Abstract
Numerous short-lived and highly reactive oxygen species (ROS) such as O2·- (superoxide), ·OH (hydroxyl radical), and H2O2 (hydrogen peroxide) are continuously generated in vivo. Depending upon concentration, location and intracellular conditions, ROS can cause toxicity or act as signaling molecules. The cellular levels of ROS are controlled by antioxidant enzymes and small molecule antioxidants. As major antioxidant enzymes, superoxide dismutases (SODs), including copper-zinc superoxide dismutase (Cu/ZnSOD), manganese superoxide dismutase (MnSOD) and extracellular superoxide dismutase (ECSOD), play a crucial role in scavenging O2·-. This review focuses on the regulation of the genes (sods) coding for these enzymes with an emphasis on human genes. Current knowledge about sods structure and their regulation is summarized and depicted as diagrams.
Studies to date on genes coding for Cu/ZnSOD (sod1) are mostly focused on alteration in the coding region and their associations with Amyotrophic Lateral Sclerosis (ALS). Evaluation of nucleotide sequences reveals that regulatory elements of the sod2 gene reside in both the non-coding and coding regions. Changes associated with sod2 lead to alteration in expression levels as well as protein function. We also discuss the structural basis for the changes in SOD expression associated with pathological conditions and where more work is needed to establish the relationship between SODs and diseases.
Keywords: Superoxide Dismutase, Gene Regulation, Oxidative Stress-related Disease
Introduction
Oxidative stress caused by the imbalance between reactive oxygen species (ROS) or reactive nitrogen species (RNS) and biological antioxidant system can lead to modification of macromolecules such as DNA, lipid and protein [1-3]. Because the redox status (oxidizing/reducing conditions) of cells is involved in regulating various transcription factors/activators (e.g., AP-1, NF-κB and p53), thereby influencing cellular target gene expression and modulating cellular signaling pathways, appropriate ROS and RNS levels are necessary for normal physiological function of the living organisms [4]. However, excessive redox active species may cause DNA damage, repress the activity of cellular enzymes and induce cell death through activation of kinases and caspase cascades [5-8].
To ameliorate and cope with injury from oxidative damage and maintain redox homeostasis, aerobic organisms have developed efficient defense systems of enzymatic and non-enzymatic antioxidants. The superoxide dismutase family is specialized in eliminating superoxide anion radicals derived from extracellular stimulants, including ionizing radiation and oxidative insults, together with those primarily produced within the mitochondrial matrix as byproducts of oxygen metabolism through the electron transport chain [9]. Three distinct isoforms of SOD have been identified and characterized in mammals: copper-zinc superoxide dismutase (Cu/ZnSOD, encoded by the sod1 gene), manganese superoxide dismutase (MnSOD, encoded by the sod2 gene) and extracellular superoxide dismutase (ECSOD, encoded by the sod3 gene). These forms of SODs elicit similar functions, but characteristics of their protein structures, chromosome localizations, metal cofactor requirements, gene distributions and cellular compartmentalization are distinctly different from one another (reviewed in [10]). Genetic comparisons indicate that similarities exist in sod1 and sod3 genes in certain levels of the amino acid homology while sod2 does not share substantial amino acid homology with either sod1 or sod3 [10]. The unique features of each SOD in terms of molecular weight, cellular localization, assembly of subunits, metal cofactor requirements and ion-delivery related proteins are summarized in Table 1.
Table 1.
Summarized characteristics of three superoxide dismutases in eukaryotic cells
| Isoform | Location | MW/kDa | Assembly of subunits | Metal ion | Ion-delivery related protein |
|---|---|---|---|---|---|
| Cu/ZnSOD | cytoplasm, nucleus, mitochondrial membrane | 88 | homodimer | Cu2+ (catalytic active) | CCS (copper chaperone for SOD1) [11-13] |
| GSH (glutatione, secondary chaperone) [14] | |||||
| Zn2+ (maintain enzyme stability) | metallotioneins; ZnT (zinc transporter) [15] | ||||
| MnSOD | mitochondria matrix | 32 | homotetramer | Mn2+ (catalytic active) | Smf2p (manganese trafficking factor for mitochondrial SOD2)* [16] |
| MTM1 (manganese trafficking factor for mitochondrial SOD2)* [17,18] | |||||
| ECSOD | plasma membrane, extracellular fluids | 135 | hornotetrarneric glycoprotein | Cu2+ (catalytic active) | Atox1 (antioxidant-1, copper chaperone) [19] |
| MNK (Menkes ATPase) [20] | |||||
| Zn2+ (maintain enzyme stability) | N/A |
the metal transporter which is found in Saccharomyces cerevisiae.
Regulation of sod genes plays a pivotal role in balancing the concentration of ROS. The compartmentalization and control of SODs at both expression and activity levels contribute to the level of SOD and consequent localized ROS level [21, 22]. This review focuses on recent progress made on sod gene regulation. Given that SODs have important functions beyond the essential role of sod2 for survival in the aerobic environment, their association with diseases such as neurodegenerative diseases, pulmonary and cardiovascular dysfunction, cancer development and progression [19, 23-27] is also discussed. Knowledge of how the expression of sod genes is modulated would provide insight into the understanding of human diseases and facilitate the development of therapeutic interventions.
Genetic structures and organization of the sod genes
sod1
The chromosomal localization and characteristics of the sod1 gene have been identified in rodents [28], bovines [29] and humans [30]. The human sod1 gene is localized on chromosome 21q22 [30]. The latest information on sod genes sequences is generated from initial sequencing data and comparative analysis of whole genomic studies among different species. Sequence comparison has revealed that the sod1 gene consists of five exons interrupted by 4 introns, which is significantly similar in all these species, in terms of the size of exons particularly the coding regions (Fig.1). The variation of intron size found in different studies may be associated with gene polymorphisms in different human tissues and cell lines. Because not all information on a specific gene, e.g., the transcription initiation site, is available from online databases, original sources on characterization and organization of sod genes are used as references to evaluate the gene regulation studies. The sod1 promoter has a high GC-rich region, as well as TATA box and CCAAT box. Several putative transcription factors binding sites in the promoter region that have been verified by subsequent functional studies are summarized in Figure 1.
Figure 1. Organization of the sod1 gene.
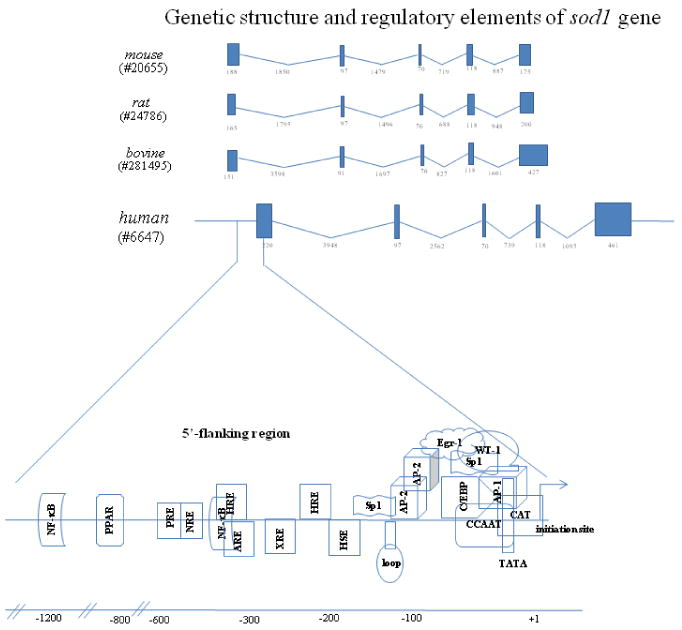
The EntrzGeneID of each gene from the NCBI database is indicated on the left. Solid boxes and the associated numbers indicate exons and the size of each exon in base pairs. Lines and numbers between each exon indicate introns and the corresponding size. The regulatory elements in the 5′ flanking region for the human sod1 are expanded and shown in the lower part. The transcription start site is indicated by an arrow and designated +1. Binding sites for transcription factors known to play a regulatory role are placed according to the location of the corresponding regulatory elements identified in published literature and are indicated by numbers on the bottom part of the figure.
sod2
Among the three SOD isoforms, sod2 has a unique genetic organization and little similarity with sod1 and sod3. In addition to the human gene, the complete genomic sequence of the sod2 gene has been isolated and characterized for other species such as mouse [31, 32], rat [33] and bovine [34]. The primary structure of sod2 genes is highly conserved and shares more than 90% sequence homology in the coding region [35], which is summarized in Figure 2. The human sod2 is located on chromosome 6q25.3. Two forms of human sod2 transcripts exist, but no significant difference exists between the lengths of translation residues. Based on molecular structure and organization of the human sod2 gene, five exons interrupted by 4 introns have been identified. The basal promoter of the sod2 gene lacks TATA and CAAT boxes but contains GC rich motifs and numerous Sp1 as well as several AP-2 consensus sequences in its proximal promoter region [36].
Figure 2. Organization of the sod2 gene.

The EntrzGeneID of each gene from the database, the size of exons and introns, and the transcriptional start site are organized as described for sod1. The regulatory elements identified in the 5′ flanking region and the second intron of the human sod2 are expanded and shown in the lower part. Corresponding numbers with positive and negative numbers indicate their location relative to the start site, which is designated +1.
sod3
The human sod3 gene is localized on chromosome 4. The sod3 gene consists of 3 exons and 2 introns. An uninterrupted coding region is present within exon 3, which is homologous to the sod1 gene [37, 38]. However, an updated database shows only two exons and excludes the 5bp exon 1 containing the transcription start site. Thus, the underlying functions of this short exon and the precise location of the first exon remain to be evaluated. Two CAAT box elements but not the classical TATA box have been found in the sod3 promoter region [38]. Information on the sod3 cDNA sequence for rat and mouse is also available [39, 40]. The full-length mouse sod3 cDNA is 82% identical to rat but only 60% identical to human ECSOD [40]. Several transcription factors binding elements are present and they are depicted in Figure 3.
Figure 3. Organization of the sod3 gene.
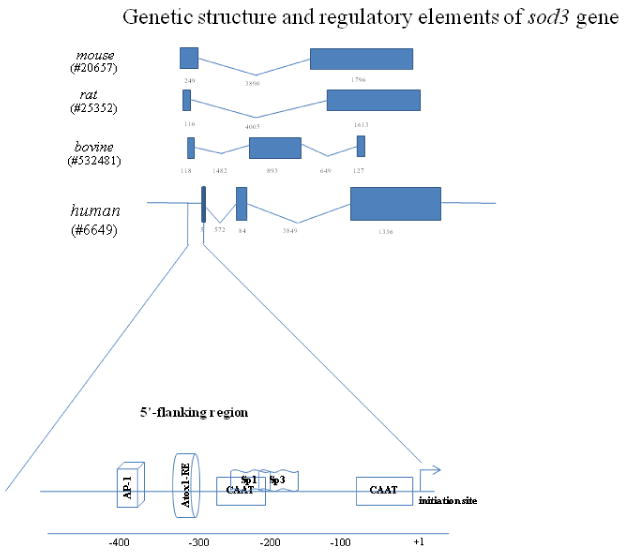
The EntrzGeneID of each gene from the database, the size of exons and introns, and the transcriptional start site are organized as described for sod1. The first exon of the human sod3 has been added according to the information extracted from the original publication. This exon was not present in the data base indicated by EntrzGeneID #6649.
Transcriptional factors involved in the regulation of the sod genes
Results of computer analysis have revealed many transcriptional regulatory elements in the proximal promoter regions of the sod genes that are binding sites for several common transcription factors. These transcriptional factors, including NF-κB, AP-1, AP-2 and Sp1, as well as C/EBP, have been shown to play important roles in regulating the constitutive or inductive expression levels of all three SODs.
Nuclear Factor-KappaB (NF-κB)
The redox-sensitive transcriptional factor NF-κB acts as a regulator of genes by serving as an “immediate responder” to harmful cellular stimuli. NF-κB responsive elements have been found in both promoter and intronic regions of all three sod genes [41-43]. Because the sod1 gene is often constitutively expressed and not as easily inducible as other superoxide dismutases, it is considered a “housekeeper gene” and is sometimes used as an internal control to compare variations in MnSOD expression level or activity [44]. Though the NF-κB site in the sod1 promoter is not very responsive to external stimuli, the PI3K/Akt pathway can activate NF-κB and upregulate Cu/ZnSOD expression [45]. The presence of the superoxide permeability pathway in endosomal membranes suggests that Cu/ZnSOD mediates O2·- dismutation at the endosomal surface and may produce the localized H2O2 required for redox activation of NF-κB [46]. Interestingly, activation of NF-κB is absent not only in sod1 overexpressing cells but also in Cu/ZnSOD deficient mice [47, 48].
