Abstract
The advent of oligonucleotide array comparative genomic hybridization (aCGH) has revolutionized diagnosis of chromosome abnormalities in the genetics clinic. This new technology also has valuable potential as a research tool to investigate larger genomic rearrangements that are typically diagnosed via routine karyotype. aCGH was used as a tool for the high resolution analysis of chromosome content in individuals with known deletions of chromosome 18. The aim of this study was to clarify the precise location of the breakpoints as well as to determine the presence of occult translocations creating additional deletions and duplications. One hundred eighty nine DNA samples from individuals with 18q deletions were analyzed. No breakpoint clusters were identified, as no more than two individuals had breakpoints within 2 Kb of each other. Only two regions of 18q were never found to be haploid, suggesting the existence of haplolethal genes in those regions. Of the individuals with only a chromosome 18 abnormality, 17% (n=29) had interstitial deletions. Six percent (n=11) had a region of duplication immediately proximal to the deletion. Eight percent (n=15) had more complex rearrangements with captured (non-18q) telomeres thus creating a trisomic region. The fifteen captured telomeres originated from a limited number of other telomeres (4q, 10q, 17p, 18p, 20q and Xq.) These data were converted into a format for ease of viewing and analysis by creating custom tracks for the UCSC Genome Browser. Taken together, these findings confirm a higher level of variability and genomic complexity surrounding deletions of 18q than has previously been appreciated.
Keywords: aCGH, 18q−, chromosome 18, chromosome abnormality, chromosome deletion
INTRODUCTION
Variations of chromosome number and content were first associated with human disease in 1959 when trisomy 21 was discovered to be the cause of Down syndrome [Lejuene et al., 1959]. Five years later, deletions of chromosome 18 were first described by de Grouchy and coworkers [1964] who believed that the exact size of the missing chromosome segment was minimally variable and the clinical presentation was well-defined and clinically apparent [de Grouchy, 1969]. However, by 1970 the number of identified cases had increased to the point that it was becoming clear that both the size of the deletion and the clinical presentation varied greatly and that there may, in fact, be a correlation between the two [Šubrt and Pokorný, 1970]. When tools such as molecular analysis and FISH became available to detect greater chromosome aberration complexity, it became clear that routine karyotyping was insufficient to fully characterize deletions of 18q [Brkanac et al, 1998].
Appreciating the molecular complexity and determining which genes are not diploid is central to the ability to elucidate genotype/phenotype correlations. These correlations will one day help us to understand the role that specific genes play, thereby enabling better predictions of clinical outcome as well as forming the basis for treatments targeting the molecular basis of differences in individuals with 18q deletions.
Identification of genotype/phenotype correlations is challenging and involves several steps. First, it is necessary to define critical regions and then to identify genes that are dosage sensitive within those regions. Second, it is necessary to rule out as candidate genes those genes on 18q that are associated with a known recessive condition. As evidenced by the normal state of carriers of such conditions, it is clear that these genes are involved in processes that are haplosufficient and therefore unlikely to contribute to the phenotype of individuals with deletions of 18q. Third, it is necessary to rule out copy number variants as potential candidate genes (or regions) in the search for genotype/phenotype correlations because by definition their hemizygosity is not sufficient to cause an abnormal phenotype. Rapid and precise molecular genotyping of affected individuals is central to the process of establishing genotype/phenotype correlations.
Here we report on the findings using oligonucleotide array comparative genomic hybridization (aCGH) in a cohort of 189 individuals with deletions of chromosome 18q, revealing an even greater level of genomic variability and complexity than previously appreciated.
METHODS
Study Participants
This study was approved by the University of Texas Health Science Center at San Antonio Institutional Review Board. Participants were eligible for the study if they had a clinically suspected or diagnosed abnormality of chromosome 18. Participants learned of the research principally through the Chromosome 18 Registry & Research Society and the internet. Medical records were collected including the karyotype of the affected individual and their parents. Blood samples were also collected from the proband and parents, if available, for DNA, chromosomes, and the establishment of cell lines. Samples were processed as previously described [Cody et al., 1997]. Informed consent was appropriately documented at all stages of the process.
Array design
Custom oligonucleotide arrays were produced by Agilent Technologies (Santa Clara, CA). The data reported here employed several array designs over the life of this longitudinal study. The current platform employs 32,000 features (60-mers) across chromosome 18 and 12,000 features across the remainder of the genome. These chromosome 18 zoom arrays were designed using the Agilent e-array software. This allowed us to generate very high resolution chromosome 18 breakpoint data for the individuals in our study. It also allows us to rule out other significant imbalances in the rest of the genome. However, as this has been an evolving project, not every individual was assayed using the same array design platform. In our analysis of 189 participants, 6 were analyzed using an 185K whole genome array, 7 were analyzed using a 44K whole genome array and 31 were analyzed with a custom 11K chromosome 18 zoom array. The remaining 145 participants were analyzed with the most current platform, a 44K chromosome 18 zoom array.
Array Hybridization and Analysis
The hybridization was performed as described in the Agilent protocol. Comparative Genomic Hybridization (CGH) uses a two sample comparative method in which the test (or patient) sample is assessed in comparison to a reference sample. The two DNA samples are labeled with different fluorophores, then mixed together and allowed to competitively hybridize to the oligonucleotides on the array slide. The reference DNA samples were from Promega (Madison, WI) and consisted of pooled same sex DNA samples. The reference sample is from the opposite sex as the test sample thus providing an internal copy number control. No second confirmatory dye swap experiment was performed in individuals in whom the initial experiment revealed a simple deletion because each study participant had a previous diagnosis of a chromosome 18 deletion. However, when our aCGH experiments detected a more complex rearrangement these results were confirmed in a dye swap experiment. In addition, in these cases we also assessed the parents' DNA samples using aCGH. For those samples that potentially defined a critical region, we also confirmed those breakpoints with BAC FISH.
Each array was scanned using the Agilent laser scanner. The scan data were extracted using Agilent Feature Extraction (version 8.1.1). Those data were then analyzed using the CGH Analytics 3.4.27 software. Data points were analyzed in continuous groups of 8 probes and log 2 ratios of sample DNA were compared to control DNA. Arrays were normalized to a median log 2 ratio of zero, except for the X and Y chromosomes. Features that were less than −1 or greater than +0.5 were identified by the grey bar. Breakpoints were determined to be between the ends of the array features on either side of the deletion breakpoint.
Custom Genome Browser Tracks
The data from the analysis of 189 participants with an 18q deletion were entered into the University of California at Santa Cruz Genome Browser (http://genome/ucsc.edu) as a set of custom tracks.
RESULTS
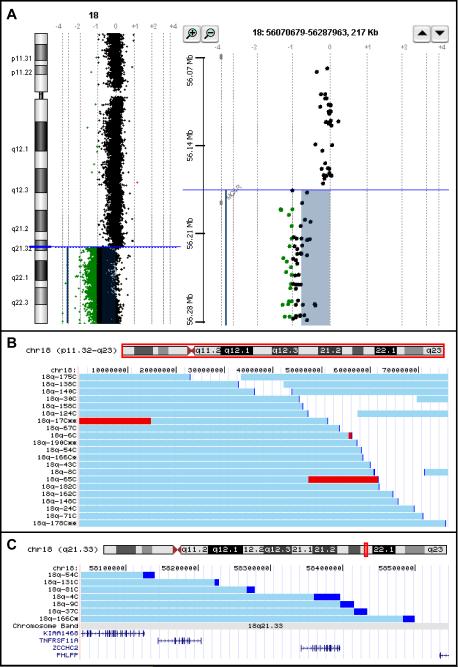
High resolution genome copy number changes were determined for 189 individuals with a clinical laboratory diagnosed or a clinically suspected chromosome 18q deletion. All samples were analyzed using an oligonucleotide microarray comparative genomic hybridization (aCGH) platform manufactured by Agilent Technologies. An example of the data from one individual is shown in Figure 1A. The panel on the left displays the data for all of chromosome 18. The panel on the right shows a zoomed in “gene view” of the breakpoint region.
Figure 1.
A. Example of aCGH data using the CHG Analytics software package from Agilent Technologies. The colored bars indicate the features that are significantly different from 0 on the log2 scale. Features exactly on the 0 line have a 1:1 red green color ratio between the test and reference DNA samples
Left panel: The “Chromosome View” from the CGH Analytics software program showing the data from an individual with an 18q deletion
Right panel: The “Gene View” from CGH Analytics showing the breakpoint in relation to the location of the genes in that region.
B. Chromosome 18 content aCGH data from 21 individuals displayed as custom tracks on the UCSC Genome Browser. The horizontal light blue bars depict the region of chromosome 18 that is present in two copies. The darker blue sections are the breakpoint regions. The red regions are present in three copies. All individuals who have retained a distal segment of chromosome 18 have interstitial deletions and not translocations to other chromosomes. Those individuals with as single asterisk by their study number have a rearrangement involving another chromosome, but that other chromosome had no net copy number change. Those individuals who have a double asterisk by their study number have a net copy number abnormality involving a non-18q region of the genome.
C. A zoomed in view of the data in Figure 2 showing the breakpoints of 7 individuals aligned with the RefSeq genes in the region.
The aCGH data from the entire cohort have been converted into custom tracks on the University of Santa Cruz Genome Browser (http://genome.ucsc.edu). The data from 21 of the 189 participants is shown in Figure 1B. These data were selected to illustrate the variety of chromosome content from our study participants. The data from all 189 participants can be viewed in the on-line supplemental material (Fig 1S). All 189 participants' chromosomes have been assessed by FISH using a commercial 18q telomere probe. The individuals who have retained a distal segment of chromosome 18 that is translocated to a different chromosome with no net copy number change of the other chromosome are indicated in Figure 1B & C by an asterisk after their participant number. Those who have interstitial deletions of chromosome 18 have no asterisk after their participant number. The individuals with an additional net copy number change that involves a non-18q chromosome region are indicated in the custom tracks by the presence of 2 asterisks after their participant number.
Interestingly, the breakpoints do not cluster within certain regions of 18q. With the exception of the two individuals with 4;18 translocations [Horbinski et al., 2008] no two unrelated individuals had identical aberrations. In addition, there was a wide variation in the size of the deletion. The person with the largest terminal deletion was hemizygous for 30.076 Mb of DNA. The person with the smallest terminal deletion was hemizygous for 0.5 Mb. However, this individual was identified through parental studies and not because of any phenotypic abnormality [South et al., 2008]. The person with the smallest terminal deletion with any clinical consequences had a 3.78 Mb deletion.
The categorical findings of the aCGH data are shown in Table I. One hundred eighty nine samples were analyzed, of which 170 involved only chromosome 18q. Of the 170 samples with abnormalities involving only 18q, 29 (17%) are interstitial and 141 (83%) are terminal deletions.
Table I.
| Aberration Description | Total | Parent with 18q− | Parent with balanced rearrangement | de novo | Parental origin of de novo | |||
|---|---|---|---|---|---|---|---|---|
| Mat | Pat | Unk. | ||||||
| 1 | Simple terminal deletions | 130 | 3 | 1 | 126 | 10 | 71 | 45 |
| 2 | 18q−/18q+ | 11 | 1 | 0 | 10 | 0 | 6 | 4 |
| 3 | Simple interstitial deletions | 29 | 2 | 1 | 26 | 2 | 12 | 12 |
| SUBTOTAL | 170 | 6 | 2 | 162 | 12 | 89 | 61 | |
| 4 | Translocation with only an18q deletion | 4 | 0 | 0 | 4 | 0 | 2 | 2 |
| 5 | 18q terminal deletion with a telomere gain of a non-18q telomere | 15 | 0 | 4 | 11 | 2 | 4 | 5 |
| TOTAL | 189 | 6 | 6 | 177 | 14 | 95 | 68 | |
Simple terminal deletion - These individuals had a net copy number loss only from 18q that extended from a breakpoint to the end of the chromosome. All of these individuals had a single 18q signal using a commercial 18q telomere FISH probe.
18q−/18q+ - These individuals had terminal deletions of 18q as well as a net copy number gain of the 18q region just proximal to their deletion.
Simple interstitial deletion - These individuals' net copy number change was within in the long arm of chromosome18. FISH with a commercial 18q telomere probe showed the 18q telomere to be located at the end of the long arm of 18q, confirming an interstitial deletion without an inversion or translocation.
Translocation with only an 18q deletion - These individuals' only net copy number change was a deletion of an interstitial portion of 18q. Their 18q telomere was exchanged with one from another chromosome. These individuals are identified in Figure 2 with an asterisk next to their study number.
18q terminal deletion with a telomere gain of a non-18q telomere - This group of individuals have an unbalanced translocation or a captured telomere that involved the gain of a non-18q telomere. They therefore have a segmental trisomy in addition to 18q−. These individuals are identified in Figure 2 with a double asterisk next to their study number.
Among the terminal deletions, 11 of the 140 (8%) have a segmental duplication immediately proximal to their hemizygous region. These duplications varied in size from 58.06 Kb to 14.3 Mb of DNA. Two samples with larger duplications were assessed for their location and orientation using two color FISH. The results for both cases indicated that the duplication was in an inverted orientation (data not shown). The finding that the duplicated regions are inverted is consistent with similar results involving chromosome 1p deletions [Ballif et al., 2003].
One hundred seventy seven of the 189 cases analyzed (94%) were found to be de novo events based on normal parental karyotypes. Regardless of the type of 18q deletion, only about 12% of the de novo events arose on the maternally inherited chromosome. Of those individuals with an inherited chromosome 18 abnormality, 6 (3% of the total) were inherited from a parent who also had an 18q deletion and in every case that parent was the mother. The other 6 (3% of the total) were the consequence of a parent with a balanced rearrangement. In those cases, 2 of the 6 resulted in a child with an 18q deletion and 4 resulted in the child having an 18q deletion as well as the gain of a non-18q telomere.
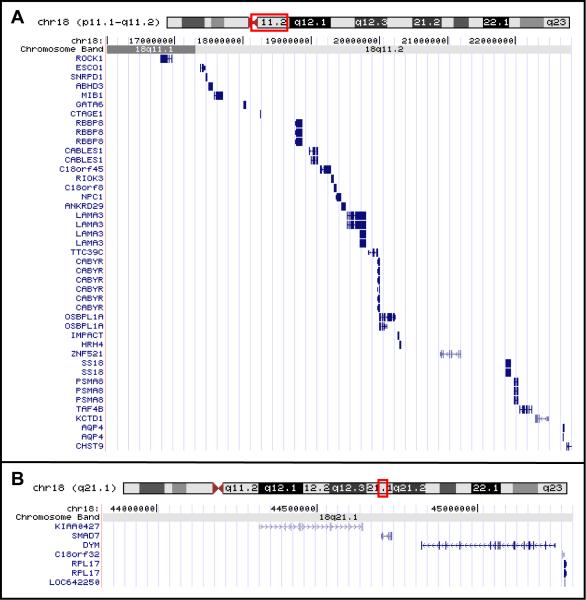
Additionally, two regions have not yet been found in a hemizygous state in any of the 189 individuals assessed. These two regions and the genes in those regions are illustrated in Figure 2. These regions are between the centromere and 22,826,284 bp and between 43,832,732 bp and 45,297,446 bp.
Figure 2.
UCSC Genome Browser depictions of the RefSeq genes in the two regions of chromosome 18 that have not been found to be hemizygous.
We also identified 19 individuals (10%) who had more complex chromosome 18 abnormalities involving another chromosome. Four of these individuals' only copy number abnormality involved chromosome 18, the other chromosome involved in the rearrangement showed no detectable net copy number change. These individuals are indicated by an asterisk after their participant number in Figures 1B and 1C. Fifteen of those 19 people had terminal deletions of 18q with a gain of a non-chromosome 18q telomere. These individuals are identified by a double asterisk next to the participant number in Figures 1B and 1C. These captured telomeres involved a limited number of chromosomes. Four were from 18p, three from 4q, three from 17p, two from Xq, two from 20q and one from 10q,
DISCUSSION
Ultimately, high resolution genotyping and subsequent identification of genotype/phenotype correlations will inform prognosis and treatment options for people with 18q deletions. Here we report the first step in this process: the use of rapid high resolution genotyping for determining which genes are not diploid in each individual with an 18q deletion. Oligonucletotide microarray comparative genomic hybridization (aCGH) facilitates this goal. In order to visualize and analyze the data, we have converted them to custom tracks on the University of California Santa Cruz Genome Browser. The data conversion to the genome browser allows us to view the data from all participants at a resolution varying from a whole chromosome view to a base pair view. This format also allows the data to be aligned with all of the other genome annotation tracks such as RefSeq Genes, Self Chain etc. To illustrate this, Figure 1C shows the chromosome content of 7 individuals aligned with the genes in that region. This will be an important tool for addressing many questions about correlations with both genotype and phenotype data.
The region of hemizygosity in our study population is highly variable. This leads us to conclude that a specific genomic architecture does not exist in 18q that creates hotspots for chromosome breaks. The presence of terminal, interstitial and more complex abnormalities without breakpoint clustering is consistent with findings reported in deletions of 1p [Gajecka et al., 2007], 4p [Maas et al., 2008], 5p [Zhang et al., 2005], 9p [Swinkels et al., 2008 and Hauge et al., 2008], 11q [Grossfeld et al., 2004], and 13q [Quélin et al., 2008]. Our group reported previously on breakpoint clustering in individuals with 18p terminal deletions [Shaub et al., 2002]. However, those clustered breakpoints were all within the centromeric region.
We do appreciate that in this study we are measuring the healing point on the chromosome, not necessarily the precise breakpoint. The breakpoint and the healing point may not be the same in cases with de novo telomeres. The possibility exists that double stranded breaks do arise in a common region or a limited number of regions, but that unprotected chromosome ends lead to DNA erosion until the end is stabilized with the addition of a telomere. Since de novo telomere addition requires no more than a few base pair homology with telomerase [Ballif et al., 2003] telomeres can be acquired at numerous places along a chromosome arm. The only instance in which a break and heal presumably occur virtually simultaneously is in chromosome translocations. In our dataset it is interesting that only the two unrelated individuals with 4;18 translocations have identical breakpoints on both chromosomes. However, the other individuals with translocations (“captured” telomeres) all have unique chromosome 18 breakpoints similar to those individuals with terminal or interstitial deletions. This suggests that predisposing genomic architecture is not a prerequisite for chromosome deletions.
The finding that there are a limited number of chromosomes which provide a captured telomere is interesting. The non-random nature of this finding may be due to the proximity of chromosome regions within the nucleus during meiosis. Alternatively, it may be the result of a limited number of trisomies that are viable in conjunction with an 18q deletion. The impact that the trisomic telomere region has on the 18q− phenotype is currently under investigation.
There are only 2 regions of 18q which have to date not been found to be hemizygous. We hypothesize that these regions may contain genes that are prenatally lethal when hemizygous. These regions and the genes they contain are shown in Figure 2. In the data reported by Feenstra and co-workers [2007] two similar and overlapping regions can be identified. Their patient 1 narrows the proximal region we describe to between the centromere and 18.9 Mb; a region containing 8 genes. There is gene dosage information on only one of those genes; RBBP8, (retinoblastoma binding protein 8). The heterozygous targeted knock-out mice for RBBP8 have a shortened lifespan and die with multiple tumors. Interestingly, the null embryos fail to form an egg cylinder meaning that RBBP8 is essential very early in embryonic development [Chen et al., 2005].
The potential hemizygous lethal region that we describe in the middle of the long arm between 43.83 and 45.30 Mb is within a region also not shown to be hemizygous by Feenstra et al. This region contains only 5 genes. One gene DYM (dymeclin) is an unlikely candidate as it is associated with the rare autosomal recessive condition Dyggve-Melchior-Clausen syndrome (DMC) [OMIM:223800]. There is also a known copy number variation within this gene [Wong et al., 2007], which is not surprising since it is associated with a recessive condition. An interesting candidate for a dosage sensitive lethal gene is SMAD7 which negatively modulates members of the TGF-β superfamily of growth and differentiation factors. The only current mouse knock-out model has a deletion in exon 1, leading to partial gene function [Li et al., 2006]. Thus no truly hemizygous mouse model exists that would test this hypothesis.
The growing appreciation of the existence of copy number variations in the human genome may be helpful in defining regions carrying genes that are in haplosufficient pathways or processes. However, caution should be used when making assumptions about haplosufficiency based on these studies since the phenotyping of the “normal” subjects is generally superficial at best. Multiple tracks on the UCSC Genome Browser contain data from numerous databases such as the Database of Genomic Variants [Iafrate et al., 2004] that can be viewed for any region of interest.
The results of this study provide additional insight into the underlying genomic architecture of chromosome 18. Excluding two regions previously discussed, there have been individuals reported with segmental aneusomy involving all regions of chromosome 18. We have speculated that this finding is due to the low density of genes on chromosome 18 reducing the probability of an individual aneusomic region generating too many haploinsufficient phenotypes to sustain life. This hypothesis would lead to the prediction that there might be more chromosome 18 copy number variations (i.e. without phenotypic effect) in more regions and in more individuals than for other chromosomes because the paucity of genes would reduce the chance of negative selection against CNVs.
Of interest, 88% of the de novo deletions were of paternal origin. This number varied no more than 2 percentage points regardless of the type of deletion. We anticipated that there may be mechanistic difference between terminal deletions and interstitial deletions, leading to parental origin differences. However, the data do not support such a difference. In addition, the size and location of the deletions did not differ between those located on maternally derived chromosomes versus those located on paternally derived chromosomes. Therefore, we do not have reason to suspect there are viability differences between those individuals with maternally derived deletions and those with paternally derived deletions. Because the number of individuals with maternal deletions is small and because every aberration is unique, it is difficult to determine if there are imprinting effects on the phenotype. However, when assessing the phenotype data we are taking parental origin into account and have not yet seen any evidence of such an effect.
Unfortunately, few of the recent molecular studies of chromosome deletions report parental origin findings with which to compare our own results. In contrast to our finding, Gajecka and co-workers [2007] found that 60% of 1p deletions occur on the maternal chromosome. Whether this is due to mechanistic differences or to imprinting and therefore viability differences is unknown at this time and is an interesting area for future investigation.
One interesting observation is that of the children in this cohort who have a parent who also has an 18q deletion, all of the parents were mothers. To our knowledge there has not been a male with an 18q deletion who has fathered children. Male infertility has not been previously reported for this syndrome; therefore, this is an area under current investigation.
Lastly, the findings of this study suggest that 18q− cannot be considered a “syndrome” in the truest sense of the word. A syndrome is defined as a cluster of characteristics with a single cause. However, no two individuals have the same aberration, with the exception of the cases discussed in Horbinski [2008]. Also, no region of the chromosome is hemizygous in every study participant. Attempts to identify a critical region for “the 18q− syndrome” are not possible because there is no common underlying defect, much less a common set of phenotypic characteristics. However, the identification of the critical regions for specific phenotypic components and ultimately the causal genes is a feasible goal. This work represents the first steps in this process.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank the families who participated in this research not only for their participation but for their tireless efforts at raising funds and awareness. This work was supported by the Chromosome 18 Registry and Research Society, the MacDonald family and the NIH (R01HD045907 to JDC and M01-RR-001346 for the Frederic C. Bartter General Clinical Research Center, San Antonio, Texas).
REFERENCES
- Ballif BC, Yu W, Shaw CA, Kashork C, Shaffer LG. Monosomy 1p36 breakpoint junctions suggest pre-meiotic breakage-fusion-bridge cycles are involved in generating terminal deletions. Hum Mol Genet. 2003;12:2153–2165. doi: 10.1093/hmg/ddg231. [DOI] [PubMed] [Google Scholar]
- Brkanac Z, Cody JD, Leach RJ, DuPont BR. Identification of cryptic rearrangements in patients with 18q− deletion syndrome. Am J Hum Genet. 1998;62:1500–1506. doi: 10.1086/301854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P-L, Liu F, Cai S, Lin X, Li A, Chen Y, Gu B, Lee EY-HP, Lee W-H. Inactivation of CtIP leads to early embryonic lethality mediated by G restraint and tumorigenesis by haploid insufficiency. Mol Cell Biol. 2005;25:3525–3542. doi: 10.1128/MCB.25.9.3535-3542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody JD, Brkanac Z, Pierce JF, Plaetke R, Ghidoni PD, Kaye CI, Leach RJ. Preferential loss of the paternal alleles in the 18q− syndrome. Am J Med Genet. 1997;69:280–286. doi: 10.1002/(sici)1096-8628(19970331)69:3<280::aid-ajmg12>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- de Grouchy J, Royer P, Salmon C, Lamy M. Deletion partielle des bras longs du chromosome 18. Path et Biol. 1964;12:579–582. [PubMed] [Google Scholar]
- de Grouchy The 18p-, 18q− and 18r syndromes. Birth Defects: OAS. 1969;5:74–87. [Google Scholar]
- Gajecka M, Mackay KL, Shaffer LG. Monosomy 1p36 deletion syndrome. Am J Med Genet C. 2007;145C:346–356. doi: 10.1002/ajmg.c.30154. [DOI] [PubMed] [Google Scholar]
- Grossfeld PD, Mattina T, Lai Z, Favier R, Jones KL, Cotter F, Jones C, 11q Consortium The 11q terminal deletion disorder: a prospective study of 110 cases. Am J Med Genet. 2004;129A:51–61. doi: 10.1002/ajmg.a.30090. [DOI] [PubMed] [Google Scholar]
- Hauge X, Raca G, Cooper S, May K, Spiro R, Adam M, Martin CL. Detailed characterization of, and clinical correlations in, 10 patients with distal deletions of chromosome 9p. Genet Med. 2008;10:599–611. doi: 10.1097/GIM.0b013e31817e2bde. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horbinski C, Carter EM, Heard PL, Santhanoori M, Hu J, Vockley J, Gunn S, Hale DE, Cody JD. Molecular and clinical characterization of a recurrent cryptic unbalanced t(4q;18q) resulting in an 18q deletion and 4q duplication. Am J Med Genet. 2008;22:2898–2904. doi: 10.1002/ajmg.a.32557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Lejeune J, Gautier M, Turpin R. Etude des chromosomes somatiques de neuf enfants mongoliens. C. R. Acad. Sci. 1959;248:1721–1722. [PubMed] [Google Scholar]
- Li R, Rosendahl A, Brodin G, Cheng AM, Ahgren A, Sundquist C, Kulkarni S, Pawson T, Heldin CH, Heuchel RL. Deletion of exon I of SMAD7 in mice results in altered B cell responses. J Immunol. 2006;176:6777–6784. doi: 10.4049/jimmunol.176.11.6777. [DOI] [PubMed] [Google Scholar]
- Linnankivi T, Tienari P, Somer M, Kähkönen M, Lönnqvist T, Valanne L, Pihko H. 18q deletions: clinical, molecular, and brain MRI findings of 14 individuals. Am J Med Genet Part A. 2006;140A:331–339. doi: 10.1002/ajmg.a.31072. [DOI] [PubMed] [Google Scholar]
- Maas NMC, van Buggenhout G, Hannes F, Thienpont B, Sanlaville D, Kok K, Midro A, Andrieux J, Anderlid B-M, Schoumans J, Hordijk R, Devriendt K, Fryns J-P, Vermeesch JR. Genotype-phenotype correlation in 21 patients with Wolf-Hirschhorn syndrome using high resolution array comparative genome hybridization (CGH) J Med Genet. 2008;45:71–80. doi: 10.1136/jmg.2007.052910. [DOI] [PubMed] [Google Scholar]
- Quélin C, Bendavid C, Dubourg C, de la Rochebrochard C, Lucas J, Henry C, Jaillard S, Loget P, Loeuillet L, Lacombe D, Rival J-M, David V, Odent S, Pasquier L. Twelve new patients with 13q deletion syndrome: genotype-phenotype analyses in progress. Eur J Med Genet. 2008 doi: 10.1016/j.ejmg.2008.10.002. doi:10.1016/j.ejmg.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Schaub RL, Reveles XT, Baillargeon JG, Leach RJ, Cody JD. Molecular characterization of 18p deletions: identification of a breakpoint cluster. Genet Med. 2002;4:15–19. doi: 10.1097/00125817-200201000-00003. [DOI] [PubMed] [Google Scholar]
- South ST, Rope AF, Lamb AN, Aston E, Glaus N, Whitby H, Maxwell T, Zhu XL, Brothman AR. Expansion in size of a terminal deletion: a paradigm shift for parental follow-up studies. J Med Genet. 2008;45:391–395. doi: 10.1136/jmg.2008.057315. [DOI] [PubMed] [Google Scholar]
- Šubrt I, Pokorný J. Familial occurrence of 18q−. Hummangenetik. 1970;10:181–187. doi: 10.1007/BF00295518. [DOI] [PubMed] [Google Scholar]
- Swinkels MEM, Simons A, Smeets DF, Vissers LE, Veltman JA, Pfundt R, de Vries BBA, Faas BHW, Schrander-Stumpel CTRM, McCann E, Sweeney E, May P, Draaisma JM, Knoers NV, van Kessel AG, van Ravenswaaij-Arts CMA. Clinical and cytogenetic characterization of 13 Dutch patients with deletion 9p syndrome: delineation of the critical region for a consensus phenotype. Am J Med Genet A. 2008;146A:1530–1538. doi: 10.1002/ajmg.a.32310. [DOI] [PubMed] [Google Scholar]
- Wong KK, deLeeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, Horsman DE, MacAulay C, Ng RT, Brown CJ, Eichler EE, Lam WL. A comprehensive analysis of common copy-number variations in the human genome. Am J Hum Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Snijders A, Segraves R, