellow.
No other potential conflicts of interest relevant to this article were reported.
We acknowledge Richard Woodman for his statistical advice and expertise.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Resnikoff S, Pascolini D, Etya'ale D, Kocur I, Pararajasegaram R, Pokharel GP, Mariotti SP: Global data on visual impairment in the year 2002. Bull World Health Organ 2004; 82: 844– 851 [PMC free article] [PubMed] [Google Scholar]
- 2.Kempen JH, O'Colmain BJ, Leske MC, Haffner SM, Klein R, Moss SE, Taylor HR, Hamman RF: The prevalence of diabetic retinopathy among adults in the United States. Arch Ophthalmol 2004; 122: 552– 563 [DOI] [PubMed] [Google Scholar]
- 3.National Health and Medical Research Council. Management of Diabetic Retinopathy: Clinical Practice Guidelines Canberra, NHMRC; 2008 [Google Scholar]
- 4.Roy MS, Klein R, O'Colmain BJ, Klein BE, Moss SE, Kempen JH: The prevalence of diabetic retinopathy among adult type 1 diabetic persons in the United States. Arch Ophthalmol 2004; 122: 546– 551 [DOI] [PubMed] [Google Scholar]
- 5.Klein R, Klein BE, Moss SE, Cruickshanks KJ: The Wisconsin Epidemiologic Study of Diabetic Retinopathy. XV. The long-term incidence of macular edema. Ophthalmology 1995; 102: 7– 16 [DOI] [PubMed] [Google Scholar]
- 6.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, Lowe CE, Szeszko JS, Hafler JP, Zeitels L, Yang JH, Vella A, Nutland S, Stevens HE, Schuilenburg H, Coleman G, Maisuria M, Meadows W, Smink LJ, Healy B, Burren OS, Lam AA, Ovington NR, Allen J, Adlem E, Leung HT, Wallace C, Howson JM, Guja C, Ionescu-Tirgoviste C, Simmonds MJ, Heward JM, Gough SC, Dunger DB, Wicker LS, Clayton DG: Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 2007; 39: 857– 864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P: A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007; 445: 881– 885 [DOI] [PubMed] [Google Scholar]
- 8.Brownlee M: Biochemistry and molecular cell biology of diabetic complications. Nature 2001; 414: 813– 820 [DOI] [PubMed] [Google Scholar]
- 9.Diabetes Control and Complications Trial Research Group. The relationship of glycemic exposure (HbA1c) to the risk of development and progression of retinopathy in the Diabetes Control and Complications Trial. Diabetes 1995; 44: 968– 983 [PubMed] [Google Scholar]
- 10.Stratton IM, Adler AI, Neil HA, Matthews DR, Manley SE, Cull CA, Hadden D, Turner RC, Holman RR: Association of glycaemia with macrovascular and microvascular complications of type 2 diabetes (UKPDS 35): prospective observational study. BMJ 2000; 321: 405– 412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein R, Klein BE, Moss SE, Davis MD, DeMets DL: The Wisconsin Epidemiologic Study of Diabetic Retinopathy. II. Prevalence and risk of diabetic retinopathy when age at diagnosis is less than 30 years. Arch Ophthalmol 1984; 102: 520– 526 [DOI] [PubMed] [Google Scholar]
- 12.Jerneld B, Algvere P: Relationship of duration and onset of diabetes to prevalence of diabetic retinopathy. Am J Ophthalmol 1986; 102: 431– 437 [DOI] [PubMed] [Google Scholar]
- 13.The Diabetes Control and Complications Trial Research Group. Clustering of long-term complications in families with diabetes in the Diabetes Control and Complications Trial. Diabetes 1997; 46: 1829– 1839 [PubMed] [Google Scholar]
- 14.Hallman DM, Huber JC, Jr, Gonzalez VH, Klein BE, Klein R, Hanis CL: Familial aggregation of severity of diabetic retinopathy in Mexican Americans from Starr County, Texas. Diabetes Care 2005; 28: 1163– 1168 [DOI] [PubMed] [Google Scholar]
- 15.Arar NH, Freedman BI, Adler SG, Iyengar SK, Chew EY, Davis MD, Satko SG, Bowden DW, Duggirala R, Elston RC, Guo X, Hanson RL, Igo RP, Jr, Ipp E, Kimmel PL, Knowler WC, Molineros J, Nelson RG, Pahl MV, Quade SR, Rasooly RS, Rotter JI, Saad MF, Scavini M, Schelling JR, Sedor JR, Shah VO, Zager PG, Abboud HE: Heritability of the severity of diabetic retinopathy: the FIND-Eye study. Invest Ophthalmol Vis Sci 2008; 49: 3839– 3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monti MC, Lonsdale JT, Montomoli C, Montross R, Schlag E, Greenberg DA: Familial risk factors for microvascular complications and differential male-female risk in a large cohort of American families with type 1 diabetes. J Clin Endocrinol Metab 2007; 92: 4650– 4655 [DOI] [PubMed] [Google Scholar]
- 17.Hietala K, Forsblom C, Summanen P, Groop PH: Heritability of proliferative diabetic retinopathy. Diabetes 2008; 57: 2176– 2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DerSimonian R, Laird N: Meta-analysis in clinical trials. Control Clin Trials 1986; 7: 177– 188 [DOI] [PubMed] [Google Scholar]
- 19.DerSimonian R, Kacker R: Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials 2007; 28: 105– 114 [DOI] [PubMed] [Google Scholar]
- 20.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE: Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet 2007; 39: 17– 23 [DOI] [PubMed] [Google Scholar]
- 21.Egger M, Davey Smith G, Schneider M, Minder C: Bias in meta-analysis detected by a simple, graphical test. BMJ 1997; 315: 629– 634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sterne JA, Egger M: Funnel plots for detecting bias in meta-analysis: guidelines on choice of axis. J Clin Epidemiol 2001; 54: 1046– 1055 [DOI] [PubMed] [Google Scholar]
- 23.Funatsu H, Yamashita H: Pathogenisis of diabetic retinopathy and the renin-angiotensin system. Ophthalmic and Physiological Optics 2003; 23: 495– 501 [DOI] [PubMed] [Google Scholar]
- 24.Wagner J, Jan Danser AH, Derkx FH, de Jong TV, Paul M, Mullins JJ, Schalekamp MA, Ganten D: Demonstration of renin mRNA, angiotensinogen mRNA, and angiotensin converting enzyme mRNA expression in the human eye: evidence for an intraocular renin-angiotensin system. Br J Ophthalmol 1996; 80: 159– 163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The EUCLID Study Group. Randomised placebo-controlled trial of lisinopril in normotensive patients with insulin-dependent diabetes and normoalbuminuria or microalbuminuria. Lancet 1997; 349: 1787– 1792 [PubMed] [Google Scholar]
- 26.Moravski CJ, Kelly DJ, Cooper ME, Gilbert RE, Bertram JF, Shahinfar S, Skinner SL, Wilkinson-Berka JL: Retinal neovascularization is prevented by blockade of the renin-angiotensin system. Hypertension 2000; 36: 1099– 1104 [DOI] [PubMed] [Google Scholar]
- 27.Fujisawa T, Ikegami H, Kawaguchi Y, Hamada Y, Ueda H, Shintani M, Fukuda M, Ogihara T: Meta-analysis of association of insertion/deletion polymorphism of angiotensin I-converting enzyme gene with diabetic nephropathy and retinopathy. Diabetologia 1998; 41: 47– 53 [DOI] [PubMed] [Google Scholar]
- 28.Wiwanitkit V: Angiotensin-converting enzyme gene polymorphism is correlated to diabetic retinopathy: a meta-analysis. J Diabetes Complications 2008; 22: 144– 146 [DOI] [PubMed] [Google Scholar]
- 29.Kador PF, Kinoshita JH: Role of aldose reductase in the development of diabetes-associated complications. Am J Med 1985; 79: 8– 12 [DOI] [PubMed] [Google Scholar]
- 30.Hohman TC, Nishimura C, Robison G: Aldose reductase and polyol in cultured pericytes of human retinal capillaries. Exp Eye Res 1989; 48: 55– 60 [DOI] [PubMed] [Google Scholar]
- 31.Buzney SM, Frank RN, Varma SD, Tanishima T, Gabbay KH: Aldose reductase in retinal mural cells. Invest Ophthalmol Visual Sci 1977; 16: 392– 396 [PubMed] [Google Scholar]
- 32.Akagi Y, Kador PF, Kuwabara T, Kinoshita JH: Aldose reductase localisation in human retinal mural cells. Invest Ophthalmol Visual Sci 1983; 24: 1516– 1519 [PubMed] [Google Scholar]
- 33.Engerman RL, Kern TS: Experimental galactosemia produces diabetic-like retinopathy. Diabetes 1984; 33: 97– 100 [DOI] [PubMed] [Google Scholar]
- 34.Aiello LP, Avery RL, Arrigg PG, Keyt BA, Jampel HD, Shah ST, Pasquale LR, Thieme H, Iwamoto MA, Park JE, Nguyen HV, Aiello LM, Ferrara N, King GL: Vascular endothelial growth factor in ocular fluid of patients with diabetic retinopathy and other retinal disorders. N Engl J Med 1994; 331: 1480– 1487 [DOI] [PubMed] [Google Scholar]
