Abstract
OBJECTIVE
Members of the family of bone morphogenetic proteins (BMPs) are important regulators of adipogenesis. We examined the role of the BMP receptor 1A gene (BMPR1A) in the pathophysiology of human obesity.
RESEARCH DESIGN AND METHODS
We measured BMPR1A mRNA expression in paired samples of visceral and subcutaneous adipose tissue from 297 subjects and sequenced the BMPR1A in 48 nonrelated white subjects. Twenty-one representative variants including HapMap tagging single nucleotide polymorphisms (SNPs) were then genotyped for association studies in German whites (n = 1,907). For replication analyses, we used a population of Sorbs from Germany (n = 900) and German childhood cohorts (n = 1,029 schoolchildren and 270 obese children).
RESULTS
mRNA expression of the BMPR1A was significantly increased in both visceral and subcutaneous adipose tissue of overweight and obese subjects compared with lean subjects (P < 0.05). In a case-control study, four SNPs (rs7095025, rs11202222, rs10788528, and rs7922846) were nominally associated with obesity (adjusted P < 0.05). For three SNPs (rs7095025, rs11202222, and rs10788528), the association with obesity was confirmed in the independent cohort of Sorbs (adjusted P < 0.005). Consistent with this, BMPR1A SNPs were nominally associated with obesity-related quantitative traits in nondiabetic subjects in both adult cohorts. Furthermore, homozygous carriers of the obesity risk alleles had higher BMPR1A mRNA expression in fat than noncarriers.
CONCLUSIONS
Our data suggest that genetic variation in the BMPR1A may play a role in the pathophysiology of human obesity, possibly mediated through effects on mRNA expression.
Recent advances in the field of genetics of complex diseases succeeded in identification of novel genes associated with human obesity (1–6). However, in contrast to fat mass and obesity associated (FTO) that turned out to be the first robustly replicated susceptibility signal for polygenic obesity (1,2), several other genes such as GAD2 and INSIG2 failed to “survive” replication efforts following initial studies (7,8). Therefore, along with the highly powered genome-wide association studies highlighting both expected and unexpected genes/variants, the candidate gene approach focusing on genes with plausible functional relevance can still substantially contribute to a better understanding of the etiology of complex disorders such as obesity and its sequelae.
Bone morphogenetic proteins (BMPs) are members of the transforming growth factor-β (TGF-β) superfamily and are involved in control of multiple key steps of embryonic development and differentiation (9–11). BMPs have been shown to have different roles in adipogenesis depending on the differentiation stage, the concentrations of BMP, as well as the presence of other extracellular and intracellular factors (12–15). Cellular responses to BMPs have been shown to be mediated by the formation of a hetero-oligomeric complex of the type 1 and type 2 BMP receptors (16–18). Among different isoforms, three type 1 receptors (BMPR1A/ALK3, BMPR1B/ALK6, and ACVR1A/ALK2) and three type 2 receptors (BMPR2, ACTR2A, and ACTR2B) mediate most of the effects of BMPs (9). Of the different BMPR isoforms, BMPR1A is particularly interesting to adipocyte biology since it has been shown to specialize in adipocyte differentiation in vitro (19). Therefore, BMPR1A seems to be a convincing candidate gene possibly involved in the pathogenesis of human obesity and type 2 diabetes.
Here, we examined the role of BMPR1A in the pathophysiology of human obesity. We measured mRNA expression of BMPR1A in paired samples of visceral and subcutaneous adipose tissue and examined its relation with anthropometric traits such as BMI, waist-to-hip ratio (WHR), and measures of glucose metabolism. Further, we investigated whether genetic variation within the BMPR1A might affect adipose tissue BMPR1A mRNA expression. Twenty-one representative variants including HapMap tagging single nucleotide polymorphisms (SNPs) were initially genotyped for association studies in German whites. For replication analyses, we used a population of Sorbs from Germany and German children cohorts (schoolchildren and obese children).
RESEARCH DESIGN AND METHODS
Adult cohorts: Leipzig cohort.
A total of 941 patients with type 2 diabetes and 966 nondiabetic subjects were recruited at the University Hospital in Leipzig, Germany. The nondiabetic subjects included 330 men and 666 women (mean age 49 ± 14 years; mean BMI 29.2 ± 6.6 kg/m2; mean WHR 0.94 ± 0.17; mean fasting plasma glucose 5.31 ± 0.59 mmol/l; mean fasting plasma insulin 120 ± 218 pmol/l) and patients with type 2 diabetes included 485 men and 456 women (mean age 64 ± 11 years; mean BMI 29.9 ± 5.8 kg/m2) (data are given as arithmetic means ± SD). In addition, oral glucose tolerance tests (OGTTs) and fasting plasma insulin measurements were performed in all nondiabetic subjects as previously described elsewhere (20). In a subgroup of 423 nondiabetic subjects, insulin sensitivity was assessed with hyperinsulinemic-euglycemic clamps and plasma leptin measurements were carried out.
Tissue studies.
Paired samples of visceral and subcutaneous adipose tissue were obtained from a subgroup of 297 white men (n = 143) and women (n = 154), who underwent open abdominal surgery (described in detail elsewhere) (20); age range 16–99 years and BMI 18–62 kg/m2. In addition to above-mentioned clinical parameters, in these subjects abdominal visceral and subcutaneous fat area was calculated using computed tomography scans at the level of L4-L5 and percentage body fat was measured by dual-energy X-ray absorptiometry.
Assays and measures of obesity and glucose metabolism.
Fasting plasma insulin was measured with an enzyme immunometric assay for the IMMULITE automated analyzer (Diagnostic Products, Los Angeles, CA). Plasma leptin levels were assessed by radioimmunoassay (Linco Research, St. Charles, MO). The OGTT was performed after an overnight fast with 75 g standardized glucose solution (Glucodex Solution 75 g; Merieux, Montreal, Canada) and insulin sensitivity was assessed with the hyperinsulinemic-euglycemic clamp method as described elsewhere (21,22).
Sorbs.
The cohort was derived from the self-contained Sorbian population in Germany. Extensive phenotyping included standardized questionnaires for past medical history and family history, collection of anthropometric data (weight, height, WHR), and OGTT. Insulin was measured with the AutoDELFIA Insulin assay (PerkinElmer Life and Analytical Sciences, Turku, Finland). Nine hundred Sorbian subjects (726 subjects with normal glucose tolerance [NGT], 72 subjects with impaired glucose tolerance [IGT], and 102 cases with type 2 diabetes) were available for the present study (mean age 48 ± 16 years; mean BMI 27.2 ± 5.0 kg/m2; mean WHR 0.87 ± 0.10; mean fasting plasma glucose 5.53 ± 1.16 mmol/l; mean fasting plasma insulin 45 ± 102 pmol/l). For the estimation of the association with glucose and insulin levels, only nondiabetic subjects were included (definition according to the American Diabetes Association criteria) (23). Because the complete family structure of all subjects is not known, the estimated effect sizes might be biased by cryptic relatedness in this sample. All studies were approved by the ethics committee of the University of Leipzig, and all subjects gave written informed consent before taking part in the study.
