Messenger RNA degradation is a vital contributor to the control of gene expression that generally involves removal of a poly(A) tail in both prokaryotes and eukaryotes. In a thought-provoking study in this issue of Genes & Development, Mullen and Marzluff (2008) present data supporting a novel mechanism of mRNA decay. They discovered that histone mRNAs, which are unique in that they are never polyadenylated in mammalian cells, degrade by a cell cycle-regulated mechanism that involves addition of a short oligo(U) tail at the 3′ end. Interestingly, this oligo(U) tract is recognized by the Lsm1–7 complex, which then appears to feed the transcript into the standard mRNA decay pathways. These findings are exciting because they invoke parallels with prokaryotic mRNA decay, which requires polyadenylation immediately prior to degradation and involves an Lsm homolog, Hfq. Moreover, recent studies have identified other oligouridylated RNAs and several poly(U) polymerases, implying that this may be a more widespread mechanism for turnover of RNA.
Messenger RNAs in both prokaryotes and eukaryotes have an interesting problem in that they need to be resistant to decay to be translated but must eventually undergo degradation to allow appropriate regulation of gene expression. At first glance, it appears that these two kingdoms have developed opposite solutions to the problem; in bacteria, polyadenylation induces decay, whereas in eukaryotic cells a poly(A) tail protects the transcript from nucleases, and its removal is the first step in degradation (Dreyfus and Regnier 2002; Edmonds 2002). However, a closer look reveals that a poly(A) tail, through its lack of structure, serves as a primer for decay in both eukaryotes and prokaryotes. The poly(A) tail simply has acquired additional roles in eukaryotic cells.
Polyadenylation essentially creates an ssRNA-binding platform at the 3′ end of RNAs in order to initiate decay. Results described below indicate that this ssRNA platform can be in the form of either oligo(U) or oligo(A). While the addition of these two homopolymeric tails may accomplish the same endpoint (recruitment of degradative enzymes), they may not be totally interchangeable as each can be regulated differently at the level of synthesis, recruit different regulatory poly(U)- or poly(A)-binding factors, or attract different degradative enzymes to the transcript. This strategy for initiating decay via 3′ unstructured extensions may have favored the evolution of the 3′ end of RNAs to focus on transcript function rather than on maintaining sequences/structures that allow for eventual decay of the transcript. It also creates a ready means for the cell to degrade any unwanted transcripts without regard to sequence or structure. In this perspective, we first describe parallels between the role of the poly(A) tail, poly(A) polymerase (PAP), and poly(A) removal in initiating mRNA degradation in prokaryotic and eukaryotic cells. Next we discuss the functions of noncanonical poly(A)/(U) polymerases in mRNA metabolism. We then go on to describe how a newly discovered 3′ modification—oligouridylation—may achieve some of the same outcomes.
Multiple ways to add and remove poly(A)
Bacterial polyadenylation and mRNA decay
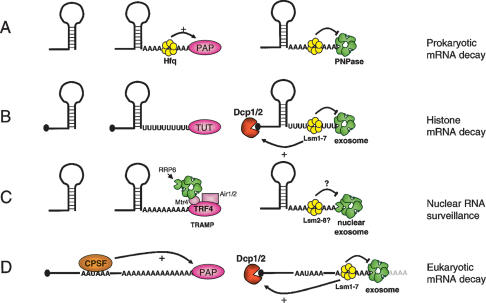
In Escherichia coli, most mRNAs have a stem–loop structure at their 3′ ends that forms the Rho-independent transcription terminator (Condon 2007). Decay is generally initiated by endonucleolytic cleavage by RNase E, a component of the bacterial degradosome, which also contains a 3′–5′ exonuclease (PNPase), enolase, and a helicase (RhlB) (Carpousis 2007). Polyadenylation by poly(A) polymerase PAP1 plays an important role in the degradation of the 3′ fragment by generating a single-stranded toehold for the PNPase component of the degradosome (Fig. 1A). PNPase is an RNase PH-domain protein with processive 3′–5′ phosphorylytic exonucleolytic activity. Once PNPase associates with the poly(A) tail, rapid deadenylation ensues, and degradation can continue into the RNA body. Secondary structures can inhibit PNPase activity, but this may be alleviated to some extent by the RhlB helicase component of the degradosome. In addition, RNase R and RNase II, hydrolytic processive 3′–5′ exonucleases, can also rapidly degrade polyadenylated mRNAs (Mohanty and Kushner 2000; Cheng and Deutscher 2005). In the event that secondary structure does stall decay, the fragment can be subjected to additional rounds of polyadenylation and 3′–5′ decay as necessary. Polyadenylation and decay in bacteria are regulated in part by Hfq, an Sm-like protein that forms a hexameric ring (Fig. 1A; Mohanty et al. 2004). Hfq binds to short oligo(A) tracts and enhances the processivity of PAP1. It has also been shown to inhibit 3′–5′ decay by PNPase and RNase II (Folichon et al. 2003).