- 35.Schlingemann RO, van Hinsbergh VW: Role of vascular permeability factor/vascular endothelial growth factor in eye disease. Br J Ophthalmol 1997; 81: 501– 512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrara N: Role of vascular endothelial growth factor in regulation of physiological angiogenesis. Am J Physiol Cell Physiol 2001; 280: C1358– C1366 [DOI] [PubMed] [Google Scholar]
- 37.Ribatti D: The crucial role of vascular permeability factor/vascular endothelial growth factor in angiogenesis: a historical review. Br J Haematol 2005; 128: 303– 309 [DOI] [PubMed] [Google Scholar]
- 38.Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE: Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine 2000; 12: 1232– 1235 [DOI] [PubMed] [Google Scholar]
- 39.Takagi H, Watanabe D, Suzuma K, Kurimoto M, Suzuma I, Ohashi H, Ojima T, Murakami T: Novel role of erythropoietin in proliferative diabetic retinopathy. Diabetes Res Clin Pract 2007; 77( Suppl. 1): S62– S64 [DOI] [PubMed] [Google Scholar]
- 40.Katsura Y, Okano T, Matsuno K, Osako M, Kure M, Watanabe T, Iwaki Y, Noritake M, Kosano H, Nishigori H, Matsuoka T: Erythropoietin is highly elevated in vitreous fluid of patients with proliferative diabetic retinopathy. Diabetes Care 2005; 28: 2252– 2254 [DOI] [PubMed] [Google Scholar]
- 41.Aiello LP, Pierce EA, Foley ED, Takagi H, Chen H, Riddle L, Ferrara N, King GL, Smith LE: Suppression of retinal neovascularization in vivo by inhibition of vascular endothelial growth factor (VEGF) using soluble VEGF-receptor chimeric proteins. Proc Natl Acad Sci U S A 1995; 92: 10457– 10461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qaum T, Xu Q, Joussen AM, Clemens MW, Qin W, Miyamoto K, Hassessian H, Wiegand SJ, Rudge J, Yancopoulos GD, Adamis AP: VEGF-initiated blood-retinal barrier breakdown in early diabetes. Invest Ophthalmol Vis Sci 2001; 42: 2408– 2413 [PubMed] [Google Scholar]
- 43.Li H, Forstermann U: Nitric oxide in the pathogenesis of vascular disease. J Pathol 2000; 190: 244– 254 [DOI] [PubMed] [Google Scholar]
- 44.Vallance P, Leiper J: Blocking NO synthesis: how, where and why? Nat Rev Drug Discov 2002; 1: 939– 950 [DOI] [PubMed] [Google Scholar]
- 45.Albrecht EW, Stegeman CA, Heeringa P, Henning RH, van Goor H: Protective role of endothelial nitric oxide synthase. J Pathol 2003; 199: 8– 17 [DOI] [PubMed] [Google Scholar]
- 46.Ando A, Yang A, Mori K, Yamada H, Yamada E, Takahashi K, Saikia J, Kim M, Melia M, Fishman M, Huang P, Campochiaro PA: Nitric oxide is proangiogenic in the retina and choroid. J Cell Physiol 2002; 191: 116– 124 [DOI] [PubMed] [Google Scholar]
- 47.Tsai DC, Chiou SH, Lee FL, Chou CK, Chen SJ, Peng CH, Kuo YH, Chen CF, Ho LL, Hsu WM: Possible involvement of nitric oxide in the progression of diabetic retinopathy. Ophthalmologica 2003; 217: 342– 346 [DOI] [PubMed] [Google Scholar]
- 48.Yilmaz G, Esser P, Kociok N, Aydin P, Heimann K: Elevated vitreous nitric oxide levels in patients with proliferative diabetic retinopathy. Am J Ophthalmol 2000; 130: 87– 90 [DOI] [PubMed] [Google Scholar]
- 49.Zintzaras E, Chatzoulis DZ, Karabatsas CH, Stefanidis I: The relationship between C677T methylenetetrahydrofolate reductase gene polymorphism and retinopathy in type 2 diabetes: a meta-analysis. J Hum Genet 2005; 50: 267– 275 [DOI] [PubMed] [Google Scholar]
- 50.Tong Z, Yang Z, Patel S, Chen H, Gibbs D, Yang X, Hau VS, Kaminoh Y, Harmon J, Pearson E, Buehler J, Chen Y, Yu B, Tinkham NH, Zabriskie NA, Zeng J, Luo L, Sun JK, Prakash M, Hamam RN, Tonna S, Constantine R, Ronquillo CC, Sadda S, Avery RL, Brand JM, London N, Anduze AL, King GL, Bernstein PS, Watkins S, Jorde LB, Li DY, Aiello LP, Pollak MR, Zhang K: Promoter polymorphism of the erythropoietin gene in severe diabetic eye and kidney complications. Proc Natl Acad Sci U S A 2008; 105: 6998– 7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.