Childhood cohorts: Leipzig schoolchildren cohort.
This cohort is part of the Leipzig Schoolchildren Project that investigated anthropometric and clinical parameters in 2,675 children aged 6–17 years from 1999 to 2000 (24). DNA was available from 1,029 children (488 boys and 541 girls; mean age 12 ± 3 years; mean BMI-SDS 0.11 ± 0.97). The BMI was standardized referring to national reference data (25), and children with a BMI ≥1.88 SDS (= 97th centile) were considered obese. A total of 715 children and adolescents with BMI between −1.0 SDS and 1.0 SDS (337 boys and 378 girls; mean age 12 ± 3 years; mean BMI-SDS −0.02 ± 0.54) were selected from the 1,029 schoolchildren to serve as the healthy normal-weight control group to the Leipzig obese children cohort.
Leipzig obese children cohort.
The 270 white children and adolescents (131 boys and 139 girls, aged 12 ± 4 years; BMI-SDS 2.68 ± 0.57) were recruited from the obesity clinic at the University Hospital for Children & Adolescents, Leipzig, Germany. All obese children had a detailed metabolic work-up including an OGTT as described in detail elsewhere (26). The studies were approved by the ethical committee of the University of Leipzig.
Analysis of human BMPR1A mRNA expression.
Human BMPR1A expression was measured by quantitative real-time RT–PCR by using SYBR Green methodology, and fluorescence was detected on an ABI PRISM 7500 sequence detector (Applied Biosystems, Darmstadt, Germany) as described in detail elsewhere (20). The following primers were used: human BMPR1A (NCBI accession no. NM_004329.2) 5′ tagttcgctgaaccaataaagg 3′ (sense) and 5′ gtcagaaaatggagtaacctta 3′ (antisense). SYBR Green I fluorescence emissions were monitored after each cycle. Human BMPR1A mRNA expression was calculated relative to the mRNA expression of 18S rRNA, determined by a premixed assay on demand for human 18S rRNA (PE Biosystems, Darmstadt, Germany). To avoid unspecific primer-dimer amplification, the secondary structure option was checked and amplification of specific transcripts was confirmed by melting curve profiles at the end of each PCR (cooling the sample to 68°C and heating slowly to 95°C with measurement of fluorescence). The specificity of the PCR was further verified by subjecting the amplification products to agarose gel electrophoresis.
Sequencing of the BMPR1A.
Sequencing of the BMPR1A was performed using the Big Dye Terminator (Applied Biosystems, Foster City, CA) on an automated DNA capillary sequencer (ABI PRISM 3100 Avant; Applied Biosystems). Sequence information and PCR conditions for all oligonucleotide primers used for variant screening are available upon request.
Genotyping of BMPR1A SNPs.
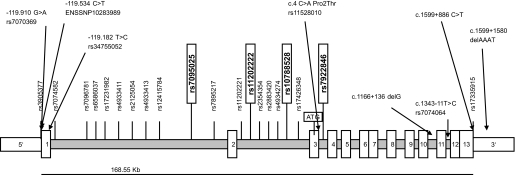
Genotyping of the 21 representative SNPs (Fig. 3) was done using the TaqMan SNP Genotyping assay according to the manufacturer's protocol (Applied Biosystems). To assess genotyping reproducibility, a random ∼5% selection of the sample was regenotyped in all SNPs; all genotypes matched initial designated genotypes.
FIG. 3.
Genetic structure and SNPs analyzed in BMPR1A. Eight genetic variants (presented horizontally) were found by direct sequencing of the BMPR1A and 20 tagging polymorphisms (vertically shown) were selected from the HapMap database (www.hapmap.org). Twenty-one (HapMap tagging SNPs and the c.2147 + 886C >T) of the 28 SNPs were representative of their linkage disequilibrium groups and were genotyped in German whites for association with obesity and related phenotypes. Four of 21 representative SNPs (in vertical boxes) showed significant association with obesity and subphenotypes and were additionally genotyped in the Sorbian and children cohorts for replication. Positions of variants are relative to the first translation initiation site and are based on the mRNA sequence NM_004329.2 (NCBI GenBank). ENSSNP10283989 denotes a variant from the Ensembl database (www.ensemble.org).
Statistical analyses.
Before statistical analysis, nonnormally distributed parameters were logarithmically transformed to approximate a normal distribution. Differences in genotype frequencies between the obese or diabetic case and healthy control subjects were compared using logistic regression analyses. Multivariate linear relationships were assessed by generalized linear regression models. All analyses were done under the additive model and the presented P values are adjusted for age and sex (and BMI for glucose traits). Differences in mRNA expression between visceral and subcutaneous adipose tissue were assessed using the paired Student's t test.
P values <0.05 were considered to provide evidence for association and are presented without correction for multiple hypothesis testing. Only two-sided P values are provided. The analysis of associations with quantitative traits was restricted to nondiabetic subjects to avoid diabetes status or treatment masking potential effects of the variants on these phenotypic traits.
To obtain the combined effect of our two cohorts, we performed a meta-analysis by using the metan command in STATA based on the estimated effect sizes of each study and their standard error for quantitative traits. The meta-analysis was performed in a fixed-effects model by using the Mantel-Haenszel method.
Statistical analyses were performed using SPSS version 15.0.1 (SPSS; Chicago, IL) and STATA (version 9.0) (StataCorp LP, College Station, TX).
RESULTS
Visceral and subcutaneous BMPR1A mRNA expression and obesity.
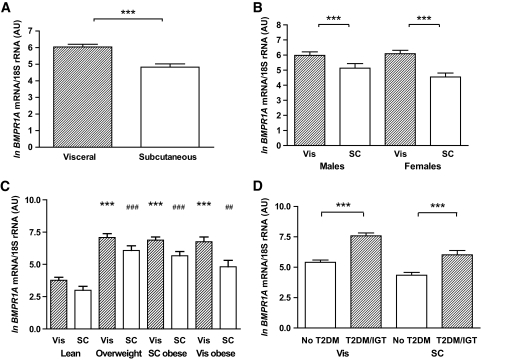
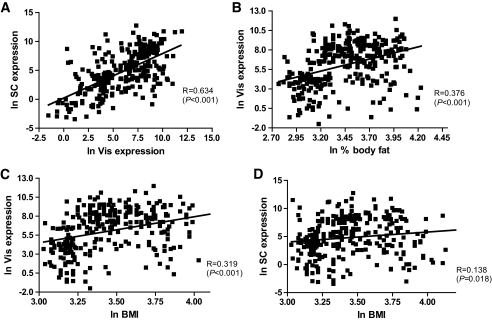
Analysis of 297 paired samples of visceral and subcutaneous adipose tissue showed significantly higher BMPR1A transcript levels in visceral compared with subcutaneous fat (Fig. 1A), independent of sex (Fig. 1B). There were no differences in either visceral or subcutaneous BMPR1A expression between men and women (Fig. 1B). To investigate the expression according to body fat mass or fat distribution, we performed additional analyses in subgroups of lean (BMI <25 kg/m2), overweight (25 kg/m2 < BMI <30 kg/m2), and obese (BMI >30 kg/m2) subjects. Based on computed tomography scans measurement (L4-L5) of abdominal visceral and subcutaneous fat areas, obese subjects were further categorized as predominantly visceral or subcutaneous obese as defined by a ratio of visceral/subcutaneously fat area >0.5 (20). BMPR1A mRNA expression was significantly increased in both visceral and subcutaneous adipose tissue of 51 overweight and 166 obese (101 subcutaneously obese and 65 visceral obese) compared with 80 lean subjects (P < 0.001) (Fig. 1C). Further, we asked whether impaired glucose metabolism in patients with either IGT (n = 24) or type 2 diabetes (type 2 diabetes; n = 65) is associated with altered BMPR1A expression in different fat depots. Subgroup analyses demonstrated that IGT and type 2 diabetic subjects were indistinguishable with regard to BMPR1A mRNA measurements; therefore, these groups were studied together. Patients with IGT/type 2 diabetes had significantly higher BMPR1A expression in both visceral and subcutaneous fat compared with NGT (n = 208) subjects (Fig. 1D). In addition, we detected a significant correlation between visceral and subcutaneous BMPR1A mRNA expression in the entire study cohort (Fig. 2A), as well as in each of the obesity subgroups (data not shown).
FIG. 1.
BMPR1A mRNA expression in visceral (Vis) and subcutaneous (SC) adipose tissue in lean obese and type 2 diabetic (T2DM) subjects. The data are means ± SE; the mRNA data were ln transformed to achieve normal distribution; BMPR1A mRNA levels in (A) entire study population (n = 297); (B) men (n = 143) and women (n = 154); (C) subgroups of lean (BMI <25 kg/m2; n = 80), overweight (25 kg/m2 < BMI <30 kg/m2; n = 51), subcutaneous obese (n = 101) and visceral obese (n = 65) (BMI >30 kg/m2); (D) subjects with NGT (n = 208) and with IGT (n = 24); or with type 2 diabetes (n = 65). ***Indicates statistical significance at P < 0.001; (C) *** indicates statistical significance at P < 0.001 when compared with visceral expression in lean subjects; and statistically significant at ##P < 0.01 and ###P < 0.001, respectively, when compared with subcutaneous expression in lean subjects.
FIG. 2.
A: Correlations between visceral (Vis) and subcutaneous (SC) BMPR1A mRNA expression, (B) Vis BMPR1A mRNA expression and percent of body fat, (C) Vis BMPR1A mRNA expression and BMI, (D) subcutaneous BMPR1A mRNA expression and BMI. Data are ln transformed to achieve normal distribution.
Correlation of BMPR1A mRNA expression with parameters of obesity, glucose metabolism, and insulin sensitivity.
In 297 subjects, univariate regression analysis revealed significant positive correlations between visceral BMPR1A mRNA expression and BMI, percent of body fat, waist, WHR, fasting and 2-h plasma glucose, and fasting plasma insulin (Table 1; Fig. 2). Subcutaneous BMPR1A mRNA expression also correlated with BMI, percent of body fat, waist, WHR, and fasting plasma insulin. There was an inverse correlation between visceral and subcutaneous BMPR1A mRNA expression and glucose uptake during the steady state of a hyperinsulinemic-euglycemic clamp (Table 1). The correlations remained unchanged also after excluding subjects with IGT and type 2 diabetes (data not shown). The correlation between BMPR1A mRNA expression in fat and glucose infusion rate during the steady state of an hyperinsulinemic-euglycemic clamp remained significant even upon adjusting for age, sex, and percent of body fat (Table 2).
TABLE 1.
Linear regression analyses of visceral and subcutaneous BMPR1A mRNA expression with anthropometric and metabolic parameters (n = 297)
| Vis BMPR1A mRNA R (P) | SC BMPR1A mRNA R (P) | |
|---|---|---|
| Age (years) | 0.108 (0.063) | 0.105 (0.070) |
| BMI (kg/m2) | 0.319 (<0.001) | 0.138 (0.018) |
| Percent of body fat | 0.376 (<0.001) | 0.208 (<0.001) |
| Waist circumference (cm) | 0.555 (<0.001) | 0.373 (<0.001) |
| WHR | 0.428 (<0.001) | 0.214 (<0.001) |
| Fasting plasma glucose (mmol/l) | 0.233 (<0.001) | 0.094 (0.104) |
| Fasting plasma insulin (pmol/l) | 0.523 (<0.001) | 0.347 (<0.001) |
| Plasma glucose (2-h) (mmol/l) | 0.304 (<0.001) | 0.110 (0.137) |
| GIR | −0.504 (<0.001) | −0.406 (<0.001) |
GIR, glucose infusion rate during the steady state of the hyperinsulinemic-euglycemic clamp; SC, subcutaneous; Vis, visceral.
TABLE 2.
Multivariate linear regression analyses of visceral and subcutaneous BMPR1A mRNA expression with anthropometric and metabolic parameters (n = 297)
| Vis BMPR1A mRNA β-coefficient (P) | SC BMPR1A mRNA β-coefficient (P) | |
|---|---|---|
| Model 1 | ||
| Age (years) | 0.02 (0.053) | 0.021 (0.088) |
| Sex | 0.156 (0.631) | −0.537 (0.178) |
| Model 2 | ||
| Age (years) | 0.029 (0.003) | 0.027 (0.032) |
| Sex | 0.016 (0.960) | −0.616 (0.119) |
| BMI (kg/m2) | 3.732 (<0.001) | 2.190 (0.004) |
| Model 3 | ||
| Age (years) | 0.023 (0.017) | 0.021 (0.083) |
| Sex | −0.194 (0.532) | −0.725 (0.067) |
| Percent of body fat | 3.284 (<0.001) | 2.374 (<0.001) |
| Model 4 | ||
| Age (years) | −0.010 (0.286) | −0.017 (0.176) |
| Sex | −0.162 (0.599) | −0.828 (0.039) |
| Percent of body fat | 2.969 (<0.001) | 2.185 (0.002) |
| GIR | −1.692 (<0.001) | −1.861 (<0.001) |
GIR, glucose infusion rate; SC, subcutaneous; Vis, visceral.