Figure 1.
3′ Single-stranded extensions lead to similar degradation pathways in prokaryotes and eukaryotes. (A) Prokaryotic mRNAs degrade from the 3′ end through polyadenylation by PAP1 and subsequent 3′–5′ exonucleolytic decay mediated either by PNPase as shown here, or other 3′–5′ exonucleases. Hfq, an Sm-like protein, associates with the 3′ poly(A) tract and modulates PAP and PNPase activity. (B) Histone mRNAs undergo oligouridylation by a cytoplasmic terminal uridyl transferase at the end of S phase. This leads to association of Lsm11–7, and recruitment of the decapping and 5′–3′ decay machinery. Decay also occurs 3′–5′ by the exosome. How Lsm1–7 association influences exosome activity is unclear, although there is evidence for an inhibitory role (see the text). (C) In yeast, aberrant nuclear RNAs undergo polyadenylation by the TRAMP complex, which contains the Trf4 noncanonical PAP as well as a helicase (Mtr4) and an RNA-binding protein (Air1 or Air2). TRAMP recruits the nuclear exosome (containing Rrp6) to the polyadenylated RNA leading to rapid degradation. It is not clear whether the nuclear Lsm complex (Lsm2–8) influences this process. (D) Eukaryotic mRNAs undergo polyadenylation in the nucleus in a cotranscriptional process involving the CPSF and nuclear PAP, among other factors. In the cytoplasm, these mRNAs undergo degradation that is initiated by removal of the majority of the poly(A) tail (not shown). Lsm1–7 is thought to associate with the remaining 3′ oligo(A) tract and recruit the decapping machinery to induce 5′–3′ decay. In addition, the exosome degrades the transcript 3′–5′. The interplay between the exosome and Lsm1–7 is not clear, although evidence suggests that Lsm inhibits exosome activity.
Eukaryotic polyadenylation and mRNA decay
In eukaryotic cells, the major polyadenylation pathway is a cotranscriptional process performed by nuclear PAP (a homolog of the bacterial enzyme) in conjunction with Cleavage Polyadenylation Specificity Factor (CPSF), which recognizes the canonical AAUAAA polyadenylation signal (Fig. 1D; Edmonds 2002). The poly(A) tail reaches ∼200 nucleotides in mammalian cells and, through its interaction with poly(A)-binding proteins, is required for efficient export and translation of mRNAs. Removal of the poly(A) tail down to an oligo(A) remnant is the first step in decay of the vast majority of transcripts. This is performed in the cytoplasm by a variety of poly(A)-specific 3′–5′ exonucleases including CCR4-NOT, PAN2/3, and PARN (Yamashita et al. 2005). Following shortening of the poly(A) tail to an oligo(A) form, a complex of seven Sm-like proteins (Lsm1–7) associates, probably recognizing the short single-stranded A tail (Fig. 1D; Chowdhury et al. 2007). The Lsm complex is required for decapping of the transcript, which allows subsequent 5′–3′ exonucleolytic decay by XRN1 (Tharun and Parker 2001). Lsm1–7 has also been shown to inhibit 3′ exonucleolytic trimming of the mRNA (He and Parker 2001; Tharun et al. 2005). Therefore the Lsm1–7 complex likely serves as an important post-deadenylation regulator of the traffic of mRNAs into the various pathways of decay. In an alternative pathway, the mRNA body is degraded 3′–5′ by the exosome, a large complex of proteins with homology with exonucleases, which can be considered analogous to the bacterial degradosome. The exosome comprises a core of six RNase PH domain proteins similar in structure to PNPase but lacking catalytic activity. An RNase R homolog, Rrp44/Dis3, contributes catalytic activity (Liu et al. 2006; Dziembowski et al. 2007), and a helicase, Mtr4, has been identified as an accessory factor of the core exosome (de la Cruz et al. 1998; Schilders et al. 2007).
Other PAPs in eukaryotes
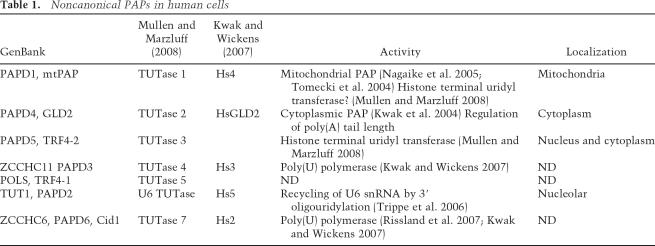
In the last few years, additional eukaryotic PAPs with novel roles in mRNA metabolism have come to light (Read et al. 2002; Kwak et al. 2004; Stevenson and Norbury 2006; Trippe et al. 2006; Kwak and Wickens 2007; Rissland et al. 2007). At least four of these (GLD2, TRF4/5, mtPAP, and U6 TUTase) have characterized functions in mediating specific cellular processes in either yeast or mammalian cells. Others have not yet been assigned a specific function, although some may not be PAPs at all, as they exhibit a preference for UTP rather than ATP.
GLD2, a cytoplasmic PAP
First, a cytoplasmic PAP, termed GLD2, has been shown to function in the cytoplasm as a regulator of poly(A) tail length and consequently as a determinant of translational efficiency (Wang et al. 2002). GLD2 is related to the nuclear PAP in that it has a DNA polymerase β nucleotidyl transferase domain, but it has diverged considerably and lacks an RNA-binding domain. During oocyte development in Xenopus, many mRNAs are exported from the nucleus with a long poly(A) tail that is shortened upon entry to the cytoplasm. The transcripts remain translationally silent until maturation, when polyadenylation occurs and translation is activated. Importantly, transcripts regulated in this way contain a cytoplasmic polyadenylation element (CPE) and associate with a complex containing CPEB (an RNA-binding protein), CPSF, symplekin (a scaffolding protein), GLD2, and the PARN deadenylase (Barnard et al. 2004; Kim and Richter 2006). During the phase when transcripts have short poly(A) tails, both PARN and GLD2 are bound, but PARN activity predominates. Upon maturation, CPEB is phosphorylated and PARN dissociates, allowing GLD2 activity to lengthen the poly(A) tail. In this instance, although the poly(A) tail itself is acting as a translational enhancer, the presence of the PAP and the deadenylase in the same complex is reminiscent of the close association between polymerization and degradation of poly(A) in prokaryotes.
TRAMP—an RNA surveillance mechanism
The second eukaryotic PAP activity is predominantly nuclear and plays a role very much like the bacterial PAP. In Saccharomyces cerevisiae, the only two noncanonical PAPs are encoded by the TRF4 and TRF5 genes. Trf4p and Trf5p form a complex, TRAMP, which also contains an RNA helicase (Mtr4p) and one of two RNA-binding proteins Air1p or Air2p (LaCava et al. 2005; Vanacova et al. 2005). TRAMP is a component of the nuclear RNA surveillance machinery that targets aberrant rRNAs, snoRNAs, and cryptic unstable nuclear transcripts for degradation. It appears that polyadenylation by TRAMP leads to recruitment of the nuclear exosome and rapid 3′–5′ decay of the entire transcript (Fig. 1C). In the nucleus, an additional 3′–5′ exonuclease of the DEDDx family, Rrp6, is a major catalytic component of the exosome and is required for this nuclear RNA surveillance pathway. Whether the nuclear Lsm complex (Lsm2–8) is involved in this mechanism has not been examined, but given the clear parallels with bacterial decay, and the involvement of 3′ poly(A), it seems quite possible. Although this mechanism likely exists in mammals (West et al. 2006), the specific PAP involved has not been pinpointed and cannot be easily recognized by homology alone. In Schizosaccharomyces pombe, Cid14, a homolog of the TRF4/5 PAP of the TRAMP complex, has been recently associated with the modification of transcripts arising from heterochromatic regions and the Transcriptional Gene Silencing branch of the RNAi system (Buhler et al. 2007). The TRAMP complex, therefore, appears to target an impressive array of independent RNAs and illustrates the broad potential of these noncanonical PAPs to influence the cellular ribonome.
Mitochondrial PAP
The final relatively well-characterized eukaryotic PAP is a human mitochondrial enzyme, mtPAP, which is involved in polyadenylation of mitochondrial mRNAs (Tomecki et al. 2004; Nagaike et al. 2005). Like GLD2 and the TRF proteins, mtPAP lacks an RNA-binding domain, suggesting that it may require an as-yet-unidentified partner. In human mitochondria, poly(A) appears to serve as a stabilizer, reminiscent of nuclear-encoded mRNAs, although in other organisms mitochondrial mRNA decay is more similar to the prokaryotic pathway in that polyadenylation initiates degradation (Slomovic et al. 2005). Removal of poly(A) in mammalian mitochondria is performed by PNPase as in prokaryotes. In addition, many mitochondrial mRNAs lack a stop codon but terminate in uridine; thus polyadenylation generates the UAA stop codon required for translation termination. Therefore, the mtPAP enzyme appears to have an impact on gene expression at multiple levels.
U6 TUTase—not a PAP at all
One noncanonical PAP, U6 TUTase, is an extremely specific enzyme that adds up to three uridines to the 3′ end of U6 snRNA (Trippe et al. 2003). No other substrate for U6 TUTase has been identified, and the modification appears to be essential, as knockdowns are lethal (Trippe et al. 2006). The role of this modification seems to be to allow recycling of the snRNA in the event that its 3′ end becomes damaged during splicing.
PAPs in Pombe
In S. pombe there are six noncanonical PAPs that have been reviewed in detail elsewhere (Stevenson and Norbury 2006). It is worth noting, however, that the Cid12 protein is part of the RNA-dependent RNA polymerase complex that is recruited by the RITS complex during RNAi-dependent heterochromatin formation and gene silencing in S. pombe (Motamedi et al. 2004). This function may well be shared with the Caenorhabditis elegans RDE-3 protein (Chen et al. 2005) and the Chlamydomonas reinhardtii MUT68 (Ibrahim et al. 2006). In addition, two S. pombe PAPs, Cid1p and Cid13p, have been linked with DNA replication, perhaps targeting specific mRNAs (Stevenson and Norbury 2006). Furthermore, Cid1 has recently been shown to exhibit a preference for UTP over ATP and, of particular interest for this article, appears to add a 3′-terminal uridine to polyadenylated actin mRNAs during S-phase arrest (Rissland et al. 2007).
Thus, in eukaryotic cells, there are several characterized noncanonical PAPs, each with a specialized function, and some with a preference for addition of poly(U) rather than poly(A). Close examination of the human genome has uncovered seven noncanonical PAPs in total (Stevenson and Norbury 2006). Several different nomenclatures are currently in use, so to avoid confusion, we attempted to clarify the names and functions in Table 1. It has proven difficult to assign functions to all seven human proteins on the basis of homology alone. The finding by Mullen and Marzluff (2008) that two noncanonical PAPs influence the oligouridylation and decay of histone mRNAs is an exciting step toward characterizing the functions of these intriguing enzymes in mammalian cells.
Table 1.
Noncanonical PAPs in human cells
Oligouridylation of histone mRNAs
Until now, terminal oligouridylation had been a rather rarely observed RNA modification in mammalian cells, although there has been ample evidence for it in trypanosomes. Examples of previously reported 3′ U tract modification included the products of miRNA-mediated cleavage (Shen and Goodman 2004) and a recycling mechanism for U6 snRNA (Trippe et al. 2006). As mentioned above, several noncanonical PAPs have been shown recently to prefer UTP over ATP (Kwak and Wickens 2007; Rissland et al. 2007). However, there was no demonstrated direct link between 3′ oligouridylation and mRNA degradation. In their study, Mullen and Marzluff (2008) have forged this link, showing that oligouridylation by noncanonical PAPs is a vital step in the cell cycle-regulated degradation of the replication-dependent histone mRNAs.
In mammalian cells, histone mRNAs are unique in that they lack introns and are not polyadenylated. Instead, their 3′ ends are formed by endonucleolytic cleavage mediated by U7 snRNP and two components of the canonical 3′ end formation machinery, CPSF73, and symplekin (Dominski and Marzluff 2007). The mature 3′ end of a histone mRNA consists of a stem–loop structure with a short 3′ ACC overhang. The 3′ stem–loop associates with the stem–loop-binding protein (SLBP), which is required for efficient 3′ processing and translation of the transcript. Importantly, histone mRNAs are expressed only during DNA replication and must be rapidly degraded at the end of S phase. However, as the major pathway of mRNA decay initiates with deadenylation, it was not clear how histone mRNA turnover proceeds. One link with characterized mRNA decay pathways was uncovered by Kaygun and Marzluff (2005), when it was discovered that the nonsense-mediated decay (NMD) factor Upf1 interacts with SLBP and is essential for histone mRNA degradation. However, in mammalian cells NMD is thought to be a deadenylation-dependent mechanism (Yamashita et al. 2005), so this observation shed little light on the pathway of histone mRNA degradation.
Mullen and Marzluff (2008) now show that near the end of S phase, or when DNA replication is inhibited, histone mRNAs acquire 3′-terminal oligo(U) tracts (Fig. 1B). Using an RNA end circularization/cloning approach, they were also able to trap decay intermediates that were shortened at both the 3′ and 5′ ends, suggesting that oligouridylation leads to both 5′–3′ and 3′–5′ decay of the transcripts. Finally, oligo(U) tracts were detected at the 3′ end of transcripts that appeared to have been shortened previously by 3′–5′ exonucleolytic activity—suggesting that perhaps multiple rounds of oligouridylation and subsequent 3′–5′ decay occur to facilitate decay through secondary structure, much as is seen during mRNA decay in prokaryotes.
Consistent with the above observations, depletion of Lsm1, exosome components, or components of the 5′–3′ decay pathway by RNAi all resulted in stabilization of histone mRNAs, suggesting that both of the standard mRNA decay pathways are involved. As the Lsm1–7 complex efficiently binds to 3′ oligo(U) tracts, and this association has recently been shown to enhance decapping in vitro (Song and Kiledjian 2007), a relatively simple model has been put forward (Fig. 1B). At the end of S phase, histone transcripts become oligouridylated and associate with the Lsm complex. This leads to decapping and 5′–3′ decay, as well as 3′–5′ decay.
The next obvious question was which enzyme is responsible for uridylation. Six of the seven putative noncanonical PAPs (termed TUTases by Mullen and Marzluff [2008]; Table 1) were depleted individually (depletion of the seventh, U6 TUTase, is lethal) by RNAi. In this way, two of the enzymes, mtPAP (TUTase 1) and PAPD5 (TUTase 3), were shown to be required for histone mRNA decay. Whether these enzymes are both directly involved is somewhat debatable, as endogenous mtPAP localizes exclusively to the mitochondria (Tomecki et al. 2004; Nagaike et al. 2005), and RNAi-mediated depletion of this enzyme has a clear effect on mitochondrial polyadenylation. It remains possible, however, that a small fraction of the protein is cytoplasmic, and an associated factor could perhaps alter the nucleotide specificity. Collectively, these data strongly suggest a role for terminal uridylation in the regulated decay of histone mRNAs in mammalian cells.
Questions for the future
Many interesting questions come to mind regarding these intriguing findings. First, how does Upf1 association with the histone transcript induce oligouridylation? The simplest mechanism would involve direct recruitment of the TUTase through protein–protein interactions. There is no existing evidence for such an interaction, but this is an easy hypothesis to test. Alternatively, UPF1, which has helicase activity (Czaplinski et al. 1995), may remodel the mRNP structure such as to allow the TUTase to access the mRNA 3′ end.
Second, given that histone mRNAs are polyadenylated in lower eukaryotes but exhibit the same regulated decay at the end of S phase, is there any evidence for a conserved mechanism in the regulation of polyadenylated and nonpolyadenylated histone transcripts? Several tantalizing observations in the literature suggest that this may, indeed, be the case. Deletions of the S. cerevisiae Trf4, Trf5, or Rrp6 genes result in elevated levels of histone mRNAs (Canavan and Bond 2007; Reis and Campbell 2007). Moreover, Trf4 mutants exhibit specific cell cycle-specific defects consistent with a role in maintaining genome stability. Finally, two noncanonical PAPs in S. pombe, Cid1 and Cid13, have been linked with the DNA replication pathways (Stevenson and Norbury 2006). Therefore, while it is not clear exactly how histone mRNA decay is regulated in fungi, it is very possible that a noncanonical PAP is involved in a fashion similar to that shown by Mullen and Marzluff (2008) for mammals.
Third, does this pathway of terminal uridylation-mediated mRNA decay for histones apply to other transcripts? There is evidence for oligouridylation of mRNA fragments following miRNA-directed cleavage in plants and mouse cells (Shen and Goodman 2004), and siRNAs and miRNAs themselves experience oligouridylation in an Arabidopsis hen1 mutant (Li et al. 2005). These results strongly suggest that this oligouridylation pathway of RNA decay may be more generally used in the cell than is currently appreciated. There is perhaps even a remote possibility that the majority of mRNA decay could actually involve an oligouridylation step following deadenylation, or even prior to it. As oligouridylated transcripts are very labile and were not anticipated to exist, it is not clear that they would have been detected before. Furthermore, the majority of cDNA libraries are oligo(dT) primed, and therefore oligouridylated transcripts would not be represented. The approach of Mullen and Marzluff (2008) to detect oligouridylated RNAs worked well in part because of the synchronized decay of a large number of histone transcripts at the end of S phase. However, now that we know oligouridylation occurs, it should be fairly straightforward to determine how widespread it is. Reverse transcription with oligo(dA) primers followed by PCR should yield interesting information, and decay intermediates can clearly be enriched by depleting the exosome and/or the 5′–3′ decay enzymes. The fact that some of the histone degradation intermediates had oligo(U) tails at an internal site further suggests that many RNAs, or at least their fragments, may experience this type of decay. In particular, as the function of oligouridylation as well as polyadenylation appears to be to provide a toehold for a 3′ exonuclease and/or a binding site for the Lsm complex, many structured RNAs such as rRNAs and tRNAs could degrade through this kind of mechanism. Finally, it is interesting to speculate that viral transcripts, such as those of flaviviruses (e.g., Hepatitis C, Dengue, and West Nile viruses), that lack a poly(A) tail but have strong secondary structures at the 3′ terminus (Gritsun and Gould 2007) may be susceptible to decay via terminal uridylation/polyadenylation via this pathway. This could serve as an interesting innate antiviral defense mechanism that has not been appreciated to date.
Finally, with regard to the actual mechanism of decay, one question that arises is how does uridylation promote both 5′–3′ and 3′–5′ decay pathways? Association of Lsm complex with the 3′ end of a transcript has been previously shown to enhance decapping (Tharun and Parker 2001; Song and Kiledjian 2007), so the induction of the 5′–3′ decay pathway is not unexpected. However, Lsm mutants in yeast exhibit unusual 3′ trimming of aberrantly stabilized mRNAs (He and Parker 2001), implying that the Lsm complex usually blocks 3′ exonuclease activity. Moreover, 3′ oligo(U) tracts inhibit 3′ exonuclease activity in vitro (Ford and Wilusz 1999; Song and Kiledjian 2007). There are two possible explanations: Lsm and the exosome may compete for the single-stranded platform provided by the oligo(U) tract, or Lsm may dissociate following decapping to allow the exosome to access the 3′ end. Further work is required to distinguish between these two pathways as well as to answer the other interesting questions posed above.
Conclusions and perspective
The existence of an oligouridylation pathway for degradation of the structured histone mRNAs brings to light important parallels with bacterial mRNA degradation. Oligouridylation, like polyadenylation in prokaryotes, provides a single-stranded platform for association of 3′–5′ exonucleases—PNPase and RNase R in bacteria, the exosome in eukaryotes. Also, an Sm-like complex—Hfq in bacteria, Lsm in eukaryotes—recognizes the unstructured 3′ end and regulates the degradation process. This cytoplasmic mRNA decay mechanism bears a resemblance to the nuclear mRNA surveillance pathway mediated by another noncanonical PAP, Trf4/5. This nuclear pathway also degrades structured transcripts by addition of an unstructured homopolymeric stretch, this time poly(A), and recruitment of the exosome. Involvement of Lsm in nuclear RNA surveillance has not been examined, although the nuclear Lsm2–8 complex has been implicated in decapping of nuclear pre-mRNAs (Kufel et al. 2004).
In conclusion, the discovery of this novel pathway of mRNA degradation has certainly uncovered many new possibilities and should lead to some exciting revelations in the near future.
Acknowledgments
We are grateful to members of the Wilusz Laboratory for discussion and comments on the manuscript. Research in the Wilusz Laboratory is supported by awards from NIH (GM072481 to J.W.) and the Muscular Dystrophy Association (to C.J.W.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1634508
References
- Barnard D.C., Ryan K., Manley J.L., Richter J.D. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Buhler M., Haas W., Gygi S.P., Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- Canavan R., Bond U. Deletion of the nuclear exosome component RRP6 leads to continued accumulation of the histone mRNA HTB1 in S-phase of the cell cycle in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:6268–6279. doi: 10.1093/nar/gkm691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis A.J. The RNA degradosome of Escherichia coli: An mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- Chen C.C., Simard M.J., Tabara H., Brownell D.R., McCollough J.A., Mello C.C. A member of the polymerase β nucleotidyltransferase superfamily is required for RNA interference in C. elegans. Curr. Biol. 2005;15:378–383. doi: 10.1016/j.cub.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Cheng Z.F., Deutscher M.P. An important role for RNase R in mRNA decay. Mol. Cell. 2005;17:313–318. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- Chowdhury A., Mukhopadhyay J., Tharun S. The decapping activator Lsm1p–7p–Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C. Maturation and degradation of RNA in bacteria. Curr. Opin. Microbiol. 2007;10:271–278. doi: 10.1016/j.mib.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Czaplinski K., Weng Y., Hagan K.W., Peltz S.W. Purification and characterization of the Upf1 protein: A factor involved in translation and mRNA degradation. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Kressler D., Tollervey D., Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Marzluff W.F. Formation of the 3′ end of histone mRNA: Getting closer to the end. Gene. 2007;396:373–390. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus M., Regnier P. The poly(A) tail of mRNAs: Bodyguard in eukaryotes, scavenger in bacteria. Cell. 2002;111:611–613. doi: 10.1016/s0092-8674(02)01137-6. [DOI] [PubMed] [Google Scholar]
- Dziembowski A., Lorentzen E., Conti E., Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- Edmonds M. A history of poly A sequences: From formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- Folichon M., Arluison V., Pellegrini O., Huntzinger E., Regnier P., Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003;31:7302–7310. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford L.P., Wilusz J. 3′-Terminal RNA structures and poly(U) tracts inhibit initiation by a 3′ → 5′ exonuclease in vitro. Nucleic Acids Res. 1999;27:1159–1167. doi: 10.1093/nar/27.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsun T.S., Gould E.A. Origin and evolution of 3′UTR of flaviviruses: Long direct repeats as a basis for the formation of secondary structures and their significance for virus transmission. Adv. Virus Res. 2007;69:203–248. doi: 10.1016/S0065-3527(06)69005-2. [DOI] [PubMed] [Google Scholar]
- He W., Parker R. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics. 2001;158:1445–1455. doi: 10.1093/genetics/158.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim F., Rohr J., Jeong W.J., Hesson J., Cerutti H. Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science. 2006;314:1893. doi: 10.1126/science.1135268. [DOI] [PubMed] [Google Scholar]
- Kaygun H., Marzluff W.F. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Richter J.D. Opposing polymerase–deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Kufel J., Bousquet-Antonelli C., Beggs J.D., Tollervey D. Nuclear pre-mRNA decapping and 5′ degradation in yeast require the Lsm2–8p complex. Mol. Cell. Biol. 2004;24:9646–9657. doi: 10.1128/MCB.24.21.9646-9657.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J.E., Wickens M. A family of poly(U) polymerases. RNA. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J.E., Wang L., Ballantyne S., Kimble J., Wickens M. Mammalian GLD-2 homologs are poly(A) polymerases. Proc. Natl. Acad. Sci. 2004;101:4407–4412. doi: 10.1073/pnas.0400779101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Li J., Yang Z., Yu B., Liu J., Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Greimann J.C., Lima C.D. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Mohanty B.K., Kushner S.R. Polynucleotide phosphorylase, RNase II and RNase E play different roles in the in vivo modulation of polyadenylation in Escherichia coli. Mol. Microbiol. 2000;36:982–994. doi: 10.1046/j.1365-2958.2000.01921.x. [DOI] [PubMed] [Google Scholar]
- Mohanty B.K., Maples V.F., Kushner S.R. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- Motamedi M.R., Verdel A., Colmenares S.U., Gerber S.A., Gygi S.P., Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Mullen T.E., Marzluff W.F. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes & Dev. 2008 doi: 10.1101/gad.1622708. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaike T., Suzuki T., Katoh T., Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J. Biol. Chem. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- Read R.L., Martinho R.G., Wang S.W., Carr A.M., Norbury C.J. Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc. Natl. Acad. Sci. 2002;99:12079–12084. doi: 10.1073/pnas.192467799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis C.C., Campbell J.L. Contribution of Trf4/5 and the nuclear exosome to genome stability through regulation of histone mRNA levels in Saccharomyces cerevisiae. Genetics. 2007;175:993–1010. doi: 10.1534/genetics.106.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland O.S., Mikulasova A., Norbury C.J. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol. Cell. Biol. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilders G., Van D.E., Pruijn G.J. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 2007;35:2564–2572. doi: 10.1093/nar/gkm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Goodman H.M. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- Slomovic S., Laufer D., Geiger D., Schuster G. Polyadenylation and degradation of human mitochondrial RNA: The prokaryotic past leaves its mark. Mol. Cell. Biol. 2005;25:6427–6435. doi: 10.1128/MCB.25.15.6427-6435.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M.G., Kiledjian M. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA. 2007;13:2356–2365. doi: 10.1261/rna.765807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson A.L., Norbury C.J. The Cid1 family of non-canonical poly(A) polymerases. Yeast. 2006;23:991–1000. doi: 10.1002/yea.1408. [DOI] [PubMed] [Google Scholar]
- Tharun S., Parker R. Targeting an mRNA for decapping: Displacement of translation factors and association of the Lsm1p–7p complex on deadenylated yeast mRNAs. Mol. Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- Tharun S., Muhlrad D., Chowdhury A., Parker R. Mutations in the Saccharomyces cerevisiae LSM1 gene that affect mRNA decapping and 3′ end protection. Genetics. 2005;170:33–46. doi: 10.1534/genetics.104.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R., Dmochowska A., Gewartowski K., Dziembowski A., Stepien P.P. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 2004;32:6001–6014. doi: 10.1093/nar/gkh923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippe R., Richly H., Benecke B.J. Biochemical characterization of a U6 small nuclear RNA-specific terminal uridylyltransferase. Eur. J. Biochem. 2003;270:971–980. doi: 10.1046/j.1432-1033.2003.03466.x. [DOI] [PubMed] [Google Scholar]
- Trippe R., Guschina E., Hossbach M., Urlaub H., Luhrmann R., Benecke B.J. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S., Wolf J., Martin G., Blank D., Dettwiler S., Friedlein A., Langen H., Keith G., Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Eckmann C.R., Kadyk L.C., Wickens M., Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- West S., Gromak N., Norbury C.J., Proudfoot N.J. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol. Cell. 2006;21:437–443. doi: 10.1016/j.molcel.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Yamashita A., Chang T.C., Yamashita Y., Zhu W., Zhong Z., Chen C.Y., Shyu A.B. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]