Abstract
Acyl-CoA thioesterases (ACOTs) catalyze the hydrolysis of CoA esters to free CoA and carboxylic acids and have important functions in lipid metabolism and other cellular processes. Type I ACOTs are found only in animals and contain an α/β hydrolase domain, through currently no structural information is available on any of these enzymes. We report here the crystal structure at 2.1 Å resolution of human mitochondrial ACOT2, a type I enzyme. The structure contains two domains, N and C domains. The C domain has the α/β hydrolase fold, with the catalytic triad Ser294-His422-Asp388. The N domain contains a seven-stranded β-sandwich, which has some distant structural homologs in other proteins. The active site is located in a large pocket at the interface between the two domains. The structural information has significant relevance for other type I ACOTs and related enzymes.
Keywords: lipid metabolism, bile acid-CoA:amino acid N-acyltransferase, α/β hydrolase
INTRODUCTION
Acyl-CoA thioesterases (ACOTs), also known as acyl-CoA hydrolases and acyl-CoA thioester hydrolases, catalyze the hydrolysis of CoA esters of various compounds, including saturated, unsaturated, and branched fatty acids, bile acids, dicarboxylic acids, and prostaglandins [1–3]. These enzymes have important functions in lipid metabolism and in regulating the levels of free CoA and various CoA esters in the cell, and have been found in the cytosol, mitochondria, and peroxisomes [2, 3].
Two types of ACOTs have been identified so far. Type II enzymes have the so-called ‘hot-dog’ fold [3–5], and their sequences are unrelated to those of type I ACOTs. A Mycobacterium ACOT shares the ‘hot-dog’ fold, although its catalytic machinery and catalytic mechanism are distinct [6].
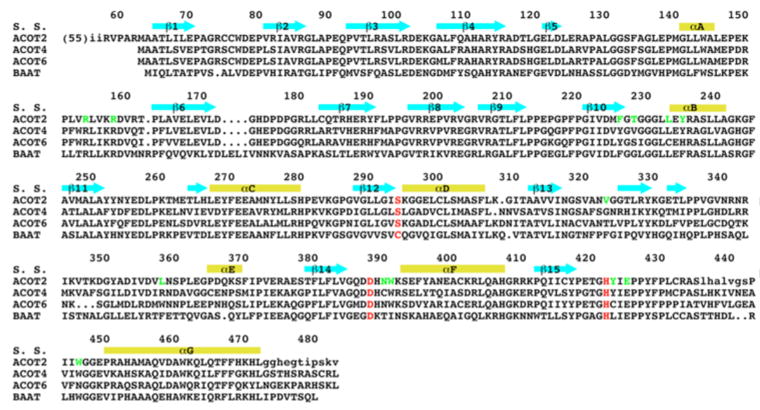
While type II ACOTs are present in most living organisms, type I ACOTs have only been found in animals. Four type I ACOTs are encoded in the human genome (ACOT1, ACOT2, ACOT4, and ACOT6) while the mouse genome contains six type I ACOTs (ACOT1-6) [3, 7]. These enzymes have conserved amino acid sequences (Fig. 1), but they do display distinct substrate preferences [2, 3]. The enzymes are believed to contain an α/β hydrolase catalytic domain in the C-terminal region, with a Ser-His-Asp catalytic triad [8]. A bile acid-CoA:amino acid N-acyltransferase (BAAT) also shares significant amino acid sequence conservation with the type I ACOTs (Fig. 1) [3], although it contains a Cys-His-Asp catalytic triad [9]. Currently, no structural information is available for any of the type I ACOTs.
Figure 1. Sequence conservation of ACOTs.
Alignment of the amino acid sequences of human ACOT2, murine ACOT4, murine ACOT6, and human bile acid-CoA:amino acid N-acyltransferase (BAAT). The secondary structure elements in the ACOT2 structure are labeled. The residues in the catalytic triad are highlighted in red, and other residues in the active site region of ACOT2 are shown in green. The first 55 residues of ACOT2 (including the mitochondrial targeting sequence) are not shown, and residues disordered in the current structure are shown in lower case.
We report here the crystal structure of human mitochondrial ACOT (ACOT2, a type I enzyme) at 2.1 Å resolution. The structure contains two domains, N and C domains. The C domain has the α/β hydrolase fold, with the catalytic triad Ser294-His422-Asp388. The N domain contains a seven-stranded β-sandwich, which has some distant structural homologs in other proteins. The active site is located in a large pocket at the interface between the two domains.
Materials and Methods
Protein expression and purification
Residues 46-483 of human ACOT2 were sub-cloned into the pET26b vector (Novagen). The expression construct introduced a hexa-histidine tag at the C-terminus. The plasmid was transformed into E. coli BL21 (DE3) Rosetta cells (Novagen) and the protein was induced and over-expressed at 20 °C for 12–14 hours by the addition of 0.4 mM IPTG (Sigma). The soluble protein was purified by nickel-agarose affinity chromatography (Qiagen) and gel-filtration chromatography (S-300, GE Healthcare). The purified protein was concentrated to 20 mg/ml in a buffer containing 20 mM Tris (pH 8.5), 250 mM NaCl, 5% glycerol (v/v), and 5 mM DTT.
The selenomethionyl protein was produced with the same protocol as the native protein except that the cells were cultivated in minimal media and supplemented with amino acids to inhibit endogenous methionine biosynthesis [10].
Protein crystallization
Both selenomethionyl and native human ACOT2 formed rod-shaped crystals at 4 °C by the sitting-drop vapor diffusion method. The reservoir solution contained 300 mM ammonium citrate tribasic (pH 7.0) and 22% (w/v) PEG 3350. The crystal quality was aided by microseeding. The crystals were cryo-protected by the introduction of 20% (v/v) ethylene glycol, and frozen in liquid nitrogen for data collection at 100 K. The crystals belong to space group P3121, with cell parameters of a=b=124.6 Å, and c=132.0 Å. There are two molecules of ACOT2 in the asymmetric unit.
Data collection and processing
A single-wavelength anomalous diffraction data set was collected on an SX-165 CCD (Rayonix) at the 24-ID beamline of the Advanced Photon Source (APS) at Argonne National Laboratory. The diffraction images were processed and scaled with the HKL package [11]. The diffraction data were collected at the selenium absorption peak (wavelength 0.979 A).
Structure determination and refinement
Sixteen selenium sites were located with the program BnP [12], and the reflection phases were calculated with Solve/Resolve [13], which also automatically placed about 50% of the residues. The atomic model was built with the program Coot [14], and the structure refinement was carried out with CNS initially [15], and followed by Refmac [16], incorporating TLS refinement. The crystallographic information is summarized in Table 1.
Table 1.
Summary of crystallographic information
| Resolution range (Å) | 30-2.1 |
| Number of observations | 744,081 |
| Rmerge (%)a | 5.8 (37.1) |
| I/σI | 21.0 (3.8) |
| Redundancy | 5.6 (5.6) |
| Number of reflections | 62,312 |
| Completeness (%) | 95 (90) |
| R factor (%) | 19.4 (20.3) |
| Free R factor (%) | 23.6 (24.2) |
| Residues in most favored region of the Ramachandran plot (%) | 91 |
| rmsd in bond lengths (Å) | 0.010 |
| rmsd in bond angles (°) | 1.2 |
The numbers in parenthesis are for the highest resolution shell.
RESULTS AND DISCUSSION
Overall structure of ACOT2
The crystal structure of the free enzyme of human mitochondrial acyl-CoA thioesterase (ACOT2) has been determined at 2.1 Å resolution (Table 1). The atomic model has excellent agreement with the crystallographic data as well as expected bond lengths, bond angles and other geometric parameters. More than 90% of the residues are located in the most favored region of the Ramachandran plot. The current model contains residues 58-435 and 443-472 for the first ACOT2 molecule, residues 57-436 and 442-472 for the second ACOT2 molecule, and 442 waters. The two ACOT2 molecules have essentially the same conformation, with a root-mean-squared distance of 0.3 Å for their equivalent Cα atoms. Purified ACOT2 is monomeric in solution, based on gel-filtration chromatography.
The structure of ACOT2 consists of two domains, N and C domains (Fig. 2). The N domain, covering residues 58-194, contains a β-sandwich, with one sheet having three short β-strands and the other sheet having four long β-strands. The topology of this β-sandwich is similar to that in several other structures, identified with the program DaliLite [17]. These include the chromoprotein antibiotic neocarzinostatin [18] and macromomycin [19], a small domain in α2-macroglobulin [20], the complement component C3 and the related thioester-containing proteins in insects [21]. However, the structural similarity is rather limited, as evidenced by the high rms distances among the structures (2.5 to 4 Å for equivalent Cα atoms), low degree of sequence conservation (6–16% amino acid identity), and the relatively low Z values (6 to 7) from DaliLite.
Figure 2. Schematic drawing in stereo of the structure of human ACOT2.
The β-strands are shown in cyan, α-helices in yellow, and connecting loops in magenta. The side chains of the residues in the catalytic triad are shown (in gray for carbon atoms). Created with PyMOL [22].
The C domain, covering residues 195-472, has the α/β hydrolase fold, with a central, mostly-parallel eight-stranded β-sheet that is surrounded by five α-helices (Fig. 2). A unique feature of this domain in ACOT2 is the presence of several extended loops as well as a β-sheet of three short strands on the surface. Residues 436-442 in one of these loops are missing in the current structure, possibly due to disorder.
The active site of ACOT2
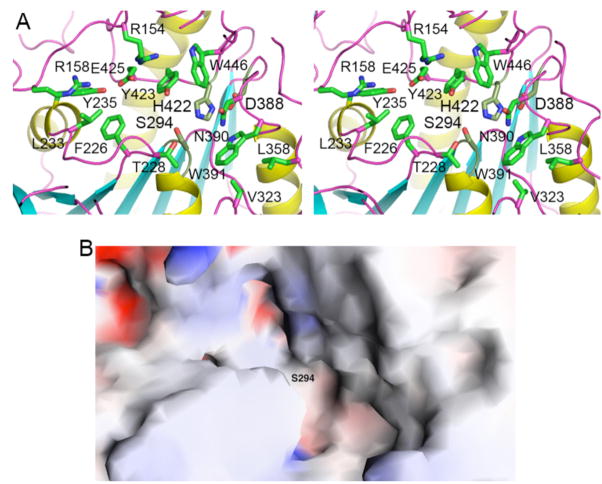
The active site of ACOT2 is located at the interface between the N and C domains (Fig. 2). Like in other α/β hydrolases, the catalytic triad, residues Ser294, His422 and Asp388, are located near the C-terminal end of the parallel β-strands in the central β-sheet. The catalytic nucleophile Ser294 is situated in a tight turn between a β-strand and the following helix (Figs. 2, 3A), and its main chain is in a strained conformation (located in a generously allowed region of the Ramachandran plot). His422 is located in an extensive loop connecting the last β-strand and the α-helix at the C-terminus. Asp388 is located in a shorter loop connecting a β-strand and the following helix (Fig. 2). Its side chain is completely buried in the structure, and is also hydrogen-bonded to the side chain of the highly conserved Ser393 residue.
Figure 3. Active site of ACOT2.
(A). Schematic drawing in stereo of the active region of ACOT2. The side chains of the catalytic triad are shown in gray for carbon atoms, and those of other side chains in green. (B). Molecular surface of the active site region of ACOT2. The location of the catalytic nucleophile Ser294 is labeled. Created with Grasp [23].
The catalytic nucleophile is located in the center of a large depression in the surface of ACOT2 (Fig. 3B). Several long loops, from both the N and C domains, contribute to the formation of this active site (Figs. 2, 3A). The residues in the active site region are generally conserved among the ACOTs (Fig. 1). The disordered segment in the current structure, residues 436-442, is located near this region (Fig. 2) and could form a lid over the active site when the substrate is bound.
The ligand-binding region of neocarzinostatin is the exposed surface of the large β-sheet of the β-sandwich [18]. The equivalent surface of the N domain in ACOT2 is located far from the active site (Fig. 2) and is rather hydrophilic in nature. It is therefore unlikely to be involved in substrate binding in ACOT2. Efforts at determining the binding mode of (acyl-)CoA to the enzyme have so far not been successful.
Implications for other ACOTs
The ACOTs have conserved amino acid sequences (Fig. 1) and therefore our structural observations on ACOT2 are likely to be highly relevant for other family members as well. It may be expected that all of these enzymes have the two domain organization, with the active site located at their interface.
BAAT is also likely to have the same structure based on its sequence conservation with the ACOTs (Fig. 1). The Cys nucleophile is important for the N-acyltransferase activity, while a mutant carrying a Ser nucleophile has greatly enhanced bile acid-CoA thioesterase (ACOT) activity [9].
While human ACOT6 is homologous to murine ACOT6, it lacks the first 270 amino acid residues [7]. Our structure suggests that human ACOT6 would be missing the N domain as well as the first 4 strands of the C domain (Fig. 1). The remaining 4 strands and 3 helices could still form a stable structure (Fig. 2), and the catalytic triad is intact in this protein. If human ACOT6 is catalytically active, its substrate preference could be very different from those of other ACOTs, as the absence of the N domain would result in an open active site.
Acknowledgments
We thank Thierry Auperin and Rona Ramsay for helpful discussions. This research is supported in part by a grant from the NIH (DK067238) to LT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hunt MC, Yamada J, Maltais LJ, Wright MW, Podesta EJ, Alexson SEH. A revised nomenclature for mammalian acyl-CoA thioesterases/hydrolases. J Lipid Res. 2005;46:2029–2032. doi: 10.1194/jlr.E500003-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Hunt MC, Alexson SEH. The role acyl-CoA thioesterases play in mediating intracellular lipid metabolism. Prog Lipid Res. 2002;41:99–130. doi: 10.1016/s0163-7827(01)00017-0. [DOI] [PubMed] [Google Scholar]
- 3.Hunt MC, Alexson SEH. Novel functions of acyl-CoA thioesterases and acyltransferases as auxiliary enzymes in peroxisomal lipid metabolism. Prog Lipid Res. 2008;47:405–421. doi: 10.1016/j.plipres.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Derewenda U, Dauter Z, Smith S, Derewenda ZS. Crystal structure of the Escherichia coli thioesterase II, a homolog of the human Nef binding enzyme. Nat Struct Biol. 2000;7:555–559. doi: 10.1038/76776. [DOI] [PubMed] [Google Scholar]
- 5.Forwood JK, Thakur AS, Guncar G, Marfori M, Mouradov D, Meng W, Robinson J, Huber T, Kellie S, Martin JL, Hume DA, Kobe B. Structural basis for recruitment of tandem hotdog domains in acyl-CoA thioesterase 7 and its role in inflammation. Proc Natl Acad Sci USA. 2007;104:10382–10387. doi: 10.1073/pnas.0700974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang F, Langley R, Gulten G, Wang L, Sacchettini JC. Identification of a type III thioesterase reveals the function of an operon crucial for Mtb virulence. Chem Biol. 2007;14:543–551. doi: 10.1016/j.chembiol.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Hunt MC, Rautanen A, Westin MAK, Svensson LT, Alexson SEH. Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional evolution results in a reduced number of human peroxisomal ACOTs. FASEB J. 2006;20:1855–1864. doi: 10.1096/fj.06-6042com. [DOI] [PubMed] [Google Scholar]
- 8.Huhtinen K, O’Byrne J, Lindquist PJG, Contreras JA, Alexson SEH. The peroxisome proliferator-induced cytosolic type I acyl-CoA thioesterase (CTE-I) is a serine-histidine-aspartic acid a/b hydrolase. J Biol Chem. 2002;277:3424–3432. doi: 10.1074/jbc.M109040200. [DOI] [PubMed] [Google Scholar]
- 9.Sfakianos MK, Wilson L, Sakalian M, Falany CN, Barnes S. Conserved residues in the putative catalytic triad of human bile acid coenzyme A:amino acid N-acyltransferase. J Biol Chem. 2002;277:47270–47275. doi: 10.1074/jbc.M207463200. [DOI] [PubMed] [Google Scholar]
- 10.Doublie S, Kapp U, Aberg A, Brown K, Strub K, Cusack S. Crystallization and preliminary X-ray analysis of the 9 kDa protein of the mouse signal recognition particle and the selenomethionyl-SRP9. FEBS Lett. 1996;384:219–221. doi: 10.1016/0014-5793(96)00316-x. [DOI] [PubMed] [Google Scholar]
- 11.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 12.Weeks CM, Miller R. The design and implementation of SnB v2.0. J Appl Cryst. 1999;32:120–124. [Google Scholar]
- 13.Terwilliger TC. SOLVE and RESOLVE: Automated structure solution and density modification. Meth Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 14.Emsley P, Cowtan KD. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 15.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 16.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 17.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim KH, Kwon BM, Myers AG, Rees DC. Crystal structure of neocarzinostatin, an antitumor protein-chromophore complex. Science. 1993;262:1042–1046. doi: 10.1126/science.8235619. [DOI] [PubMed] [Google Scholar]
- 19.van Roey P, Beerman TA. Crystal structure analysis of auromomycin apoprotein (macromomycin) shows importance of protein side chains to chromophore binding selectivity. Proc Natl Acad Sci USA. 1989;86:6587–6591. doi: 10.1073/pnas.86.17.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doan N, Gettins PGW. Human a2-macroglobulin is composed of multiple domains, as predicted by homology with complement component C3. Biochem J. 2007;407:23–30. doi: 10.1042/BJ20070764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baxter RHG, Chang CI, Chelliah Y, Blandin S, Levashina EA, Deisenhofer J. Structural basis for conserved complement factor-like function in the antimalarial protein TEP1. Proc Natl Acad Sci USA. 2007;104:11615–11620. doi: 10.1073/pnas.0704967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeLano WL. The PyMOL molecular viewer. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 23.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]