The induction of sod2 in response to oxidative stress has been well established in organisms, tissues and cells growing under various stress conditions. Stimuli such as ionizing radiation [49, 50], 12-O-tetradecanoylphorbol-13-acetate (TPA) [51], interferon-gamma (IFN-γ) [52] and proinflammatory cytokines, such as tumor necrosis factor α (TNF α) [53, 54], interleukin-1beta (IL-1 β) [55], interleukin-4 (IL-4) and interleukin-6 (IL-6), can rapidly modulate sod2 gene transcription [56]. Previous studies from our laboratory and by other investigators have demonstrated that the stimulus-dependent MnSOD mRNA level elevation is controlled at the transcription level [57]. In one of these studies, NF-κB was identified as the most crucial transcriptional factor regulating MnSOD induction [49]. Interestingly, functional studies have demonstrated that while the NF-κB site located within the second intron of the sod2 gene is necessary for cytokine-mediated induction of MnSOD expression [43, 58-60], p50, a member of the NF-κB family, elicits a negative role in sod2 expression [61].
The regulatory effects of different stimuli such as cytokines, vasoactive factors, nitric oxide, cyclic nucleotides and angiotensin II on sod3 have been reviewed previously [20, 62]. A putative NF-κB motif in the human sod3 promoter region has been proposed as the functional transcriptional binding site contributing to induction and coregulation of nitric oxide synthase (iNOS) and ECSOD [38, 42, 63]. However, the location of this regulatory element has not been functionally identified. An additional NF-κB site has been identified in the second intron, but its regulatory role in ECSOD expression remains unclear [42].
Specificity Protein 1 (Sp1)
Sp1 is a zinc-finger protein that acts as a transcription factor by binding directly to DNA through three consecutive zinc-finger domains in the C-terminus and enhances gene transcription with one of the two glutamine-rich domains [64-66]. The existence of multiple GC boxes is the identifiable characteristic of the Sp1-dependent promoter. Thus, the GC-rich motif contained within the three sod gene promoters suggests a common regulatory role of Sp1 in the expression of SODs [67, 68].
Sp1 is a zinc-finger protein that acts as a transcription factor by binding directly to DNA through three consecutive zinc-finger domains in the C-terminus and enhances gene transcription with one of the two glutamine-rich domains [64-66]. The existence of multiple GC boxes is the identifiable characteristic of the Sp1-dependent promoter. Thus, the GC-rich motif contained within the three sod gene promoters suggests a common regulatory role of Sp1 in the expression of SODs [67, 68].
Sp1 is necessary for both basal transcription and TPA-induced transcription via non-canonical binding sites of the sod1 proximal promoter region [44, 69]. Consistently, sod1 basal promoter activity has been significantly elevated by ectopically overexpressing Sp1 [70]. Studies have shown that DNA binding and promoter activity can be completely abolished by mutations in the Sp1/Egr-1 site [70]. Thus, Sp1 can directly activate sod1 by binding to DNA. Sp1 can also interact with other proteins to enhance the expression of sod1. For example, the expression of Cu/ZnSOD in both neuronal and non-neuronal cell lines is modulated by Sp1 activity through direct interaction with neuronal nitric oxide synthase (nNOS) in the cytosol as well as in the nucleus [71].
In the 5′-flanking region of sod2, transcription factor Sp1 is essential for not only the constitutive but also the inducible expression of MnSOD [67, 72]. The importance of the Sp1 binding site on endogenous sod2 promoter activity has been confirmed by potentially unbiased PIN*POINT (ProteIN POsition Identification with Nuclease Tail) analysis in vivo [73]. Multiple Sp1 binding elements are needed to induce sod2 expression via the proximal promoter and the intronic enhancer element. A unique DNA looping structure in the 5′-flanking region formed by direct interactions between distant and local Sp1 is able to synergistically activate transcription in vivo [74]. This unique single-strand structure of the sod2 promoter creates the required structure for RNA-binding proteins, such as nucleophosmin (NPM). NPM binds to an 11G single-strand loop structure in the sod2 promoter region and integrates Sp1 and NF- κB responses. Disruption of this loop structure impairs constitutive and inductive transcriptions [75]. The binding of NPM to this loop structure enables Sp1, NF- κB and other important transcription factors, such as p53, to interact and exert a positive or negative effect on the expression of sod2 [76]. This complex relationship between transcription factors may explain the alteration of sod2 expression as a disease progresses.
It has been demonstrated that binding of Sp1/Sp3 transcription factors to the human sod3 gene proximal promoter region is essential for trichostatin A (TSA) dependent and basal transcription of sod3 because deletion of the Sp1 binding site significantly abolishes this activation [77]. This role of Sp1/Sp3 has also been demonstrated in lung fibroblasts [78]. Thus, it is unclear whether the presence of Sp1 is absolutely essential or other members of the Sp family are able to compensate for Sp1 function for the expression of sod3.
Activator Protein 1 (AP-1)
AP-1 is a homo- or hetero-dimeric protein composed of proteins belonging to the c-Fos, c-Jun and Fra families. AP-1 acts as a transcriptional regulator to modulate signal transduction processes involved in cell proliferation and transformation [79]. Studies of the redox regulation of AP-1 oncogenes have been excellently reviewed [4]. The expression of c-fos and c-jun genes is responsive to a variety of stimuli, including cytokines, growth factors and oxidative stress. However, the increased DNA binding capacity of AP-1 could cause a reduction in Cu/ZnSOD [71]. The activity of AP-1 is also subject to redox regulation. Thus, alteration in sod genes expression also modulates AP-1 activity [80, 81]. In a skin cancer model, overexpression of the sod2 gene in transgenic mice results in reduced tumor incidence by suppressing AP-1 activation whereas MnSOD deficiency enhances AP-1 and p53 levels, as well as increases proliferation and apoptosis events [82]. Increased MnSOD in response to oxidative stress during hepatitis C replication is regulated by two distinct signaling pathways involving p38 MAPK and JNK via AP-1 [83]. Therefore, AP-1 could activate the expression of sod genes and could be activated by the expression of SODs.
Activating Protein 2 (AP-2)
AP-2 is a family of closely related transcription factors consisting of AP-2alpha, AP-2beta, AP-2gamma, AP-2delta and AP-2epsilon [84]. In addition to directly binding to the cis-element in the target gene, AP-2 is able to crosstalk with other transcriptional factors to alter the expression of a specific gene [85]. Ginsenoside Rb2, purified from the panaxadiol fractions of Panax ginseng extracts, is able to increase sod1 transcription through the AP-2 site [86]. On the other hand, AP-2 plays a negative role in the constitutively low expression of MnSOD by suppressing Sp1-dependent transcription [67, 87, 88]. A methylated AP-2 binding site in the sod2 promoter reduces AP-2 DNA binding and relieves transcriptional repression of MnSOD [89]. Thus, the binding of AP-2 to the sod2 promoter may have a negative effect on MnSOD expression. Consistent with this possibility, a low Sp1/AP-2 ratio plays a role in dysregulating the promoter activity of the sod2 gene. Consequently, the loss of AP-2 activity in several types of human cancer may explain a high level of MnSOD in these cancers.
CCAAT-Enhancer-Binding Proteins (C/EBP)
C/EBP proteins consist of six members, C/EBP α to C/EBP ζ, which can interact with the CCAAT box motif present in many gene promoters. The binding of C/EBPs to DNA requires that homo- or hetero- dimerization be formed within the various members of the C/EBP family or with other transcription factors [90]. C/EBP-related factors are necessary for basal sod1 transcription [69]. Of the known C/EBP family members, C/EBPα and C/EBPβ play similar roles in stimulating the human sod1 gene. C/EBPα also plays a major role in activating the transcription of the rat sod1 gene [91, 92]. The C/EBP binding site located in the sod2 intronic enhancer region elicits the supportive role of MnSOD induction in response to cytokine stimulation. Various C/EBP isoforms, C/EBPβ/LAP*, C/EBPβ/LAP, C/EBPβ/LIP and C/EBPδ, perform distinct functions in MnSOD transcription [58, 93]. A C/EBPβ-binding site located between -242 and -178 in the promoter region of the sod3 gene has been identified by deletion analysis of the sod3 promoter-luciferase construct. This C/EBPβ-binding site is also involved in the induction of ECSOD mRNA and protein levels, as well as confers resistance to insulin [94].
Unique Transcriptional Factors
The five transcriptional factors discussed above share relatively common effects on the regulation of all three superoxide dismutase genes. Because of the structural characteristics and organization of each gene sequence, several transcription factors play a unique regulatory role in the expression of superoxide dismutase isoforms. For example, arachidonic acid induces the rat sod1 gene through the peroxisome proliferator-responsive element (PPRE) in the 5′-flanking sequence [95]. The transcription factors Elk1 and YY1, which bind to the positive and negative regulatory elements in the upstream region of the rat sod1 gene, respectively, coordinate the expression of rat sod1 [96]. The anticancer drug mitomycin C inhibits sod1 gene transcription through p53-mediated transcriptional repression [97]. In addition, xenobiotics such as TCDD can stimulate induction of sod1 by either the Nrf2 protein or the Ah receptor protein through the antioxidant responsive element (ARE) and xenobiotic responsive element (XRE), respectively [98]. This information suggests possible mechanisms for the adverse effect of xenobiotics on human health. FOXO3a, a member of the family of Forkhead transcription factors, can bind to the 5′-flanking region and mediate the expression of MnSOD resulting in protection of quiescent cells from oxidative stress [99]. Increased age-related Akt activity might be responsible for the phosphorylation and inactivation of FOXO3a, which in turn down-regulates MnSOD transcription [100]. Several putative FOXO transcription factor binding sites starting at positions -1376, -1231 and -822 have also been identified in the human sod1 gene which suggests dual regulation by the pair NF-κB/FOXO [45]. Knocking down peroxisome proliferator-activated receptor γ (PPAR γ) could also down-regulate MnSOD at the mRNA and protein levels and reduce the protective capability of MnSOD against oxidative damage in cardiomyocytes [101]. It is possible that a pathway crosstalk may exist between the nuclear receptor- regulated signaling cascade and the regulation of sod2 gene expression.
The enzyme ECSOD contains copper and its activity directly correlates with the copper concentration and the copper chaperone antioxidant-1 (Atox1) [102]. Surprisingly, Atox1 not only acts as a copper chaperone involved in copper delivery to ECSOD at the trans-Golgi network but also positively regulates sod3 gene transcription. For ECSOD to be active at a high level, it requires both the copper chaperone and transcription factor functions of Atox1 [18, 102]. Menkes ATPase, a copper transporter, is also required for the activation of sod3, and modulated AngII induces hypertension and endothelial function by regulating ECSOD activity and vascular superoxide anion production [103, 104]. In addition to being regulated by copper containing enzymes and transporters, the sod3 gene is uniquely regulated negatively or positively by transcriptional repressors and enhancers. Repressors of sod3 gene expression include myeloid zinc finger 1 and a gut-enriched Kruppel-like factor. Activators of sod3 transcription include Ets family members such as Elf-1 and GA-binding protein α and β that actively interact with the mouse sod3 purine-rich proximal promoter region to regulate the cell-type specific expression of mouse ECSOD [105]. Knowledge of positive and negative roles of these transcription factors may facilitate our understanding of sods expression under pathological conditions.
Epigenetic regulation of the sod genes
Epigenetic regulation refers to heritable changes in the level of gene expression not related to the underlying DNA sequence. Though cancer is clearly a genetic disease, either hereditary or somatic, epigenetic modulation may affect or contribute to carcinogenesis. It is well documented that malignant cancer often exhibits altered expression and activity of MnSOD compared with normal counterparts, but the reasons for this alteration are poorly defined. Doman's group published a series of papers that demonstrate the role of epigenetic regulation in increasing cytosine methylation and decreasing histone acetylation to create a repressive chromatin structure associated with epigenetic silencing of sod2 expression in human breast cancer cells and other transformed cells [106-108]. The relative expression patterns of MnSOD are inversely related to the methylation status of the sod2 promoter in several pancreatic carcinoma cell lines. The methyltransferase inhibitor is able to reverse the hypermethylation status of CpG sites to restore MnSOD levels [109]. Similar observations indicating that the sod2 gene is epigenetically silenced as a result of promoter hypermethylation have also been reported by others using KAS 6/1 human multiple myeloma cells [110].). In addition, early growth responsive-1 (Egr-1) interacts with Sp1 and histone deacetylase-1 (HDAC1) in the proximal promoter region of sod2 to suppress histone acetylation and further inactivate MnSOD expression [111]. These publications provide insights into the possibility of an epigenetic mechanism for the regulation of the sod2 gene.
Studies of the methylation status of the sod1 gene in ALS patients have revealed that the promoter of the sod1 gene was largely unmethylated in the subjects studied. Epigenetic silencing of sod1 is therefore unlikely to be a common mechanism in ALS [112]. Though no difference in the methylation levels of CpG sites across the promoter region of the sod3 gene has been found, it is possible that the activity of the sod3 gene proximal promoter element may also be modulated by epigenetic processes [113]. Hypomethylation of the rabbit sod3 gene has been associated with the development of atherosclerosis, suggesting that epigenetic modulation of sod3 may be involved in the development of atherosclerotic lesions [114].
Post-transcriptional Regulation of the sod genes
It has been well-documented that specific gene expression is regulated not only at the transcriptional level but also at the post-transcriptional level by changes in mRNA stability, mRNA translation and post-translational modification. Thus, increased mRNA level may not be sufficient to compensate for a compromised translational deficiency. The presence of post-transcriptional regulation and RNA silencing pathways provides conserved mechanisms by which target gene expression could be rapidly modulated. Post-transcriptional processes such as mRNA processing, export and microRNA modulation form a complex regulatory network contributing to target gene expression patterns.
Because of the presence of the typical splice junction after the sod2 gene is transcribed, human MnSOD mRNA is processed into two mRNA species of approximately 1 and 4kb size. A ∼280-nucleotide fragment within the MnSOD mRNA coding region determines the stability of messenger RNA in the absence of ribosome transit, providing a mechanism for both basal and stimulus-dependent post-transcriptional regulation of MnSOD [115]. The Alu-like element contained in MnSOD mRNA 3′UTR acts as probable microRNA targets [116, 117]. Moreover, mRNA binding with proteins and other RNAs may function as a critical regulator in post-transcriptional processes. Thus, predicting and identifying appropriate RNA structural motifs could provide insight into the regulatory mechanisms of gene expression [118]. A special, developmentally regulated MnSOD RNA-binding protein that is redox sensitive exists in rat lung and forms complexes with a fragment of the 3′UTR region of MnSOD mRNA [119]. RNA in polymers acts to inhibit MnSOD binding protein activity and prevent it from participating in post-transcriptional control of mature MnSOD protein product and activity [120, 121]. On the other hand, the partially conserved 3′UTR cis-element in the sod2 gene and RNA-protein binding activity are required for improving the translation of MnSOD. A translational enhancer in this 3′UTR cis-element, designated MnSOD-response element, is involved in the interaction with MnSOD RNA-binding protein to enhance translation efficiency and to increase translation of a heterologous RNA when it is positioned in a post-transcriptional reporter construct [122, 123]. Interestingly, tyrosine phosphorylation of this cytosolic MnSOD binding protein (MnSOD-BP) can modulate MnSOD protein level even though the identity of MnSOD-BP remains unknown [124]. The post-transcriptional mechanism will be another potentially important level of regulation defending mitochondria against oxidative stress-mediated injury.
Compared to the number of studies about sod2, studies of regulation mechanisms, such as post-transcriptional modulation of sod1 and sod3, are relatively rare. Diverse 3′UTR lengths of sod1 mRNA are associated with human Cu/ZnSOD enzyme activity [125]. Mutations in the 3′UTR of sod1 in CNS tissue are associated with motor neuron disease [126]. The 10bp deletion in 3′UTR of the sod3 transcript, which may be responsible for the alteration of ECSOD RNA half-life, has been described, but the significance and identity of the regulatory element remain unclear [127]. Given the potentially important role of ECSOD in defending against extracellular superoxide, additional studies of sod3 gene activation and mRNA modification and processing should be useful for understanding the processes of superoxide dismutase-dependent diseases.
Biomedical implications
Cu/ZnSOD (sod1)
Cu/ZnSOD was the first superoxide dismutase in eukaryote to be characterized and it has been found in cytoplasm, nucleus, microsomes and also in the mitochondrial intermembrane space [9, 128]. The discovery of the Cu/ZnSOD mutation in ALS, a fatal, adult-onset neurodegenerative disease primarily affecting motor neurons in the brain, brainstem, and spinal cord, has attracted broad attention [129]. Most current Cu/ZnSOD studies are focused on identification of the mutations in the coding region of the sod1 gene and how the mutated sod1 gene causes ALS and other neurodegenerative diseases. The focus at the protein level is due, in part, to the finding that ALS mice are not deficient in Cu/ZnSOD activity, but rather expression of mutant protein may lead to gain of toxic function. In addition, the promoter of the sod1 gene is mostly unmethylated in familial ALS (fALS) patients, so epigenetic silencing of sod1 is therefore unlikely to be a common mechanism in fALS [112]. The discovery of 11 missense sod1 gene mutations in 13 familial ALSs by Rosen, et al. in 1993 provided the first molecular connection between Cu/ZnSOD and the pathogenesis of fALS [130]. At present, over 150 mutations distributed in coding regions of the sod1 gene, affecting over 70 positions, have been reported [131]. Updated information about these mutations is available at the ALS online database (http://alsod.iop.kcl.ac.uk/Als/index.aspx) [132]. The majority of known mutations are missense mutations distributed among five exons of the sod1 gene. A newly identified mutation with a defect in the TATA box of the sod1 promoter has been reported, but genetic analysis has revealed that it is not a disease-causing mutation or susceptibility factor for ALS [133]. Several mutations of highly diverse properties cause the same clinical outcomes, but the precise mechanism of how each mutation in the sod1 gene leads to neurodegeneration remains unclear. The Ala4Val mutation in exon1 in humans is the most frequent mutation found from both extensive screening results of the sod1 coding region and clinical experiences in the U.S. [130]. However, the type and relative frequency of each mutation vary in the world population. Although mutant Cu/ZnSOD-catalyzed oxidative reactions and/or misfolding are being proposed as driving fALS pathogenesis [131], mice deficient for sod1 are viable and appear to develop without obvious motor abnormalities, suggesting that “gain-of-function” mutations in sod1 may be a major factor causing fALS. A number of transgenic rodent models with different point mutations in the sod1 gene have been developed. Rodents carrying the G93A sod1 gene mutation are the most extensively studied despite the fact that this mutation is rarely detected in humans. Whether the information harvested from animal models can be appropriately applied to humans requires careful genetic comparison between different species. It has been found that accumulation and aggregation of insoluble Cu/ZnSOD exist in motor neurons of fALS patients and transgenic animal models with designed sod1 mutations such as G93A and G37R. These mutations cause inhibition of axonal transport and mitochondrial dysfunction [134, 135]. Excellent reviews of the catalytic activity of mutant Cu/ZnSOD, in vivo metallation state and stability, as well as oxidative modification related to sod1-associated fALS, have been written [131, 136].
For the Cu/ZnSOD enzyme, the copper ion functions in the oxidized and reduced alternation while the Zn2+ helps maintain enzyme stability [11, 137]. The copper chaperone for SOD (CCS), which has been suggested as a robust, sensitive and specific biomarker of copper status, delivers copper to the Cu/ZnSOD apoenzyme [12]. The CCS-knockout mice serve as a resource for studying alternative copper source for Cu/ZnSOD [138]. It has been shown that CCS facilitates the stepwise conversion of the disulfide-reduced immature Cu/ZnSOD to the active disulfide-containing enzyme [139, 140]. Thus, copper related post-translational regulation may affect enzyme activation, folding and aggregation leading to aberrant protein function in diseases, especially in fALS. In addition to copper zinc being a multipurpose trace element, it contributes to Cu/ZnSOD structure and modulates the folding free energy surface of the enzyme [14]. The potential role of zinc-binding proteins, such as metallothioneins and zinc transporter (ZnT), in delivering metal ions to enzymes has been suggested but remains to be established. Appropriate copper and zinc accessibility to Cu/ZnSOD can influence the structural geometry and its redox activity, as well as the protein function. Decreased zinc affinity of ALS-mutant SOD can lead to elevation of peroxynitrite-mediated tyrosine nitration [141]. Zinc-deficiency in either wild-type or ALS-mutant SOD exhibits nitric oxide-dependent neurons apoptosis, a process which can be prevented by zinc repletion [142]. Structural analysis of constitutively zinc-deficient SOD protein generated by mutation in zinc-binding pocket identifies asymmetric dimers with weakened dimer interface, enhanced redox properties of the copper and thiol-dependent aggregation of zinc-deficient SOD [143]. However, whether supplement of zinc will be beneficial to ALS patient is unknown. Zn supplementation in ALS mice has either protective or toxic effect depending on the levels of dietary Zinc [144, 145]. Therefore, considering this paradoxically dose-dependent effect, Zinc supplementation in ALS patient will require information on the zinc status of individual patient.
In addition to fALS, alteration of Cu/ZnSOD expression level or catalytic activity has been identified in several physiological situations such as aging and age-associated diseases (reviewed in [11, 146, 147]). Sod1 knock-out mice have been shown to suffer from physiological impairment including reduced female fertility, macular degeneration and death from liver tumors [131, 136]. Considering the important role of Cu/ZnSOD in these diseases, and the fact that the majority of ALS is sporadic, future studies should include understanding the regulation of sod1 gene expression and post-translational modifications.
MnSOD (sod2)
Abnormal cellular redox status has been associated with many types of diseases. Among three SOD isoforms, MnSOD is the only SOD that has proved to be essential for the survival of aerobic organisms [148]. The physiological role of MnSOD as a cytoprotective enzyme has been clearly confirmed by the extremely short life-span of MnSOD knockout mice, which died shortly after birth with dilated cardiomyopathy and neurodegeneration [149, 150]. The importance of MnSOD beyond the need for survival in the aerobic environment has also been well-established. For example, in the development of cancer, which involves either the activation of oncogenes or the inactivation of the tumor suppressor gene, the levels of ROS and superoxide dismutase are regulated reciprocally. It has been demonstrated that, at an early stage of cancer development, oxidative stress and relatively low levels of antioxidant enzymes result in DNA damage and cell injury. Because MnSOD plays a critical role in the defense against oxidant-induced injury and apoptosis of rapidly growing cancer cells, it is considered a unique tumor suppressor protein [151]. The tumor suppressive effect of MnSOD has been demonstrated in numerous cell types with malignant phenotype via modulating redox-related transcriptional factors (reviewed in [152]). However, after cancer has progressed, the expression of MnSOD can be higher in aggressive cancer compared to benign counterparts. These findings have been reported for advanced cancer tissues and blood samples of leukemia. Significantly higher MnSOD levels in malignant ovarian cancer tissue compared to normal ovarian epithelium and benign lesions have also been identified in a large number of samples using comparative tissue microarray analysis [153, 154]. ROS partially renders cancer cells more dependent on the function of superoxide dismutase to protect itself from damage caused by increased amounts of superoxide radicals. The necessity to alleviate ROS stress, coupled to the loss of MnSOD suppressors as the cancer progresses, provides a mechanistic explanation for a high MnSOD level in some cancer types at advanced stages. Due to the pivotal role of mitochondria and MnSOD in regulating cell death and cancer development, alteration of MnSOD activity and levels could be a potential target for therapeutic intervention.
A number of studies have identified the association of sod2 genetic polymorphisms with various diseases including type II diabetes and hypertension [159-162]. The presence of MnSOD single nucleotide polymorphisms (SNPs) and the potential effects of these SNPs on human MnSOD have been briefly reviewed [163] and are summarized in Table 2. Mutations detected in the sod2 promoter region reveal the possibility for decreased expression of MnSOD in several human cancer cells [155]. Among these mutations, C to T transition at -102 and an A insertion at -93 interrupting the single-strand loop structure have been suggested as reasons for reduced levels of MnSOD activity in some tumor cell lines. These mutations create an extra DNA binding site for the transcription factor AP-2 and alter sequence-specific interaction between DNA-protein and protein-protein in the transcription initiation complex [75, 155]. C to G transversion at -38 modulates an AP-2 dependent dysregulation of sod2 gene expression [85].
Table 2.
Identified valiants of the human sod2 gene
| Mutation/SNP | Position | Physiological function | Reference |
|---|---|---|---|
| non-coding region | |||
| C to T | -102 (promoter) | down-regulate sod2 expression by interrupting the single-loop structure | [75,155] |
| insertion A | -93 (promoter) | down-regulate sod2 expression by interrupting the single-loop structure | [75,155] |
| C to G | -38 (promoter) | AP-2 dependent dysregulation of sod2 expression | [85,155] |
| coding region | |||
| GCT (Ala) to GTT (Val) | 16 *(presequence) | MnSOD mitochondrial targeting sequence | [156] |
| ATA (Ile) to ACA (Thr) | 58 (exon 3) | reduce MnSOD activity by destabilizing the tetrarneric interface | [157] |
| CTT (Leu) to TTT (Phe) | 60 (exon 3) | deficiency in MnSOD activity | [158] |
Based on codon position
The newly synthesized polypeptide for MnSOD requires that it be transported across two mitochondrial membranes into the mitochondrial matrix where the enzyme is converted to an active form. This transport activity is mediated by the presence of a signal sequence within the N-terminal of the polypeptide. An extensively investigated cytosine to thymine (C to T) single nucleotide polymorphism in the sod2 mitochondrial targeting sequence, which causes the substitution of alanine (GCT) with valine (GTT) at codon 16, can disrupt the secondary α-helix structure of MnSOD and affect the localization and efficiency of mitochondrial transport of MnSOD enzyme [156]. Sutton et al. have reported that the MnSOD Ala variant generates 30-40% more active MnSOD enzyme and allows more efficient MnSOD import into the mitochondrial matrix than its Val counterpart, suggesting that the homozygous AA genotype may have higher MnSOD activity than its VV counterpart [164]. This target sequence polymorphism of human MnSOD gene and its association with cancer risk have been recently reviewed [165]. With respect to the AA genotype having a beneficial role in higher MnSOD activity, indeed, the sod2 homozygous variant genotype (Val/Val) has been associated with a greater risk of pancreatic cancer compared with the Ala allele. The occurrence of the VV variant also enhances the risk of non-small cell lung carcinoma in the presence of p53 and XRCC1 polymorphism [166, 167]. However, there is significant variability in results regarding the association of Ala16Val polymorphism with increased risk of disease. Little overall association has been found between MnSOD polymorphism and other diseases, such as asthma, Parkinson's disease, lung cancer and prostate cancer [162, 168-171]. Some studies demonstrate that the AA genotype elicits increased cancer risk. The Finnish study of male heavy smokers found that the MnSOD AA polymorphism was significantly associated with higher risk of prostate cancer, especially for tumors with higher Gleason score [172]. S. H. Olson and colleagues also found that carriers of the AA genotype had greater risk for ovarian carcinoma. Similar results have been described in breast cancer studies [173]. These reports indicate that this polymorphism may be functionally neutral. However, these studies do not delineate the levels of corresponding factors, such as the loss of p53 or AP-2. The variability of sample characteristics and disease stages may contribute to the discrepancies among studies even on the same type of disease [170, 172]. Thus, extensive, population-based, age-matched, case-control studies will be required to thoroughly investigate Ala16Val polymorphism so as to provide more reliable information for prevention, treatment and surveillance.
Another potentially important polymorphic substitution for isoleucine or threonine is found at amino acid 58 in the mature human MnSOD protein. The Thr variant is less stable and more susceptible to inactivation by S-thiolation reaction than the Ile counterpart due to its stability at the tetrameric interface [157]. Consistent with the tumor suppressive role of MnSOD, human malignant breast cancer cells overexpressing Ile58 MnSOD have 3-fold higher MnSOD activity and a greater tumor-suppressive effect than cells overexpressing the Thr58 counterpart at an equal MnSOD protein level [174]. Further direct testing of this possibility would be to replace cellular MnSOD with either version and then to study the effect on tumorigenesis, e.g., in a transgenic knockin model. Another relatively unique point mutation, C5782T in exon3 of sod2 producing a L60F mutation in the mature enzyme, has been found in Jurkat T lymphocytes. The L60F mutation led to a deficiency in MnSOD activity and correlated with the malignant phenotype. This finding also explains the paradoxical effects of thiol reagents on antioxidants in some leukemia cells [158]. The different roles that MnSOD plays at different cancer stages suggest that specific strategies will be needed for specific situations. Thus, understanding how the expression of MnSOD can be selectively regulated at each stage of cancer development is likely to have important implications for the prevention and treatment of cancer.
ECSOD (sod3)
This copper- and zinc-containing dismutase, extracellular superoxide dismutase (ECSOD), was first discovered in extracellular fluids including human plasma, lymph, and synovial fluid by Marklund, et al. in 1982 [175-177]. ECSOD is the least characterized SOD of the three SOD isoforms. It is a secreted hydrophobic glycoprotein usually existing as a homotetramer with an approximate molecular weight of 135,000 Da [175]. After removal of the signal peptide, mature ECSOD protein is composed of three domains: the amino-terminal domain containing glycosylation sites, an active site domain that shows strong homology with Cu/ZnSOD and a short carboxyl-terminal domain [38]. One unique characteristic of ECSOD that was identified during initial purification is its strong affinity for heparin and other heparin sulfates, the interaction of which was further localized to the C-terminal, the positively charged, heparin-binding domain of ECSOD [178, 179]. The polycationic matrix-binding domain makes ECSOD directly bind to hyaluronan and inhibit oxidant-induced degradation of this glycosaminoglycan so as to prevent inflammation in response to lung injury [180]. A common human sod3 gene variant (substitution of argine 213 with glycine R213G), which is located in the center of the carboxyl-terminal cluster of positively charged amino acid residues of the heparin-binding domain, has been described [181-183]. Due to impaired heparin and collagen binding affinities, this variant is considered to be related to increased plasma ECSOD concentration but not to enzymatic activity [181]. The significance of this polymorphism varies depending on the disease and population [184, 185]. The association between the R213G ECSOD polymorphism and clinical significance has been summarized in several reviews of ECSOD [20, 42, 183]. Additional variants of the human sod3 gene include 4 other missense mutations, Ala40Thr, Phe131Cys, Val160Leu and Arg202Leu. A silent mutation, Leu53Leu (CTG to TTG), has been found in samples of Japanese and Mediterranean populations [183-185]. However, the functional significance of most of these new variants on metabolic activity and disease incidence requires further investigation.
Interestingly, the human ECSOD polypeptide folds in two distinct ways with different disulfide bridge patterns resulting in enzymatically active (aECSOD) and inactive (iECSOD) subunits [186-188]. Thus, differential modification of different disulfide bonds, intracellular folding and proteolytic processing of protein also contribute importantly to the levels of enzymatic activity.
The expression of ECSOD and its catalytic activity have been associated with a variety of diseases including cardiovascular, neurological disorders and pulmonary diseases. Several regulatory mechanisms have been proposed [42]. It has been shown that ECSOD is a main regulator of nitric oxide (NO) bioavailability and bioactivity by modulating generation of the toxic product peroxynitrite in the vasculature [189]. ECSOD binding to protein filbulin-5 maintains vascular O2·- levels [187, 190]. Our focus is on the molecular mechanism known to regulate sod3 gene expression and the related biomedical implication. Thus, because ECSOD activity and function and methods or applications of sod3 gene transfer have all been described elegantly in other reviews, they will not be presented here [20, 42, 177, 183]. Despite the availability of several disease models related to ECSOD, the precise mechanism for the control of ECSOD expression is unknown. Understanding the molecular mechanisms involved in the regulation of sod3 gene expression and the factors involved in tissue and cell-specific expression of the sod3 gene will be important for therapeutic attempts to design novel strategies for controlling ECSOD- related tissue injury.
Conclusions and Future Directions
The essential role of SODs in the survival of aerobic organisms and prevention of pathological conditions demonstrates the significance of understanding how the expression of these genes can be regulated. At the transcription level, an individual sod gene has its own unique regulatory mechanisms and also uses common transcriptional factors. These features provide the opportunity to either selectively alter the expression of a sod gene or concurrently modulate all of them as a family. In addition to transcriptional control, epigenetic regulation and post-transcriptional modifications can also play important roles in controlling the level of functional SODs. These processes include stabilizing SOD mRNAs and regulating their translation. Considering the current explosion of microRNA studies and the roles of microRNA in diseases, future studies should also emphasize regulation of SOD translation by microRNA. Due to the importance of metal cofactors to the activity of superoxide dismutase, the processes and effects of metal ion insertion on SOD function deserve further in-depth investigations. Because the compartmental localization of each SOD is different in sub-cellular compartments, approaches to target their site-specific expression will be very important and could be used to aid the development of novel SOD-dependent therapeutic strategies.
Acknowledgments
Work for this review was supported by NIH grants CA 49797 and CA 73599.
Abbreviations
- ROS
Reactive oxygen species
- O2·-
Superoxide
- ·OH
Hydroxyl radical
- H2O2
Hydrogen peroxide
- RNS
Reactive nitrogen species
- SOD
Superoxide dismutase
- Cu/ZnSOD
Copper-zinc superoxide dismutase
- MnSOD
Manganese superoxide dismutase
- ECSOD
Extracellular superoxide dismutase
- ALS
Amyotrophic Lateral Sclerosis
- NF-κB
Nuclear Factor-KappaB
- Sp1
Specificity Protein 1
- AP-1
Activator Protein 1
- AP-2
Activating Protein 2
- C/EBP
CCAAT Enhancer-Binding Proteins
- PI3K
Phosphoinositide 3-kinases
- TPA
12-O-tetradecanoylphorbol-13-acetate
- IFN-γ
Interferon-gamma
- TNF α
Tumor necrosis factor α
- IL-1
Interleukin-1beta
- IL-4
Interleukin-4
- IL-6
Interleukin-6
- iNOS
Inducible nitric oxide synthase
- Egr-1
Early growth response factor 1
- nNOS
Neuronal nitric oxide synthase
- PIN*POINT
ProteIN POsition Identification with Nuclease Tail
- NPM
Nucleophosmin
- TSA
Trichostatin A
- MAPK
Mitogen-activated protein kinases
- JNK
c-Jun N-terminal kinases
- PPRE
Peroxisome proliferator-responsive element
- Elk-1
E-26 like protein 1
- YY1
Yin Yang 1
- ARE
Antioxidant responsive element
- XRE
Xenobiotic responsive element
- FOXO3
Forkhead box O3
- PPAR γ
Peroxisome proliferator-activated receptor γ
- Atox1
Antioxidant-1
- Ang II
Angiotensin II
- HDAC1
Histone deacetylase-1
- MnSOD-BP
MnSOD binding protein
- fALS
familial Amyotrophic Lateral Sclerosis
- ZnT
Zinc transporter
- SNP
Single nucleotide polymorphism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA damage and disease: induction, repair and significance. Mutat Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Chen AF, Davies CM, De Lin M, Fermor B. Oxidative DNA damage in osteoarthritic porcine articular cartilage. J Cell Physiol. 2008;217:828–833. doi: 10.1002/jcp.21562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie L, Zhu X, Hu Y, Li T, Gao Y, Shi Y, Tang S. Mitochondrial DNA oxidative damage triggering mitochondrial dysfunction and apoptosis in high glucose-induced HRECs. Invest Ophthalmol Vis Sci. 2008;49:4203–4209. doi: 10.1167/iovs.07-1364. [DOI] [PubMed] [Google Scholar]
- 4.Sun Y, Oberley LW. Redox regulation of transcriptional activators. Free Radic Biol Med. 1996;21:335–348. doi: 10.1016/0891-5849(96)00109-8. [DOI] [PubMed] [Google Scholar]
- 5.Cai J, Yang J, Jones DP. Mitochondrial control of apoptosis: the role of cytochrome c. Biochim Biophys Acta. 1998;1366:139–149. doi: 10.1016/s0005-2728(98)00109-1. [DOI] [PubMed] [Google Scholar]
- 6.Giannattasio S, Atlante A, Antonacci L, Guaragnella N, Lattanzio P, Passarella S, Marra E. Cytochrome c is released from coupled mitochondria of yeast en route to acetic acid-induced programmed cell death and can work as an electron donor and a ROS scavenger. FEBS Lett. 2008;582:1519–1525. doi: 10.1016/j.febslet.2008.03.048. [DOI] [PubMed] [Google Scholar]
- 7.Archer SL, Gomberg-Maitland M, Maitland ML, Rich S, Garcia JG, Weir EK. Mitochondrial metabolism, redox signaling, and fusion: a mitochondria-ROS-HIF-1alpha-Kv1.5 O2-sensing pathway at the intersection of pulmonary hypertension and cancer. Am J Physiol Heart Circ Physiol. 2008;294:H570–578. doi: 10.1152/ajpheart.01324.2007. [DOI] [PubMed] [Google Scholar]
- 8.Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007;17:422–427. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 9.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 10.Parge HE, Hallewell RA, Tainer JA. Atomic structures of wild-type and thermostable mutant recombinant human Cu,Zn superoxide dismutase. Proc Natl Acad Sci U S A. 1992;89:6109–6113. doi: 10.1073/pnas.89.13.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 12.Harvey LJ, McArdle HJ. Biomarkers of copper status: a brief update. Br J Nutr. 2008;99 3:S10–13. doi: 10.1017/S0007114508006806. [DOI] [PubMed] [Google Scholar]
- 13.Carroll MC, Girouard JB, Ulloa JL, Subramaniam JR, Wong PC, Valentine JS, Culotta VC. Mechanisms for activating Cu- and Zn-containing superoxide dismutase in the absence of the CCS Cu chaperone. Proc Natl Acad Sci U S A. 2004;101:5964–5969. doi: 10.1073/pnas.0308298101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kayatekin C, Zitzewitz JA, Matthews CR. Zinc binding modulates the entire folding free energy surface of human Cu,Zn superoxide dismutase. J Mol Biol. 2008;384:540–555. doi: 10.1016/j.jmb.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luk EE, Culotta VC. Manganese superoxide dismutase in Saccharomyces cerevisiae acquires its metal co-factor through a pathway involving the Nramp metal transporter, Smf2p. J Biol Chem. 2001;276:47556–47562. doi: 10.1074/jbc.M108923200. [DOI] [PubMed] [Google Scholar]
- 16.Luk E, Carroll M, Baker M, Culotta VC. Manganese activation of superoxide dismutase 2 in Saccharomyces cerevisiae requires MTM1, a member of the mitochondrial carrier family. Proc Natl Acad Sci U S A. 2003;100:10353–10357. doi: 10.1073/pnas.1632471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luk E, Yang M, Jensen LT, Bourbonnais Y, Culotta VC. Manganese activation of superoxide dismutase 2 in the mitochondria of Saccharomyces cerevisiae. J Biol Chem. 2005;280:22715–22720. doi: 10.1074/jbc.M504257200. [DOI] [PubMed] [Google Scholar]
- 18.Itoh S, Ozumi K, Kim HW, Nakagawa O, McKinney RD, Folz RJ, Zelko IN, Ushio-Fukai M, Fukai T. Novel mechanism for regulation of extracellular SOD transcription and activity by copper: Role of antioxidant-1. Free Radic Biol Med. 2009;46:95–104. doi: 10.1016/j.freeradbiomed.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin Z, Gongora MC, Ozumi K, Itoh S, Akram K, Ushio-Fukai M, Harrison DG, Fukai T. Role of Menkes ATPase in Angiotensin II-Induced Hypertension. A Key Modulator for Extracellular Superoxide Dismutase Function. Hypertension. 2008 doi: 10.1161/HYPERTENSIONAHA.108.116467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33:337–349. doi: 10.1016/s0891-5849(02)00905-x. [DOI] [PubMed] [Google Scholar]
- 21.Forman HJ. Use and abuse of exogenous H2O2 in studies of signal transduction. Free Radic Biol Med. 2007;42:926–932. doi: 10.1016/j.freeradbiomed.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu B, Chen Y, St Clair DK. ROS and p53: a versatile partnership. Free Radic Biol Med. 2008;44:1529–1535. doi: 10.1016/j.freeradbiomed.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalmar B, Novoselov S, Gray A, Cheetham ME, Margulis B, Greensmith L. Late stage treatment with arimoclomol delays disease progression and prevents protein aggregation in the SOD1(G93A) mouse model of ALS. J Neurochem. 2008 doi: 10.1111/j.1471-4159.2008.05595.x. [DOI] [PubMed] [Google Scholar]
- 24.Joseph A, L Y, Koo HC, Davis JM, Pollack S, Kazzaz JA. Superoxide dismutase attenuates hyperoxia-induced interleukin-8 induction via AP-1. Free radical biology & medicine. 2008 doi: 10.1016/j.freeradbiomed.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Pawlak K, Domaniewski T, Mysliwiec M, Pawlak D. The kynurenines are associated with oxidative stress, inflammation and the prevalence of cardiovascular disease in patients with end-stage renal disease. Atherosclerosis. 2008 doi: 10.1016/j.atherosclerosis.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 26.Kaewpila S, Venkataraman S, Buettner GR, Oberley LW. Manganese superoxide dismutase modulates hypoxia-inducible factor-1 alpha induction via superoxide. Cancer Res. 2008;68:2781–2788. doi: 10.1158/0008-5472.CAN-07-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oberley LW. Anticancer therapy by overexpression of superoxide dismutase. Antioxid Redox Signal. 2001;3:461–472. doi: 10.1089/15230860152409095. [DOI] [PubMed] [Google Scholar]
- 28.Hsu JL, Visner GA, Burr IA, Nick HS. Rat copper/zinc superoxide dismutase gene: isolation, characterization, and species comparison. Biochem Biophys Res Commun. 1992;186:936–943. doi: 10.1016/0006-291x(92)90836-a. [DOI] [PubMed] [Google Scholar]
- 29.Schmutz SM, Cornwell D, Moker JS, Troyer DL. Physical mapping of SOD1 to bovine chromosome 1. Cytogenet Cell Genet. 1996;72:37–39. doi: 10.1159/000134156. [DOI] [PubMed] [Google Scholar]
- 30.Levanon D, Lieman-Hurwitz J, Dafni N, Wigderson M, Sherman L, Bernstein Y, Laver-Rudich Z, Danciger E, Stein O, Groner Y. Architecture and anatomy of the chromosomal locus in human chromosome 21 encoding the Cu/Zn superoxide dismutase. EMBO J. 1985;4:77–84. doi: 10.1002/j.1460-2075.1985.tb02320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones PL, Kucera G, Gordon H, Boss JM. Cloning and characterization of the murine manganous superoxide dismutase-encoding gene. Gene. 1995;153:155–161. doi: 10.1016/0378-1119(94)00666-g. [DOI] [PubMed] [Google Scholar]
- 32.DiSilvestre D, Kleeberger SR, Johns J, Levitt RC. Structure and DNA sequence of the mouse MnSOD gene. Mamm Genome. 1995;6:281–284. doi: 10.1007/BF00352417. [DOI] [PubMed] [Google Scholar]
- 33.Ho YS, Howard AJ, Crapo JD. Molecular structure of a functional rat gene for manganese-containing superoxide dismutase. Am J Respir Cell Mol Biol. 1991;4:278–286. doi: 10.1165/ajrcmb/4.3.278. [DOI] [PubMed] [Google Scholar]
- 34.Meyrick B, Magnuson MA. Identification and functional characterization of the bovine manganous superoxide dismutase promoter. Am J Respir Cell Mol Biol. 1994;10:113–121. doi: 10.1165/ajrcmb.10.1.8292376. [DOI] [PubMed] [Google Scholar]
- 35.Marlhens F, Nicole A, Sinet PM. Lowered level of translatable messenger RNAs for manganese superoxide dismutase in human fibroblasts transformed by SV 40. Biochem Biophys Res Commun. 1985;129:300–305. doi: 10.1016/0006-291x(85)91437-8. [DOI] [PubMed] [Google Scholar]
- 36.Wan XS, Devalaraja MN, St Clair DK. Molecular structure and organization of the human manganese superoxide dismutase gene. DNA Cell Biol. 1994;13:1127–1136. doi: 10.1089/dna.1994.13.1127. [DOI] [PubMed] [Google Scholar]
- 37.Hjalmarsson K, Marklund SL, Engstrom A, Edlund T. Isolation and sequence of complementary DNA encoding human extracellular superoxide dismutase. Proc Natl Acad Sci U S A. 1987;84:6340–6344. doi: 10.1073/pnas.84.18.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Folz RJ, Crapo JD. Extracellular superoxide dismutase (SOD3): tissue-specific expression, genomic characterization, and computer-assisted sequence analysis of the human EC SOD gene. Genomics. 1994;22:162–171. doi: 10.1006/geno.1994.1357. [DOI] [PubMed] [Google Scholar]
- 39.Perry AC, Jones R, Hall L. Isolation and characterization of a rat cDNA clone encoding a secreted superoxide dismutase reveals the epididymis to be a major site of its expression. Biochem J. 1993;293(Pt 1):21–25. doi: 10.1042/bj2930021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Folz RJ, Guan J, Seldin MF, Oury TD, Enghild JJ, Crapo JD. Mouse extracellular superoxide dismutase: primary structure, tissue-specific gene expression, chromosomal localization, and lung in situ hybridization. Am J Respir Cell Mol Biol. 1997;17:393–403. doi: 10.1165/ajrcmb.17.4.2826. [DOI] [PubMed] [Google Scholar]
- 41.Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NF-kappaB signaling module. Oncogene. 2006;25:6706–6716. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- 42.Fattman CL, Schaefer LM, Oury TD. Extracellular superoxide dismutase in biology and medicine. Free Radic Biol Med. 2003;35:236–256. doi: 10.1016/s0891-5849(03)00275-2. [DOI] [PubMed] [Google Scholar]
- 43.Xu Y, Kiningham KK, Devalaraja MN, Yeh CC, Majima H, Kasarskis EJ, St Clair DK. An intronic NF-kappaB element is essential for induction of the human manganese superoxide dismutase gene by tumor necrosis factor-alpha and interleukin-1beta. DNA Cell Biol. 1999;18:709–722. doi: 10.1089/104454999314999. [DOI] [PubMed] [Google Scholar]
- 44.Minc E, de Coppet P, Masson P, Thiery L, Dutertre S, Amor-Gueret M, Jaulin C. The human copper-zinc superoxide dismutase gene (SOD1) proximal promoter is regulated by Sp1, Egr-1, and WT1 via non-canonical binding sites. J Biol Chem. 1999;274:503–509. doi: 10.1074/jbc.274.1.503. [DOI] [PubMed] [Google Scholar]
- 45.Rojo AI, Salinas M, Martin D, Perona R, Cuadrado A. Regulation of Cu/Zn-superoxide dismutase expression via the phosphatidylinositol 3 kinase/Akt pathway and nuclear factor-kappaB. J Neurosci. 2004;24:7324–7334. doi: 10.1523/JNEUROSCI.2111-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mumbengegwi DR, Li Q, Li C, Bear CE, Engelhardt JF. Evidence for a superoxide permeability pathway in endosomal membranes. Mol Cell Biol. 2008;28:3700–3712. doi: 10.1128/MCB.02038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dimayuga FO, Wang C, Clark JM, Dimayuga ER, Dimayuga VM, Bruce-Keller AJ. SOD1 overexpression alters ROS production and reduces neurotoxic inflammatory signaling in microglial cells. J Neuroimmunol. 2007;182:89–99. doi: 10.1016/j.jneuroim.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beni SM, Tsenter J, Alexandrovich AG, Galron-Krool N, Barzilai A, Kohen R, Grigoriadis N, Simeonidou C, Shohami E. CuZn-SOD deficiency, rather than overexpression, is associated with enhanced recovery and attenuated activation of NF-kappaB after brain trauma in mice. J Cereb Blood Flow Metab. 2006;26:478–490. doi: 10.1038/sj.jcbfm.9600209. [DOI] [PubMed] [Google Scholar]
- 49.Eastgate J, Moreb J, Nick HS, Suzuki K, Taniguchi N, Zucali JR. A role for manganese superoxide dismutase in radioprotection of hematopoietic stem cells by interleukin-1. Blood. 1993;81:639–646. [PubMed] [Google Scholar]
- 50.Akashi M, Hachiya M, Paquette RL, Osawa Y, Shimizu S, Suzuki G. Irradiation increases manganese superoxide dismutase mRNA levels in human fibroblasts. Possible mechanisms for its accumulation. J Biol Chem. 1995;270:15864–15869. doi: 10.1074/jbc.270.26.15864. [DOI] [PubMed] [Google Scholar]
- 51.Fujii J, Taniguchi N. Phorbol ester induces manganese-superoxide dismutase in tumor necrosis factor-resistant cells. J Biol Chem. 1991;266:23142–23146. [PubMed] [Google Scholar]
- 52.Harris CA, Derbin KS, Hunte-McDonough B, Krauss MR, Chen KT, Smith DM, Epstein LB. Manganese superoxide dismutase is induced by IFN-gamma in multiple cell types. Synergistic induction by IFN-gamma and tumor necrosis factor or IL-1. Journal of Immunology. 1991;147:149–154. [PubMed] [Google Scholar]
- 53.Wong GH, Elwell JH, Oberley LW, Goeddel DV. Manganous superoxide dismutase is essential for cellular resistance to cytotoxicity of tumor necrosis factor. Cell. 1988;58:923–931. doi: 10.1016/0092-8674(89)90944-6. [DOI] [PubMed] [Google Scholar]
- 54.Wong GH, Goeddel DV. Induction of manganous superoxide dismutase by tumor necrosis factor: possible protective mechanism. Science. 1988;242:941–944. doi: 10.1126/science.3263703. [DOI] [PubMed] [Google Scholar]
- 55.Visner GA, Dougall WC, Wilson JM, Burr IA, Nick HS. Regulation of manganese superoxide dismutase by lipopolysaccharide, interleukin-1, and tumor necrosis factor. Role in the acute inflammatory response. J Biol Chem. 1990;265:2856–2864. [PubMed] [Google Scholar]
- 56.Dougall WC, Nick HS. Manganese superoxide dismutase: a hepatic acute phase protein regulated by interleukin-6 and glucocorticoids. Endocrinology. 1991;129:2376–2384. doi: 10.1210/endo-129-5-2376. [DOI] [PubMed] [Google Scholar]
- 57.St Clair DK, Porntadavity S, Xu Y, Kiningham K. Transcription regulation of human manganese superoxide dismutase gene. Methods Enzymol. 2002;349:306–312. doi: 10.1016/s0076-6879(02)49345-7. [DOI] [PubMed] [Google Scholar]
- 58.Kiningham KK, Xu Y, Daosukho C, Popova B, St Clair DK. Nuclear factor kappaB-dependent mechanisms coordinate the synergistic effect of PMA and cytokines on the induction of superoxide dismutase 2. Biochem J. 2001;353:147–156. [PMC free article] [PubMed] [Google Scholar]
- 59.Guo Z, Boekhoudt GH, Boss JM. Role of the intronic enhancer in tumor necrosis factor-mediated induction of manganous superoxide dismutase. J Biol Chem. 2003;278:23570–23578. doi: 10.1074/jbc.M303431200. [DOI] [PubMed] [Google Scholar]
- 60.Rogers RJ, Chesrown SE, Kuo S, Monnier JM, Nick HS. Cytokine-inducible enhancer with promoter activity in both the rat and human manganese-superoxide dismutase genes. Biochem J. 2000;347(Pt 1):233–242. [PMC free article] [PubMed] [Google Scholar]
- 61.Dhar SK, Xu Y, Noel T, St Clair DK. Chronic exposure to 12-O-tetradecanoylphorbol-13-acetate represses sod2 induction in vivo: the negative role of p50. Carcinogenesis. 2007;28:2605–2613. doi: 10.1093/carcin/bgm163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marklund SL. Regulation by cytokines of extracellular superoxide dismutase and other superoxide dismutase isoenzymes in fibroblasts. J Biol Chem. 1992;267:6696–6701. [PubMed] [Google Scholar]
- 63.Brady TC, Chang LY, Day BJ, Crapo JD. Extracellular superoxide dismutase is upregulated with inducible nitric oxide synthase after NF-kappa B activation. Am J Physiol. 1997;273:L1002–1006. doi: 10.1152/ajplung.1997.273.5.L1002. [DOI] [PubMed] [Google Scholar]
- 64.Dynan WS, Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- 65.Briggs MR, Kadonaga JT, Bell SP, Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986;234:47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- 66.Dynan WS, Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983;32:669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- 67.Xu Y, Porntadavity S, St Clair DK. Transcriptional regulation of the human manganese superoxide dismutase gene: the role of specificity protein 1 (Sp1) and activating protein-2 (AP-2) Biochem J. 2002;362:401–412. doi: 10.1042/0264-6021:3620401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yeh CC, Wan XS, St Clair DK. Transcriptional regulation of the 5′ proximal promoter of the human manganese superoxide dismutase gene. DNA Cell Biol. 1998;17:921–930. doi: 10.1089/dna.1998.17.921. [DOI] [PubMed] [Google Scholar]
- 69.Seo SJ, Kim HT, Cho G, Rho HM, Jung G. Sp1 and C/EBP-related factor regulate the transcription of human Cu/Zn SOD gene. Gene. 1996;178:177–185. doi: 10.1016/0378-1119(96)00383-6. [DOI] [PubMed] [Google Scholar]
- 70.Afonso V, Santos G, Collin P, Khatib AM, Mitrovic DR, Lomri N, Leitman DC, Lomri A. Tumor necrosis factor-alpha down-regulates human Cu/Zn superoxide dismutase 1 promoter via JNK/AP-1 signaling pathway. Free Radic Biol Med. 2006;41:709–721. doi: 10.1016/j.freeradbiomed.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 71.Baldelli S, Aquilano K, Rotilio G, Ciriolo MR. Glutathione and copper, zinc superoxide dismutase are modulated by overexpression of neuronal nitric oxide synthase. Int J Biochem Cell Biol. 2008;40:2660–2670. doi: 10.1016/j.biocel.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 72.Tanaka T, Kurabayashi M, Aihara Y, Ohyama Y, Nagai R. Inducible expression of manganese superoxide dismutase by phorbol 12-myristate 13-acetate is mediated by Sp1 in endothelial cells. Arterioscler Thromb Vasc Biol. 2000;20:392–401. doi: 10.1161/01.atv.20.2.392. [DOI] [PubMed] [Google Scholar]
- 73.Kuo S, Chokas AL, Rogers RJ, Nick HS. PIN*POINT analysis on the endogenous MnSOD promoter: specific demonstration of Sp1 binding in vivo. Am J Physiol Cell Physiol. 2003;284:C528–534. doi: 10.1152/ajpcell.00356.2002. [DOI] [PubMed] [Google Scholar]
- 74.Mastrangelo IA, Courey AJ, Wall JS, Jackson SP, Hough PV. DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc Natl Acad Sci U S A. 1991;88:5670–5674. doi: 10.1073/pnas.88.13.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu Y, Fang F, Dhar SK, St Clair WH, Kasarskis EJ, St Clair DK. The role of a single-stranded nucleotide loop in transcriptional regulation of the human sod2 gene. J Biol Chem. 2007;282:15981–15994. doi: 10.1074/jbc.M608979200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dhar SK, Xu Y, Chen Y, St Clair DK. Specificity protein 1-dependent p53-mediated suppression of human manganese superoxide dismutase gene expression. J Biol Chem. 2006;281:21698–21709. doi: 10.1074/jbc.M601083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zelko IN, Folz RJ. Sp1 and Sp3 transcription factors mediate trichostatin A-induced and basal expression of extracellular superoxide dismutase. Free Radic Biol Med. 2004;37:1256–1271. doi: 10.1016/j.freeradbiomed.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 78.Zelko IN, Mueller MR, Folz RJ. Transcription factors sp1 and sp3 regulate expression of human extracellular superoxide dismutase in lung fibroblasts. Am J Respir Cell Mol Biol. 2008;39:243–251. doi: 10.1165/rcmb.2007-0378OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 80.Huang CY, Fujimura M, Chang YY, Chan PH. Overexpression of copper-zinc superoxide dismutase attenuates acute activation of activator protein-1 after transient focal cerebral ischemia in mice. Stroke. 2001;32:741–747. doi: 10.1161/01.str.32.3.741. [DOI] [PubMed] [Google Scholar]
- 81.Zhou W, Zhang Y, Hosch MS, Lang A, Zwacka RM, Engelhardt JF. Subcellular site of superoxide dismutase expression differentially controls AP-1 activity and injury in mouse liver following ischemia/reperfusion. Hepatology. 2001;33:902–914. doi: 10.1053/jhep.2001.23073. [DOI] [PubMed] [Google Scholar]
- 82.Zhao Y, Oberley TD, Chaiswing L, Lin SM, Epstein CJ, Huang TT, St Clair D. Manganese superoxide dismutase deficiency enhances cell turnover via tumor promoter-induced alterations in AP-1 and p53-mediated pathways in a skin cancer model. Oncogene. 2002;21:3836–3846. doi: 10.1038/sj.onc.1205477. [DOI] [PubMed] [Google Scholar]
- 83.Qadri I, Iwahashi M, Capasso JM, Hopken MW, Flores S, Schaack J, Simon FR. Induced oxidative stress and activated expression of manganese superoxide dismutase during hepatitis C virus replication: role of JNK, p38 MAPK and AP-1. Biochem J. 2004;378:919–928. doi: 10.1042/BJ20031587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eckert D, Buhl S, Weber S, Jager R, Schorle H. The AP-2 family of transcription factors. Genome Biol. 2005;6:246. doi: 10.1186/gb-2005-6-13-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu Y, Fang F, Dhar SK, Bosch A, St Clair WH, Kasarskis EJ, St Clair DK. Mutations in the SOD2 Promoter Reveal a Molecular Basis for an Activating Protein 2-Dependent Dysregulation of Manganese Superoxide Dismutase Expression in Cancer Cells. Mol Cancer Res. 2008;6:1881–1893. doi: 10.1158/1541-7786.MCR-08-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim YH, Park KH, Rho HM. Transcriptional activation of the Cu,Zn-superoxide dismutase gene through the AP2 site by ginsenoside Rb2 extracted from a medicinal plant, Panax ginseng. J Biol Chem. 1996;271:24539–24543. doi: 10.1074/jbc.271.40.24539. [DOI] [PubMed] [Google Scholar]
- 87.Zhu C, Huang Y, Weydert CJ, Oberley LW, Domann FE. Constitutive activation of transcription factor AP-2 is associated with decreased MnSOD expression in transformed human lung fibroblasts. Antioxid Redox Signal. 2001;3:387–395. doi: 10.1089/15230860152409031. [DOI] [PubMed] [Google Scholar]
- 88.Zhu CH, Huang Y, Oberley LW, Domann FE. A family of AP-2 proteins down-regulate manganese superoxide dismutase expression. J Biol Chem. 2001;276:14407–14413. doi: 10.1074/jbc.M009708200. [DOI] [PubMed] [Google Scholar]
- 89.Huang Y, Peng J, Oberley LW, Domann FE. Transcriptional inhibition of manganese superoxide dismutase (SOD2) gene expression by DNA methylation of the 5′ CpG island. Free Radic Biol Med. 1997;23:314–320. doi: 10.1016/s0891-5849(97)00095-6. [DOI] [PubMed] [Google Scholar]
- 90.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Seo SJ, Kang SS, Cho G, Rho HM, Jung G. C/EBP alpha and C/EBPbeta play similar roles in the transcription of the human Cu/Zn SOD gene. Gene. 1997;203:11–15. doi: 10.1016/s0378-1119(97)00484-8. [DOI] [PubMed] [Google Scholar]
- 92.Kim YH, Yoo HY, Chang MS, Jung G, Rho HM. C/EBP alpha is a major activator for the transcription of rat Cu/Zn superoxide dismutase gene in liver cell. FEBS Lett. 1997;401:267–270. doi: 10.1016/s0014-5793(96)01487-1. [DOI] [PubMed] [Google Scholar]
- 93.Qiu X, Aiken KJ, Chokas AL, Beachy DE, Nick HS. Distinct functions of CCAAT enhancer-binding protein isoforms in the regulation of manganese superoxide dismutase during interleukin-1beta stimulation. J Biol Chem. 2008;283:25774–25785. doi: 10.1074/jbc.M801178200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Adachi T, Inoue M, Hara H, Suzuki S. Effects of PPARgamma ligands and C/EBPbeta enhancer on expression of extracellular-superoxide dismutase. Redox Rep. 2004;9:207–212. doi: 10.1179/135100004225005985. [DOI] [PubMed] [Google Scholar]
- 95.Yoo HY, Chang MS, Rho HM. Induction of the rat Cu/Zn superoxide dismutase gene through the peroxisome proliferator-responsive element by arachidonic acid. Gene. 1999;234:87–91. doi: 10.1016/s0378-1119(99)00176-6. [DOI] [PubMed] [Google Scholar]
- 96.Chang MS, Yoo HY, Rho HM. Positive and negative regulatory elements in the upstream region of the rat Cu/Zn-superoxide dismutase gene. Biochem J. 1999;339(Pt 2):335–341. [PMC free article] [PubMed] [Google Scholar]
- 97.Cho G, Kang S, Seo SJ, Kim Y, Jung G. The transcriptional repression of the human Cu/Zn superoxide dismutase(sod1) gene by the anticancer drug, mitomycin C(MMC) Biochem Mol Biol Int. 1997;42:949–956. doi: 10.1080/15216549700203391. [DOI] [PubMed] [Google Scholar]
- 98.Park EY, Rho HM. The transcriptional activation of the human copper/zinc superoxide dismutase gene by 2,3,7,8-tetrachlorodibenzo-p-dioxin through two different regulator sites, the antioxidant responsive element and xenobiotic responsive element. Mol Cell Biochem. 2002;240:47–55. doi: 10.1023/a:1020600509965. [DOI] [PubMed] [Google Scholar]
- 99.Kops GJ, D T, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH, Burgering BM. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 100.Li M, Chiu JF, Mossman BT, Fukagawa NK. Down-regulation of manganese-superoxide dismutase through phosphorylation of FOXO3a by Akt in explanted vascular smooth muscle cells from old rats. J Biol Chem. 2006;281:40429–40439. doi: 10.1074/jbc.M606596200. [DOI] [PubMed] [Google Scholar]
- 101.Ding G, Fu M, Qin Q, Lewis W, Kim HW, Fukai T, Bacanamwo M, Chen YE, Schneider MD, Mangelsdorf DJ, Evans RM, Yang Q. Cardiac peroxisome proliferator-activated receptor gamma is essential in protecting cardiomyocytes from oxidative damage. Cardiovasc Res. 2007;76:269–279. doi: 10.1016/j.cardiores.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 102.Jeney V, Itoh S, Wendt M, Gradek Q, Ushio-Fukai M, Harrison DG, Fukai T. Role of antioxidant-1 in extracellular superoxide dismutase function and expression. Circ Res. 2005;96:723–729. doi: 10.1161/01.RES.0000162001.57896.66. [DOI] [PubMed] [Google Scholar]
- 103.Qin Z, Itoh S, Jeney V, Ushio-Fukai M, Fukai T. Essential role for the Menkes ATPase in activation of extracellular superoxide dismutase: implication for vascular oxidative stress. FASEB J. 2006;20:334–336. doi: 10.1096/fj.05-4564fje. [DOI] [PubMed] [Google Scholar]
- 104.Qin Z, Gongora MC, Ozumi K, Itoh S, Akram K, Ushio-Fukai M, Harrison DG, Fukai T. Role of Menkes ATPase in angiotensin II-induced hypertension: a key modulator for extracellular superoxide dismutase function. Hypertension. 2008;52:945–951. doi: 10.1161/HYPERTENSIONAHA.108.116467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zelko IN, Folz RJ. Myeloid zinc finger (MZF)-like, Kruppel-like and Ets families of transcription factors determine the cell-specific expression of mouse extracellular superoxide dismutase. Biochem J. 2003;369:375–386. doi: 10.1042/BJ20021431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang Y, He T, Domann FE. Decreased expression of manganese superoxide dismutase in transformed cells is associated with increased cytosine methylation of the SOD2 gene. DNA Cell Biol. 1999;18:643–652. doi: 10.1089/104454999315051. [DOI] [PubMed] [Google Scholar]
- 107.Hitchler MJ, Wikainapakul K, Yu L, Powers K, Attatippaholkun W, Domann FE. Epigenetic regulation of manganese superoxide dismutase expression in human breast cancer cells. Epigenetics. 2006;1:163–171. doi: 10.4161/epi.1.4.3401. [DOI] [PubMed] [Google Scholar]
- 108.Hitchler MJ, Oberley LW, Domann FE. Epigenetic silencing of SOD2 by histone modifications in human breast cancer cells. Free Radic Biol Med. 2008;45:1573–1580. doi: 10.1016/j.freeradbiomed.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hurt EM, Thomas SB, Peng B, Farrar WL. Molecular consequences of SOD2 expression in epigenetically silenced pancreatic carcinoma cell lines. Br J Cancer. 2007;97:1116–1123. doi: 10.1038/sj.bjc.6604000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hodge DR, Peng B, Pompeia C, Thomas S, Cho E, Clausen PA, Marquez VE, Farrar WL. Epigenetic silencing of manganese superoxide dismutase (SOD-2) in KAS 6/1 human multiple myeloma cells increases cell proliferation. Cancer Biol Ther. 2005;4:585–592. doi: 10.4161/cbt.4.5.1704. [DOI] [PubMed] [Google Scholar]
- 111.Maehara K, Uekawa N, Isobe K. Effects of histone acetylation on transcriptional regulation of manganese superoxide dismutase gene. Biochem Biophys Res Commun. 2002;295:187–192. doi: 10.1016/S0006-291X(02)00646-0. [DOI] [PubMed] [Google Scholar]
- 112.Oates N, Pamphlett R. An epigenetic analysis of SOD1 and VEGF in ALS. Amyotroph Lateral Scler. 2007;8:83–86. doi: 10.1080/17482960601149160. [DOI] [PubMed] [Google Scholar]
- 113.Hirsch S, Ronco AM, Guerrero-Bosagna C, de la Maza MP, Leiva L, Barrera G, Llanos M, Alliende MA, Silva F, Bunout D. Methylation status in healthy subjects with normal and high serum folate concentration. Nutrition. 2008;24:1103–1109. doi: 10.1016/j.nut.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 114.Laukkanen MO, Mannermaa S, Hiltunen MO, Aittomaki S, Airenne K, Janne J, Yla-Herttuala S. Local hypomethylation in atherosclerosis found in rabbit ec-sod gene. Arterioscler Thromb Vasc Biol. 1999;19:2171–2178. doi: 10.1161/01.atv.19.9.2171. [DOI] [PubMed] [Google Scholar]
- 115.Davis CA, Monnier JM, Nick HS. A coding region determinant of instability regulates levels of manganese superoxide dismutase mRNA. J Biol Chem. 2001;276:37317–37326. doi: 10.1074/jbc.M104378200. [DOI] [PubMed] [Google Scholar]
- 116.Stuart JJ, Egry LA, Wong GH, Kaspar RL. The 3′ UTR of human MnSOD mRNA hybridizes to a small cytoplasmic RNA and inhibits gene expression. Biochem Biophys Res Commun. 2000;274:641–648. doi: 10.1006/bbrc.2000.3189. [DOI] [PubMed] [Google Scholar]
- 117.Smalheiser NR, Torvik VI. Alu elements within human mRNAs are probable microRNA targets. Trends in Genetics. 2006;22:532–536. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 118.Rabani M, Kertesz M, Segal E. Computational prediction of RNA structural motifs involved in posttranscriptional regulatory processes. Proc Natl Acad Sci U S A. 2008;105:14885–14890. doi: 10.1073/pnas.0803169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fazzone H, Wangner A, Clerch LB. Rat lung contains a developmentally regulated manganese superoxide dismutase mRNA-binding protein. J Clin Invest. 1993;92:1278–1281. doi: 10.1172/JCI116700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chung DJ, Clerch LB. RNA in polysomes is an inhibitor of manganese superoxide dismutase RNA-binding protein activity. Am J Physiol. 1997;272:L714–719. doi: 10.1152/ajplung.1997.272.4.L714. [DOI] [PubMed] [Google Scholar]
- 121.Clerch LB. Post-transcriptional regulation of lung antioxidant enzyme gene expression. Ann N Y Acad Sci. 2000;899:103–111. doi: 10.1111/j.1749-6632.2000.tb06179.x. [DOI] [PubMed] [Google Scholar]
- 122.Chung DJ, Wright AE, Clerch LB. The 3′ untranslated region of manganese superoxide dismutase RNA contains a translational enhancer element. Biochemistry. 1998;37:16298–16306. doi: 10.1021/bi980935g. [DOI] [PubMed] [Google Scholar]
- 123.Knirsch L, Clerch LB. A region in the 3′ UTR of MnSOD RNA enhances translation of a heterologous RNA. Biochem Biophys Res Commun. 2000;272:164–168. doi: 10.1006/bbrc.2000.2754. [DOI] [PubMed] [Google Scholar]
- 124.Knirsch L, Clerch LB. Tyrosine phosphorylation regulates manganese superoxide dismutase (MnSOD) RNA-binding protein activity and MnSOD protein expression. Biochemistry. 2001;40:7890–7895. doi: 10.1021/bi010197n. [DOI] [PubMed] [Google Scholar]
- 125.Kilk A, Laan M, Torp A. Human CuZn superoxide dismutase enzymatic activity in cells is regulated by the length of the mRNA. FEBS Lett. 1995;362:323–327. doi: 10.1016/0014-5793(95)00266-c. [DOI] [PubMed] [Google Scholar]
- 126.Shaw PJ, Tomkins J, Slade JY, Usher P, Curtis A, Bushby K, Ince PG. CNS tissue Cu/Zn superoxide dismutase (SOD1) mutations in motor neurone disease (MND) Neuroreport. 1997;8:3923–3927. doi: 10.1097/00001756-199712220-00016. [DOI] [PubMed] [Google Scholar]
- 127.Mirossay A, Jun S, Dory L. Cloning and characterization of two alleles of the murine extracellular superoxide dismutase gene. Biochem Biophys Res Commun. 2007;352:739–743. doi: 10.1016/j.bbrc.2006.11.081. [DOI] [PubMed] [Google Scholar]
- 128.Okado-Matsumoto A, Fridovich I. Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu,Zn-SOD in mitochondria. J Biol Chem. 2001;276:38388–38393. doi: 10.1074/jbc.M105395200. [DOI] [PubMed] [Google Scholar]
- 129.Orrell R, de Belleroche J, Marklund S, Bowe F, Hallewell R. A novel SOD mutant and ALS. Nature. 1995;374:504–505. doi: 10.1038/374504a0. [DOI] [PubMed] [Google Scholar]
- 130.Rosen DR. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;364:362. doi: 10.1038/364362c0. [DOI] [PubMed] [Google Scholar]
- 131.Turner BJ, Talbot K. Transgenics, toxicity and therapeutics in rodent models of mutant SOD1-mediated familial ALS. Prog Neurobiol. 2008;85:94–134. doi: 10.1016/j.pneurobio.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 132.Wroe R, Wai-Ling Butler A, Andersen PM, Powell JF, Al-Chalabi A. ALSOD: the Amyotrophic Lateral Sclerosis Online Database. Amyotroph Lateral Scler. 2008;9:249–250. doi: 10.1080/17482960802146106. [DOI] [PubMed] [Google Scholar]
- 133.Niemann S, Broom WJ, Brown RH., Jr Analysis of a genetic defect in the TATA box of the SOD1 gene in a patient with familial amyotrophic lateral sclerosis. Muscle Nerve. 2007;36:704–707. doi: 10.1002/mus.20855. [DOI] [PubMed] [Google Scholar]
- 134.Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 135.Furukawa Y, Kaneko K, Yamanaka K, O'Halloran TV, Nukina N. Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in the familial form of amyotrophic lateral sclerosis. J Biol Chem. 2008;283:24167–24176. doi: 10.1074/jbc.M802083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Valentine JS, Doucette PA, Zittin Potter S. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu Rev Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 137.Borgstahl GE, Parge HE, Hickey MJ, Beyer WF, Jr, Hallewell RA, Tainer JA. The structure of human mitochondrial manganese superoxide dismutase reveals a novel tetrameric interface of two 4-helix bundles. Cell. 1992;71:107–118. doi: 10.1016/0092-8674(92)90270-m. [DOI] [PubMed] [Google Scholar]
- 138.Beckman JS, Esetvez AG, Barbeito L, Crow JP. CCS knockout mice establish an alternative source of copper for SOD in ALS. Free Radic Biol Med. 2002;33:1433–1435. doi: 10.1016/s0891-5849(02)01092-4. [DOI] [PubMed] [Google Scholar]
- 139.Furukawa Y, O'Halloran TV. Posttranslational modifications in Cu,Zn-superoxide dismutase and mutations associated with amyotrophic lateral sclerosis. Antioxid Redox Signal. 2006;8:847–867. doi: 10.1089/ars.2006.8.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Carroll MC, Outten CE, Proescher JB, Rosenfeld L, Watson WH, Whitson LJ, Hart PJ, Jensen LT, Cizewski Culotta V. The effects of glutaredoxin and copper activation pathways on the disulfide and stability of Cu,Zn superoxide dismutase. J Biol Chem. 2006;281:28648–28656. doi: 10.1074/jbc.M600138200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Crow JP, Sampson JB, Zhuang Y, Thompson JA, Beckman JS. Decreased zinc affinity of amyotrophic lateral sclerosis-associated superoxide dismutase mutants leads to enhanced catalysis of tyrosine nitration by peroxynitrite. J Neurochem. 1997;69:1936–1944. doi: 10.1046/j.1471-4159.1997.69051936.x. [DOI] [PubMed] [Google Scholar]
- 142.Estevez AG, Crow JP, Sampson JB, Reiter C, Zhuang Y, Richardson GJ, Tarpey MM, Barbeito L, Beckman JS. Induction of nitric oxide-dependent apoptosis in motor neurons by zinc-deficient superoxide dismutase. Science. 1999;286:2498–2500. doi: 10.1126/science.286.5449.2498. [DOI] [PubMed] [Google Scholar]
- 143.Roberts BR, Tainer JA, Getzoff ED, Malencik DA, Anderson SR, Bomben VC, Meyers KR, Karplus PA, Beckman JS. Structural characterization of zinc-deficient human superoxide dismutase and implications for ALS. J Mol Biol. 2007;373:877–890. doi: 10.1016/j.jmb.2007.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Groeneveld GJ, de Leeuw van Weenen J, van Muiswinkel FL, Veldman H, Veldink JH, Wokke JH, Bar PR, van den Berg LH. Zinc amplifies mSOD1-mediated toxicity in a transgenic mouse model of amyotrophic lateral sclerosis. Neurosci Lett. 2003;352:175–178. doi: 10.1016/j.neulet.2003.08.062. [DOI] [PubMed] [Google Scholar]
- 145.Ermilova IP, Ermilov VB, Levy M, Ho E, Pereira C, Beckman JS. Protection by dietary zinc in ALS mutant G93A SOD transgenic mice. Neurosci Lett. 2005;379:42–46. doi: 10.1016/j.neulet.2004.12.045. [DOI] [PubMed] [Google Scholar]
- 146.McCord JM, Fridovich I. Superoxide dismutase: the first twenty years (1968-1988) Free Radic Biol Med. 1988;5:363–369. doi: 10.1016/0891-5849(88)90109-8. [DOI] [PubMed] [Google Scholar]
- 147.Oberley LW, Buettner GR. Role of superoxide dismutase in cancer: a review. Cancer Res. 1979;39:1141–1149. [PubMed] [Google Scholar]
- 148.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, Chan PH, Wallace DC, Epstein CJ. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 150.Lebovitz RM, Zhang H, Vogel H, Cartwright J, Jr, Dionne L, Lu N, Huang S, Matzuk MM. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proc Natl Acad Sci U S A. 1996;93:9782–9787. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kinnula VL, Crapo JD. Superoxide dismutases in malignant cells and human tumors. Free Radic Biol Med. 2004;36:718–744. doi: 10.1016/j.freeradbiomed.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 152.Oberley LW. Mechanism of the tumor suppressive effect of MnSOD overexpression. Biomed Pharmacother. 2005;59:143–148. doi: 10.1016/j.biopha.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 153.Devi GS, Prasad MH, Saraswathi I, Raghu D, Rao DN, Reddy PP. Free radicals antioxidant enzymes and lipid peroxidation in different types of leukemias. Clin Chim Acta. 2000;293:53–62. doi: 10.1016/s0009-8981(99)00222-3. [DOI] [PubMed] [Google Scholar]
- 154.Hu Y, Rosen DG, Zhou Y, Feng L, Yang G, Liu J, Huang P. Mitochondrial manganese-superoxide dismutase expression in ovarian cancer: role in cell proliferation and response to oxidative stress. J Biol Chem. 2005;280:39485–39492. doi: 10.1074/jbc.M503296200. [DOI] [PubMed] [Google Scholar]
- 155.Xu Y, Krishnan A, Wan XS, Majima H, Yeh CC, Ludewig G, Kasarskis EJ, St Clair DK. Mutations in the promoter reveal a cause for the reduced expression of the human manganese superoxide dismutase gene in cancer cells. Oncogene. 1999;18:93–102. doi: 10.1038/sj.onc.1202265. [DOI] [PubMed] [Google Scholar]
- 156.Rosenblum JS, Gilula NB, Lerner RA. On signal sequence polymorphisms and diseases of distribution. Proc Natl Acad Sci U S A. 1996;93:4471–4473. doi: 10.1073/pnas.93.9.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Borgstahl GE, Parge HE, Hickey MJ, Johnson MJ, Boissinot M, Hallewell RA, Lepock JR, Cabelli DE, Tainer JA. Human mitochondrial manganese superoxide dismutase polymorphic variant Ile58Thr reduces activity by destabilizing the tetrameric interface. Biochemistry. 1996;35:4287–4297. doi: 10.1021/bi951892w. [DOI] [PubMed] [Google Scholar]
- 158.Hernandez-Saavedra D, McCord JM. Paradoxical effects of thiol reagents on Jurkat cells and a new thiol-sensitive mutant form of human mitochondrial superoxide dismutase. Cancer Res. 2003;63:159–163. [PubMed] [Google Scholar]
- 159.Nakanishi S, Y K, Ohishi W, Nakashima R, Yoneda M, Nojima H, Watanabe H, Kohno N. Manganese superoxide dismutase Ala16Val polymorphism is associated with the development of type 2 diabetes in Japanese-Americans. Diabetes research and clinical practice. 2008 doi: 10.1016/j.diabres.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 160.Hirooka Y. Role of reactive oxygen species in brainstem in neural mechanisms of hypertension. Auton Neurosci. 2008;142:20–24. doi: 10.1016/j.autneu.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 161.Arsova-Sarafinovska Z, M N, Petrovski D, Banev S, Dzikova S, Georgiev V, Sikole A, Sayal A, Aydin A, Suturkova L, Dimovski AJ. Manganese superoxide dismutase (MnSOD) genetic polymorphism is associated with risk of early-onset prostate cancer. Cell biochemistry and function. 2008 doi: 10.1002/cbf.1504. [DOI] [PubMed] [Google Scholar]
- 162.Mikhak B, Hunter DJ, Spiegelman D, Platz EA, Wu K, Erdman JW, Jr, Giovannucci E. Manganese Superoxide Dismutase (MnSOD) Gene Polymorphism, Interactions with Carotenoid Levels, and Prostate Cancer Risk. Carcinogenesis. 2008 doi: 10.1093/carcin/bgn212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.St Clair D, Kasarskis E. Genetic polymorphism of the human manganese superoxide dismutase: what difference does it make? Pharmacogenetics. 2003;13:129–130. doi: 10.1097/00008571-200303000-00002. [DOI] [PubMed] [Google Scholar]
- 164.Sutton A, Khoury H, Prip-Buus C, Cepanec C, Pessayre D, Degoul F. The Ala16Val genetic dimorphism modulates the import of human manganese superoxide dismutase into rat liver mitochondria. Pharmacogenetics. 2003;13:145–157. doi: 10.1097/01.fpc.0000054067.64000.8f. [DOI] [PubMed] [Google Scholar]
- 165.Bag A, Bag N. Target sequence polymorphism of human manganese superoxide dismutase gene and its association with cancer risk: a review. Cancer Epidemiol Biomarkers Prev. 2008;17:3298–3305. doi: 10.1158/1055-9965.EPI-08-0235. [DOI] [PubMed] [Google Scholar]
- 166.Wheatley-Price P, Asomaning K, Reid A, Zhai R, Su L, Zhou W, Zhu A, Ryan DP, Christiani DC, Liu G. Myeloperoxidase and superoxide dismutase polymorphisms are associated with an increased risk of developing pancreatic adenocarcinoma. Cancer. 2008;112:1037–1042. doi: 10.1002/cncr.23267. [DOI] [PubMed] [Google Scholar]
- 167.Liu G, Zhou W, Park S, Wang LI, Miller DP, Wain JC, Lynch TJ, Su L, Christiani DC. The SOD2 Val/Val genotype enhances the risk of nonsmall cell lung carcinoma by p53 and XRCC1 polymorphisms. Cancer. 2004;101:2802–2808. doi: 10.1002/cncr.20716. [DOI] [PubMed] [Google Scholar]
- 168.Holla LI, Kankova K, Vasku A. Functional polymorphism in the manganese superoxide dismutase (MnSOD) gene in patients with asthma. Clin Biochem. 2006;39:299–302. doi: 10.1016/j.clinbiochem.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 169.Singh M, Khan AJ, Shah PP, Shukla R, Khanna VK, Parmar D. Polymorphism in environment responsive genes and association with Parkinson disease. Mol Cell Biochem. 2008;312:131–138. doi: 10.1007/s11010-008-9728-2. [DOI] [PubMed] [Google Scholar]
- 170.Li H, Kantoff PW, Giovannucci E, Leitzmann MF, Gaziano JM, Stampfer MJ, Ma J. Manganese superoxide dismutase polymorphism, prediagnostic antioxidant status, and risk of clinical significant prostate cancer. Cancer Res. 2005;65:2498–2504. doi: 10.1158/0008-5472.CAN-04-3535. [DOI] [PubMed] [Google Scholar]
- 171.Ho JC, Mak JC, Ho SP, Ip MS, Tsang KW, Lam WK, Chan-Yeung M. Manganese superoxide dismutase and catalase genetic polymorphisms, activity levels, and lung cancer risk in Chinese in Hong Kong. J Thorac Oncol. 2006;1:648–653. [PubMed] [Google Scholar]
- 172.Woodson K, Tangrea JA, Lehman TA, Modali R, Taylor KM, Snyder K, Taylor PR, Virtamo J, Albanes D. Manganese superoxide dismutase (MnSOD) polymorphism, alpha-tocopherol supplementation and prostate cancer risk in the alpha-tocopherol, beta-carotene cancer prevention study (Finland) Cancer Causes Control. 2003;14:513–518. doi: 10.1023/a:1024840823328. [DOI] [PubMed] [Google Scholar]
- 173.Olson SH, Carlson MD, Ostrer H, Harlap S, Stone A, Winters M, Ambrosone CB. Genetic variants in SOD2, MPO, and NQO1, and risk of ovarian cancer. Gynecol Oncol. 2004;93:615–620. doi: 10.1016/j.ygyno.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 174.Zhang HJ, Yan T, Oberley TD, Oberley LW. Comparison of effects of two polymorphic variants of manganese superoxide dismutase on human breast MCF-7 cancer cell phenotype. Cancer Res. 1999;59:6276–6283. [PubMed] [Google Scholar]
- 175.Marklund SL. Human copper-containing superoxide dismutase of high molecular weight. Proc Natl Acad Sci U S A. 1982;79:7634–7638. doi: 10.1073/pnas.79.24.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Marklund SL, Holme E, Hellner L. Superoxide dismutase in extracellular fluids. Clin Chim Acta. 1982;126:41–51. doi: 10.1016/0009-8981(82)90360-6. [DOI] [PubMed] [Google Scholar]
- 177.Marklund SL. Extracellular superoxide dismutase. Methods Enzymol. 2002;349:74–80. doi: 10.1016/s0076-6879(02)49322-6. [DOI] [PubMed] [Google Scholar]
- 178.Karlsson K, Lindahl U, Marklund SL. Binding of human extracellular superoxide dismutase C to sulphated glycosaminoglycans. Biochem J. 1988;256:29–33. doi: 10.1042/bj2560029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tibell LA, Sethson I, Buevich AV. Characterization of the heparin-binding domain of human extracellular superoxide dismutase. Biochim Biophys Acta. 1997;1340:21–32. doi: 10.1016/s0167-4838(97)00024-1. [DOI] [PubMed] [Google Scholar]
- 180.Gao F, Koenitzer JR, Tobolewski JM, Jiang D, Liang J, Noble PW, Oury TD. Extracellular superoxide dismutase inhibits inflammation by preventing oxidative fragmentation of hyaluronan. J Biol Chem. 2008;283:6058–6066. doi: 10.1074/jbc.M709273200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sandstrom J, Nilsson P, Karlsson K, Marklund SL. 10-fold increase in human plasma extracellular superoxide dismutase content caused by a mutation in heparin-binding domain. J Biol Chem. 1994;269:19163–19166. [PubMed] [Google Scholar]
- 182.Adachi T, Yamada H, Yamada Y, Morihara N, Yamazaki N, Murakami T, Futenma A, Kato K, Hirano K. Substitution of glycine for arginine-213 in extracellular-superoxide dismutase impairs affinity for heparin and endothelial cell surface. Biochem J. 1996;313(Pt 1):235–239. doi: 10.1042/bj3130235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Qin Z, Reszka KJ, Fukai T, Weintraub NL. Extracellular superoxide dismutase (ecSOD) in vascular biology: an update on exogenous gene transfer and endogenous regulators of ecSOD. Transl Res. 2008;151:68–78. doi: 10.1016/j.trsl.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Campo S, Sardo AM, Campo GM, D'Ascola A, Avenoso A, Castaldo M, Saitta C, Lania A, Saitta A, Calatroni A. Extracellular superoxide dismutase (EC-SOD) gene mutations screening in a sample of Mediterranean population. Mutat Res. 2005;578:143–148. doi: 10.1016/j.mrfmmm.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 185.Tamai M, Furuta H, Kawashima H, Doi A, Hamanishi T, Shimomura H, Sakagashira S, Nishi M, Sasaki H, Sanke T, Nanjo K. Extracellular superoxide dismutase gene polymorphism is associated with insulin resistance and the susceptibility to type 2 diabetes. Diabetes Res Clin Pract. 2006;71:140–145. doi: 10.1016/j.diabres.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 186.Petersen SV, Oury TD, Valnickova Z, Thogersen IB, Hojrup P, Crapo JD, Enghild JJ. The dual nature of human extracellular superoxide dismutase: one sequence and two structures. Proc Natl Acad Sci U S A. 2003;100:13875–13880. doi: 10.1073/pnas.2436143100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Petersen SV, Kristensen T, Petersen JS, Ramsgaard L, Oury TD, Crapo JD, Nielsen NC, Enghild JJ. The folding of human active and inactive extracellular superoxide dismutases is an intracellular event. J Biol Chem. 2008;283:15031–15036. doi: 10.1074/jbc.M801548200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Olsen DA, Petersen SV, Oury TD, Valnickova Z, Thogersen IB, Kristensen T, Bowler RP, Crapo JD, Enghild JJ. The intracellular proteolytic processing of extracellular superoxide dismutase (EC-SOD) is a two-step event. J Biol Chem. 2004;279:22152–22157. doi: 10.1074/jbc.M401180200. [DOI] [PubMed] [Google Scholar]
- 189.Gongora MC, Qin Z, Laude K, Kim HW, McCann L, Folz JR, Dikalov S, Fukai T, Harrison DG. Role of extracellular superoxide dismutase in hypertension. Hypertension. 2006;48:473–481. doi: 10.1161/01.HYP.0000235682.47673.ab. [DOI] [PubMed] [Google Scholar]
- 190.Nguyen AD, Itoh S, Jeney V, Yanagisawa H, Fujimoto M, Ushio-Fukai M, Fukai T. Fibulin-5 is a novel binding protein for extracellular superoxide dismutase. Circ Res. 2004;95:1067–1074. doi: 10.1161/01.RES.0000149568.85071.FB. [DOI] [PubMed] [Google Scholar]