Genetic variation in the BMPR1A.
We sequenced the BMPR1A (13 exons, exon-intron boundaries, 5′ and 3′ UTRs; 3631 bp, NM_004329.2 in the NCBI GenBank) and 1,200 bp in the 5′ region in 48 nonrelated white subjects (12 lean subjects with NGT, 12 visceral obese, 12 subcutaneously obese, 12 with type 2 diabetes). Eight genetic variants were found (Fig. 3).
In addition, 20 HapMap tagging polymorphisms (27) (www.hapmap.org) covering 100% of the variation in BMPR1A locus were genotyped in these 48 DNA samples. HapMap tagging SNPs were selected from the HapMap Phase II using the Tagger software according to the following selection criteria: minor allele frequency >0.05 and r2 >0.8 (28).
Based on linkage disequilibrium, seven of eight SNPs identified by sequencing were tagged by at least one of the HapMap tagging SNPs (linkage disequilibrium with r2 >0.8) (supplemental Table 1, available in an online appendix at http://diabetes.diabetesjournals.org/cgi/content/full/db08-1458/DC1). Linkage disequilibrium was calculated among the more common variants (minor allele frequency ≥0.05) using the EMLD statistical program (https://epi.mdanderson.org/qhuang/Software/pub.htm) (supplemental Table 1). One novel SNP (c.2147 + 886 C > T) was unique and therefore genotyped together with the 20 HapMap tagging SNPs for association studies in German whites (n = 1,907) with detailed metabolic testing. For replication analyses, we used the Sorbian cohort from Germany (n = 900) and children cohorts from Leipzig, Germany (n = 1,029 schoolchildren and 270 obese children). All SNPs were in Hardy-Weinberg equilibrium (P > 0.05).
BMPR1A variants and association with obesity: Leipzig cohort.
In a case-control study including 488 lean (BMI <25 kg/m2) and 752 obese (BMI >30 kg/m2) subjects, 4 of 21 genotyped SNPs (rs7095025, rs11202222, rs10788528, and rs7922846) were nominally associated with obesity (P < 0.05, adjusted for age and sex). Carriers of the major alleles were at higher risk of being obese (ORs ranging from 1.22 to 1.40; Table 3).
TABLE 3.
Association analyses of the BMPR1A genetic variants with obesity in the Leipzig and the Sorbian cohorts
| Major allele | Minor allele | Leipzig* |
Sorbs† |
Combined analysis |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Major allele frequency (case/control) | OR (95% CI) | P | Major allele frequency (case/control) | OR (95% CI) | P | OR (95% CI) | P | |||
| rs7095025 | C | T | 0.76/0.73 | 1.26 (1.02; 1.56) | 0.03 | 0.82/0.74 | 1.85 (1.25; 2.73) | 0.002 | 1.29 (1.08; 1.54) | 0.004 |
| rs11202222 | C | G | 0.79/0.75 | 1.26 (1.02; 1.55) | 0.03 | 0.86/0.77 | 2.09 (1.38; 3.18) | 5.2 × 10−4 | 1.40 (1.16; 1.69) | 4.4 × 10−4 |
| rs10788528 | G | A | 0.86/0.82 | 1.40 (1.10; 1.78) | 0.006 | 0.92/0.84 | 2.39 (1.45; 3.93) | 6.1 × 10−4 | 1.56 (1.27; 1.94) | 3.5 × 10−5 |
| rs7922846 | A | T | 0.73/0.70 | 1.22 (1.00; 1.48) | 0.05 | 0.77/0.72 | 1.21 (0.86; 1.71) | 0.27 | 1.23 (1.04; 1.46) | 0.016 |
P values were adjusted for age and sex. ORs indicate effect directions of the major allele in additive mode of inheritance. Combined ORs and P values represent the combined analysis of the two cohorts (Leipzig and Sorbs). The combined analyses were adjusted for age, sex, and study cohort.
*n = 752 case subjects (BMI >30 kg/m2) vs. 488 control subjects (BMI <25 kg/m2);
†n = 210 case subjects vs. 303 control subjects.
Sorbs.
The four SNPs nominally associated with obesity were additionally genotyped in the Sorbian cohort for replication. For three SNPs (rs7095025, rs11202222, and rs10788528), the association with obesity could be replicated in the Sorbian case-control study (303 lean and 210 obese subjects; adjusted P < 0.005) (Table 3).
In a combined analysis including both the Sorbian and the Leipzig cohort, rs10788528 and rs11202222 showed the strongest association with obesity (P = 4.4 × 10−4 and P = 3.5 × 10−5) (Table 3). The combined effects of the SNPs on obesity were confirmed in a meta-analysis using the metan command in STATA based on the estimated effect sizes of each study and their CIs (data not shown). The results remained unchanged also after excluding subjects with type 2 diabetes (data not shown).
BMPR1A variants and association with extended phenotypes: Leipzig cohort.
Consistent with case-control studies, we found nominal associations between BMPR1A SNPs and BMI as well as obesity–related quantitative traits in 966 subjects without type 2 diabetes. Three (rs7095025, rs11202222, and rs10788528) of the four SNPs associated with obesity in case-control studies showed nominal associations with BMI. As expected, the obesity risk alleles (major alleles) were associated with higher BMI and increased plasma leptin concentrations (Table 4 and supplemental Table 2). In addition, rs4933413, rs11202221, and rs2354354 also showed effects on BMI and plasma leptin levels (supplemental Table 2). Furthermore, several SNPs (rs17231982, rs7895217, rs2354354, rs4934274, rs7922846) were nominally associated with 2-h plasma glucose (Table 4 and supplemental Table 2).
TABLE 4.
Association of BMPR1A obesity risk variants with quantitative traits in the Leipzig cohort
| Minor allele frequency | BMI (kg/m2) | WHR | Plasma leptin (ng/ml) | Fasting plasma glucose (mmol/l) | Plasma glucose 120 min (mmol/l) | Fasting plasma insulin (pmol/l) | |
|---|---|---|---|---|---|---|---|
| rs7095025 | 0.26 | ||||||
| P | 0.006 | 0.840 | 0.0003/0.002 | 0.441/0.554 | 0.566/0.457 | 0.859/0.436 | |
| β | 0.023 | 0.002 | 0.358/0.272 | 0.005/0.004 | −0.009/−0.012 | 0.016/−0.065 | |
| SE | 0.008 | 0.009 | 0.098/0.087 | 0.007/0.007 | 0.016/0.016 | 0.091/0.084 | |
| rs11202222 | 0.22 | ||||||
| P | 0.004 | 0.730 | 0.009/0.036 | 0.177/0.244 | 0.804/0.686 | 0.825/0.449 | |
| β | 0.025 | 0.003 | 0.266/0.190 | 0.009/0.008 | −0.004/−0.007 | 0.021/−0.066 | |
| SE | 0.009 | 0.010 | 0.102/0.09 | 0.007/0.007 | 0.016/0.016 | 0.094/0.087 | |
| rs10788528 | 0.16 | ||||||
| P | 0.005 | 0.198 | 0.101/0.221 | 0.334/0.423 | 0.872/0.765 | 0.470/0.849 | |
| β | 0.028 | 0.014 | 0.203/0.134 | 0.008/0.006 | −0.003/−0.006 | 0.077/−0.019 | |
| SE | 0.010 | 0.011 | 0.124/0.109 | 0.008/0.008 | 0.019/0.019 | 0.107/0.099 | |
| rs7922846 | 0.27 | ||||||
| P | 0.13 | 0.07 | 0.125/0.239 | 0.290/0.327 | 0.009/0.01 | 0.035/0.133 | |
| β | 0.012 | 0.016 | 0.156/0.105 | 0.007/0.006 | 0.040/0.039 | 0.190/0.125 | |
| SE | 0.008 | 0.009 | 0.101/0.089 | 0.006/0.006 | 0.015/0.015 | 0.090/0.083 |
Data represent subjects without type 2 diabetes (n = 966). Parameters were ln transformed before analyses. P values were adjusted for age and sex for all variables; for the variables serum leptin, fasting plasma glucose, fasting plasma insulin, and 2-h plasma glucose, the first P value was calculated after adjusting for age and sex and the second P value after adjusting for age, sex, and BMI. β indicates effect directions of the major allele in additive mode of inheritance.
We also assessed the association of the 21 SNPs with type 2 diabetes in a case-control study including 941 case subjects with type 2 diabetes and 783 healthy control subjects with NGT from the Leipzig cohort. No significant association with type 2 diabetes was found under logistic regression analysis (P > 0.05; adjusted for age, sex, and BMI; data not shown).
Sorbs.
The four SNPs nominally associated with obesity in case-control studies in the Leipzig cohort were also tested for associations with BMI- and obesity-related traits in 798 nondiabetic Sorbian subjects. Similar to the Leipzig cohort, major alleles of the three SNPs (rs7095025, rs11202222, and rs10788528) were nominally associated with increased BMI (P < 0.01, adjusted for age and sex; Table 5 and supplemental Table 3). Moreover, these SNPs were associated with higher mean fasting plasma insulin (P < 0.05, adjusted for age and sex). However, this association did not withstand adjustment for BMI (Table 5).
TABLE 5.
Association of BMPR1A obesity risk variants with quantitative traits in the Sorbian cohort
| Minor allele frequency | BMI (kg/m2) | WHR | Fasting plasma glucose (mmol/l) | Plasma glucose 120 min (mmol/l) | Fasting plasma insulin (pmol/l) | |
|---|---|---|---|---|---|---|
| rs7095025 | 0.22 | |||||
| P | 0.007 | 0.361 | 0.100/0.455 | 0.350/0.660 | 0.005/0.08 | |
| β | 0.027 | 0.005 | 0.011/0.005 | 0.019/0.009 | 0.120/0.068 | |
| SE | 0.010 | 0.005 | 0.006/0.006 | 0.020/0.020 | 0.042/0.038 | |
| Combined β (95% CI) | 0.025 (0.012; 0.037) | 0.004 (−0.004; 0.013) | 0.005 (−0.004; 0.014) | −0.004 (−0.028; 0.021) | 0.045 (−0.022; 0.113) | |
| Combined P | 8.4 × 10−5 | 0.326 | 0.272 | 0.761 | 0.190 | |
| rs11202222 | 0.19 | |||||
| P | 0.003 | 0.131 | 0.043/0.273 | 0.669/0.910 | 0.014/0.221 | |
| β | 0.031 | 0.008 | 0.014/0.007 | 0.009/−0.002 | 0.109/0.050 | |
| SE | 0.010 | 0.005 | 0.007/0.006 | 0.021/0.021 | 0.044/0.040 | |
| Combined β (95% CI) | 0.028 (0.015; 0.041) | 0.007 (−0.002; 0.016) | 0.007 (−0.002; 0.016) | −0.005 (−0.030; 0.020) | 0.030 (−0.041; 0.101) | |
| Combined P | 3.4 × 10−5 | 0.118 | 0.103 | 0.685 | 0.413 | |
| rs10788528 | 0.13 | |||||
| P | 0.001 | 0.410 | 0.094/0.543 | 0.682/0.881 | 0.018/0.290 | |
| β | 0.038 | 0.005 | 0.013/0.004 | 0.010/−0.004 | 0.119/0.049 | |
| SE | 0.012 | 0.006 | 0.008/0.007 | 0.024/0.024 | 0.050/0.046 | |
| Combined β (95% CI) | 0.032 (0.017; 0.047) | 0.007 (−0.003; 0.017) | 0.005 (−0.005; 0.015) | −0.005 (−0.034; 0.024) | 0.037 (−0.045; 0.119) | |
| Combined P | 3.0 × 10−5 | 0.180 | 0.356 | 0.726 | 0.376 | |
| rs7922846 | 0.27 | |||||
| P | 0.118 | 0.977 | 0.247/0.644 | 0.438/0.679 | 0.833/0.424 | |
| β | 0.014 | 0.0001 | 0.007/0.003 | 0.014/0.007 | 0.008/−0.028 | |
| SE | 0.009 | 0.005 | 0.006/0.006 | 0.018/0.018 | 0.039/0.035 | |
| Combined β (95% CI) | 0.013 (0.001; 0.025) | 0.004 (−0.005; 0.012) | 0.005 (−0.004; 0.013) | 0.026 (0.003; 0.048) | −0.005 (−0.068; 0.058) | |
| Combined P | 0.031 | 0.378 | 0.289 | 0.025 | 0.879 |
Data represent subjects without type 2 diabetes (n = 798). Parameters were ln transformed before analyses. P values were adjusted for age and sex for all variables; for the variables fasting plasma glucose, fasting plasma insulin, and 2-h plasma glucose, the first P value was calculated after adjusting for age and sex and the second P value after adjusting for age, sex, and BMI. β indicates effect directions of the major allele in additive mode of inheritance. Combined β and P values represent the estimated combined effects in the two cohorts (Leipzig and Sorbs) assessed by meta-analysis. Combined P values for the variables fasting plasma glucose, fasting plasma insulin, and 2-h plasma glucose were calculated after adjusting for age, sex, and BMI; for variables BMI and WHR adjustments were made for age and sex.
Combined effect of the BMPR1A SNPs in our two study cohorts were assessed by a meta-analysis. All four SNPs nominally associated with obesity in the Leipzig cohort (rs7095025, rs11202222, rs10788528, and rs7922846) showed associations with BMI. Similar to the combined analysis of case-control studies on obesity in both cohorts, the highest P values were obtained for rs11202222 and rs10788528 (adjusted P = 3.4 × 10−5 and 3.0 × 10−5, respectively; Table 5). Furthermore, in meta-analyses, the major allele at rs7922846 was nominally associated with 2-h plasma glucose (Table 5), which was because of effects seen in the Leipzig cohort but not replicated in the Sorbs.
Association analyses in children cohorts.
In a case-control study including 715 lean and 270 obese children, no significant association of the four obesity risk variants detected in the adults cohorts was found (P > 0.05 after adjusting for age, sex, pubertal stage, and height). In line with this, the BMPR1A SNPs did not associate with measures of obesity (BMI, WHR) in either schoolchildren (n = 1,029) or obesity cohort (n = 270) (adjusted P < 0.05; data not shown).
BMPR1A variants and association with mRNA expression.
In a subgroup of 232 nondiabetic (NGT + IGT) subjects, homozygous carriers of the obesity risk alleles for rs7095025, rs11202222, rs10788528, and rs7922846 had higher BMPR1A fat mRNA expression when compared with carriers of the nonrisk alleles (supplemental Table 4). The obesity risk allele A in rs7922846 was nominally associated with increased visceral mRNA expression (P = 0.01 adjusted for age, sex, and BMI; supplemental Table 4).
DISCUSSION
BMPR1A is one of the three specific BMP receptors that are essential for the bone morphogenetic protein signaling and is widely expressed in various tissues (29). Moreover, there is evidence for a role of developmental genes in the origin of obesity and body fat distribution (30). Therefore, genes coding proteins such as BMPs or BMPRs seem to be plausible candidate gene involved in the pathogenesis of human obesity.
We describe both mRNA expression and genetic analysis of the BMPR1A and its role in the etiology of polygenic obesity. Consistent with the postulated role of BMPRs in the pathophysiology of human obesity, we found significant differences in mRNA expression in adipose tissue between lean and obese subjects. Visceral and subcutaneous mRNA expression was significantly higher in overweight and obese individuals compared with lean subjects. BMPR1A mRNA expression correlated strongly with measures of obesity such as BMI, percent of body fat, and WHR, as well as with traits of glucose and insulin metabolism. Moreover, the visceral mRNA expression positively correlated with the subcutaneous expression. Based on correlations of mRNA expression in adipose tissue with obesity and relevant metabolic traits, the BMPR1A might play a relevant role in human obesity. It was shown recently that in mesenchymal stem cells, BMP-2/4 use predominantly BMPR1A, whereas BMP-6/7 prefer activin A receptor, type I (ACVR1A) (31). According to the proposed model, BMP-2/4 induce white fat differentiation, and BMP-7 specify brown adipogenesis (15). The positive correlation in the increased expression of BMPR1A with BMI (and percent of body fat) suggests that overweight or obese people may have enhanced BMP-2/4 signaling, causing the increased adiposity.
Further support for the role of BMPR1A in obesity comes from the genetics part of our study, in which 21 representative variants including HapMap (27) tagging SNPs were genotyped for association studies in German whites with detailed metabolic testing. Three of four variants (rs7095025, rs11202222, rs10788528, and rs7922846) that were initially nominally associated with obesity in German whites (the Leipzig cohort) showed association with obesity also in a second, independent Sorbian cohort. Besides replicating the initial findings, these SNPs along with other variants in BMPR1A were nominally associated with BMI as well as traits related to glucose and insulin metabolism. Associations with glucose- and insulin-related traits appeared to be mediated through obesity as they did not withstand adjustments for BMI. Because we did not find a relationship between SNPs and measures of fat distribution including WHR, it is possible that BMPR1A genotype contributes to whole-body fat mass and not primarily to fat distribution in human obesity. However, more sophisticated measures of whole-body fat distribution are needed to address whether BMPR1A genotype plays a role in the determination of fat distribution.
Because of the strong correlation of the visceral and subcutaneous BMPR1A mRNA expression with obesity, we also asked whether the obesity risk variants have any influence on the mRNA expression. In a subgroup of nondiabetic subjects, homozygous carriers of the obesity risk alleles for rs7095025, rs11202222, rs10788528, and rs7922846 had higher BMPR1A fat mRNA expression when compared with carriers of the nonrisk allele variants. The obesity risk allele A in rs7922846 was nominally associated with increased visceral mRNA expression. These findings indicate that the SNP effects on obesity and related metabolic parameters might be mediated through their effects on mRNA expression. In our study, two BMPR1A SNPs (rs3905377 and rs7070369) located 5′ upstream of the translation start site in a putative promoter region were in strong linkage disequilibrium (r2 = 0.80, D′ = 1.0) with the obesity-associated rs7095025. Considering higher BMPR1A mRNA levels in carriers of the obesity risk alleles at these SNPs, we used the Transcription Element Search System (TESS; http://www.cbil.upenn.edu/tess) to examine transcriptional regulatory sequences surrounding these genetic variants, which might modify BMPR1A expression. The region surrounding rs3905377 (T > C) matches human transcriptional binding sites for a transcription factor IID (TFIID) for the major allele T. TFIID (alias TATA-binding protein) is required for transcription by polymerase I, II, and III (32). The region around rs7070369 (G > A) matches transcriptional binding sites for the factor E2-early promoter (EIIaE-A) for the major allele G (33). Even though these data may indicate functional relevance of the noncoding BMPR1A variants in the putative promoter region, their biological relevance remains to be explored. Alternatively, the BMPR1A haplotypes might harbor a variant, which controls BMPR1A mRNA levels but was not tested in our present study as only exons and potentially regulatory regions were sequenced. It is also noteworthy that the only amino acid changing variant identified in our study (Pro2Thr; rs11528010) is in strong linkage disequilibrium with rs7922846, which showed moderate association with obesity and 2-h plasma glucose. Because the Pro2Thr is located in the potential BMPR1A signal peptide chain (1–23 amino acids) it could affect the posttranslational transport of the BMPR1A. This remains to be clarified in in vitro studies aimed at intercellular trafficking of the BMPR1A. We are also aware that coverage of the common genetic variation in the BMPR1A locus was solely based on HapMap data along with our own sequencing of the coding region and that additional variants may have been identified by sequencing of introns. We believe that resequencing of the complete BMPR1A locus (with introns) in appropriate samples (including obese subjects) would provide a conclusive coverage of genetic variation in the BMPR1A and might consequently lead to the causal SNPs.
Children represent a particularly interesting study population for identifying primary genetic determinants involved in susceptibility to complex polygenic diseases such as obesity, because unlike in adults phenotypes they are less influenced by comorbidities and prolonged exposure to environmental factors. Lack of association of BMPR1A variants with childhood obesity indicates that the SNP effects most likely manifest in later stages of life and that these effects might be modified by gene-environment interactions. Based on our data, BMPR1A does not seem to be a candidate gene for early-onset obesity.
BMP4 and its high-affinity receptor BMPR1A are expressed in differentiating and adult β-cells, although it is not required during pancreatic development (34). Transgenic expression of BMP4 in mice β-cells enhances glucose-stimulated insulin secretion and glucose clearance (34). In our present study, no association with type 2 diabetes was found suggesting that BMPR1A does not appear to be a major player in the polygenic etiology of type 2 diabetes. This is in line with recent genome-wide association studies that did not identify any variants within the BMPR1A locus being significantly associated with type 2 diabetes (1).
In conclusion, the role of genetic variation in the BMPR1A in the pathophysiology of human obesity might be mediated through effects on mRNA expression. Even though the correlation analyses do not allow establishing a true cause relationship chain between obesity and BMPR1A mRNA expression in adipose tissue, our expression data together with genetics data suggest that BMPR1A as well as common genetic variants within the gene and its untranslated regulatory regions might be involved in the polygenic etiology of human obesity.
Supplementary Material
Acknowledgments
This work was supported by grants from Deutsche Forschungsgemeinschaft (DFG), the Clinical Research Group Atherobesity KFO 152 (projects BL 833/1-1 to M.B. and Stu192/6-1 to M.S., KO 3512/1-1 to A.K., TP5 to W.K., and KO 3880/1-1 to P.K.), the Interdisciplinary Centre for Clinical Research (IZKF) Leipzig at the Faculty of Medicine of the University of Leipzig (project N06 to P.K., D.S., B.E., K.D.; project B24 to M.B.; and B27 to M.S. and A.T.), from the German Diabetes Association (to Y.B., A.T., P.K., and A.K.), from the National Institutes of Health (U.S.) Grants R01 DK077097 and P30 DK040561 (to Y.H.T.), from the Tanita Healthy Weight Community (to Y.H.T.), and from the Deutsche Hochdruckliga (to W.K.).
This work was supported by unrestricted grants from Merck Serono, Ipsen, and Novo Nordisk (to W.K.). No other potential conflicts of interest relevant to this article were reported.
We thank all those who participated in the studies. We also thank Roy Tauscher for excellent technical assistance.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI: A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007; 316: 889– 894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dina C, Meyre D, Gallina S, Durand E, Körner A, Jacobson P, Carlsson LM, Kiess W, Vatin V, Lecoeur C, Delplanque J, Vaillant E, Pattou F, Ruiz J, Weill J, Levy-Marchal C, Horber F, Potoczna N, Hercberg S, Le Stunff C, Bougneres P, Kovacs P, Marre M, Balkau B, Cauchi S, Chevre JC, Froguel P: Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet 2007; 39: 724– 726 [DOI] [PubMed] [Google Scholar]
- 3.Meyre D, Bouatia-Naji N, Tounian A, Samson C, Lecoeur C, Vatin V, Ghoussaini M, Wachter C, Hercberg S, Charpentier G, Patsch W, Pattou F, Charles MA, Tounian P, Clement K, Jouret B, Weill J, Maddux BA, Goldfine ID, Walley A, Boutin P, Dina C, Froguel P: Variants of ENPP1 are associated with childhood and adult obesity and increase the risk of glucose intolerance and type 2 diabetes. Nat Genet 2005; 37: 863– 867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, Wichmann HE, Meitinger T, Hunter D, Hu FB, Colditz G, Hinney A, Hebebrand J, Koberwitz K, Zhu XF, Cooper R, Ardlie K, Lyon H, Hirschhorn JN, Laird NM, Lenburg ME, Lange C, Christman MF: A common genetic variant is associated with adult and childhood obesity. Science 2006; 312: 279– 283 [DOI] [PubMed] [Google Scholar]
- 5.Groves CJ, Zeggini E, Walker M, Hitman GA, Levy JC, O'Rahilly S, Hattersley AT, McCarthy MI, Wiltshire S: Significant linkage of BMI to chromosome 10p in the UK population and evaluation of GAD2 as a positional candidate. Diabetes 2006; 55: 1884– 1889 [DOI] [PubMed] [Google Scholar]
- 6.Boutin P, Dina C, Vasseur F, Dubois S, Corset L, Seron K, Bekris L, Cabellon J, Neve B, Vasseur-Delannoy V, Chikri M, Charles MA, Clement K, Lernmark A, Froguel P: GAD2 on chromosome 10p12 is a candidate gene for human obesity. Plos Biology 2003; 1: 361– 371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dina C, Meyre D, Samson C, Tichet J, Marre M, Jouret B, Charles MA, Balkau B, Froguel P: Comment on “A common genetic variant is associated with adult and childhood obesity.” Science 2007; 315. [DOI] [PubMed] [Google Scholar]
- 8.Boutin P, Froguel P: GAD2: a polygenic contribution to genetic susceptibility for common obesity? Pathologie Biologie 2005; 53: 305– 307 [DOI] [PubMed] [Google Scholar]
- 9.Kishigami S, Mishina Y: BMP signaling and early embryonic patterning. Cytokine Growth Factor Rev 2005; 16: 265– 278 [DOI] [PubMed] [Google Scholar]
- 10.Chen D, Zhao M, Mundy GR: Bone morphogenetic proteins. Growth Factors 2004; 22: 233– 241 [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto Y, Oelgeschlager M: Regulation of bone morphogenetic proteins in early embryonic development. Naturwissenschaften 2004; 91: 519– 534 [DOI] [PubMed] [Google Scholar]
- 12.Wang EA, Israel DI, Kelly S, Luxenberg DP: Bone morphogenetic protein-2 causes commitment and differentiation in C3H10T1/2 and 3T3 cells. Growth Factors 1993; 9: 57– 71 [DOI] [PubMed] [Google Scholar]
- 13.Tang QQ, Otto TC, Lane MD: Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc Natl Acad Sci U S A 2004; 101: 9607– 9611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin W, Takagi T, Kanesashi SN, Kurahashi T, Nomura T, Harada J, Ishii S: Schnurri-2 controls BMP-dependent adipogenesis via interaction with Smad proteins. Dev Cell 2006; 10: 461– 471 [DOI] [PubMed] [Google Scholar]
- 15.Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, Ahrens MJ, Dudley AT, Norris AW, Kulkarni RN, Kahn CR: New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature 2008; 454: 1000– 1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogan BL: Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev 1996; 10: 1580– 1594 [DOI] [PubMed] [Google Scholar]
- 17.Kingsley DM: The TGF-β superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev 1994; 8: 133– 146 [DOI] [PubMed] [Google Scholar]
- 18.Massague J, Weis-Garcia F: Serine/threonine kinase receptors: mediators of transforming growth factor β family signals. Cancer Surv 1996; 27: 41– 64 [PubMed] [Google Scholar]
- 19.Chen D, Ji X, Harris MA, Feng JQ, Karsenty G, Celeste AJ, Rosen V, Mundy GR, Harris SE: Differential roles for bone morphogenetic protein (BMP) receptor type IB and IA in differentiation and specification of mesenchymal precursor cells to osteoblast and adipocyte lineages. J Cell Biol 1998; 142: 295– 305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berndt J, Klöting N, Kralisch S, Kovacs P, Fasshauer M, Schön MR, Stumvoll M, Blüher M: Plasma visfatin concentrations and fat depot-specific mRNA expression in humans. Diabetes 2005; 54: 2911– 2916 [DOI] [PubMed] [Google Scholar]
- 21.Defronzo RA, Tobin JD, Andres R: Glucose clamp technique: method for quantifying insulin-secretion and resistance. Am J Physiol 1979; 237: E214– E223 [DOI] [PubMed] [Google Scholar]
- 22.Blüher M, Unger R, Rassoul F, Richter V, Paschke R: Relation between glycaemic control, hyperinsulinaemia and plasma concentrations of soluble adhesion molecules in patients with impaired glucose tolerance or type 2 diabetes. Diabetologia 2002; 45: 210– 216 [DOI] [PubMed] [Google Scholar]
- 23.American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2006; 29: S43– S48 [PubMed] [Google Scholar]
- 24.Reich A, Müller G, Gelbrich G, Deutscher K, Godicke R, Kiess W: Obesity and blood pressure-results from the examination of 2365 schoolchildren in Germany. Int J Obes Relat Metab Disord 2003; 27: 1459– 1464 [DOI] [PubMed] [Google Scholar]
- 25.Kromeyer-Hauschild K, Wabitsch M, Kunze D, Geller D, Geiss HC, Hesse V, von Hippel A, Jaeger U, Johnsen D, Korte W, Menner K, Muller G, Muller JM, Niemann-Pilatus A, Remer T, Schaefer F, Wittchen HU, Zabransky S, Zellner K, Ziegler A, Hebebrand J: Percentiles of body mass index in children and adolescents evaluated from different regional German studies. Monatsschrift Kinderheilkunde 2001; 149: 807– 818 [Google Scholar]
- 26.Böttner A, Kratzsch J, Müller G, Kapellen TM, Blüher S, Keller E, Blüher M, Kiess W: Gender differences of adiponectin levels develop during the progression of puberty and are related to serum androgen levels. J Clin Endocrinol Metabol 2004; 89: 4053– 4061 [DOI] [PubMed] [Google Scholar]
- 27.Gibbs RA, Belmont JW, Hardenbol P, Willis TD, Yu FL, Yang HM, Ch'ang LY, Huang W, Liu B, Shen Y, Tam PKH, Tsui LC, Waye MMY, Wong JTF, Zeng CQ, Zhang QR, Chee MS, Galver LM, Kruglyak S, Murray SS, Oliphant AR, Montpetit A, Hudson TJ, Chagnon F, Ferretti V, Leboeuf M, Phillips MS, Verner A, Kwok PY, Duan SH, Lind DL, Miller RD, Rice JP, Saccone NL, Taillon-Miller P, Xiao M, Nakamura Y, Sekine A, Sorimachi K, Tanaka T, Tanaka Y, Tsunoda T, Yoshino E, Bentley DR, Deloukas P, Hunt S, Powell D, Altshuler D, Gabriel SB, Qiu RZ, Ken A, Dunston GM, Kato K, Niikawa N, Knoppers BM, Foster MW, Clayton EW, Wang VO, Watkin J, Gibbs RA, Belmont JW, Sodergren E, Weinstock GM, Wilson RK, Fulton LL, Rogers J, Birren BW, Han H, Wang HG, Godbout M, Wallenburg JC, L'Archeveque P, Bellemare G, Todani K, Fujita T, Tanaka S, Holden AL, Lai EH, Collins FS, Brooks LD, Mcewen JE, Guyer MS, Jordan E, Peterson JL, Spiegel J, Sung LM, Zacharia LF, Kennedy K, Dunn MG, Seabrook R, Shillito M, Skene B, Stewart JG, Valle DL, Clayton EW, Jorde LB, Belmont JW, Chakravarti A, Cho MK, Duster T, Foster MW, Jasperse M, Knoppers BM, Kwok PY, Licinio J, Long JC, Marshall PA, Ossorio PN, Wang VO, Rotimi CN, Royal CDM, Spallone P, Terry SF, Lander ES, Lai EH, Nickerson DA, Abecasis GR, Altshuler D, Bentley DR, Boehnke M, Cardon LR, Daly MJ, Deloukas P, Douglas JA, Gabriel SB, Hudson RR, Hudson TJ, Kruglyak L, Kwok PY, Nakamura Y, Nussbaum RL, Royal CDM, Schaffner SF, Sherry ST, Stein LD, Tanaka T: The International HapMap Project. Nature 2003; 426: 789– 796 [DOI] [PubMed] [Google Scholar]
- 28.de Bakker PIW, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D: Efficiency and power in genetic association studies. Nat Genet 2005; 37: 1217– 1223 [DOI] [PubMed] [Google Scholar]
- 29.Gesta S, Tseng YH, Kahn CR: Developmental origin of fat: tracking obesity to its source. Cell 2007; 131: 242– 256 [DOI] [PubMed] [Google Scholar]
- 30.Gesta S, Bluher M, Yamamoto Y, Norris AW, Berndt J, Kralisch S, Boucher J, Lewis C, Kahn CR: Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc Natl Acad Sci U S A 2006; 103: 6676– 6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lavery K, Swain P, Falb D, Alaoui-Ismaili MH: BMP-2/4 and BMP-6/7 differentially utilize cell surface receptors to induce osteoblastic differentiation of human bone marrow-derived mesenchymal stem cells. J Biol Chem 2008; 283: 20948– 20958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lobo SM, Lister J, Sullivan ML, Hernandez N: The cloned RNA polymerase-II transcription factor-IID selects RNA polymerase-III to transcribe the human U6 gene in vitro. Genes Dev 1991; 5: 1477– 1489 [DOI] [PubMed] [Google Scholar]
- 33.Jalinot P, Devaux B, Kedinger C: The abundance and in vitro DNA-binding of 3 cellular proteins interacting with the adenovirus Ella early promoter are not modified by the ela gene-products. Mol Cell Biol 1987; 7: 3806– 3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goulley J, Dahl U, Baeza N, Mishina Y, Edlund H: BMP4-BMPR1A signaling in β cells is required for and augments glucose-stimulated insulin secretion. Cell Metabolism 2007; 5: 207– 219 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.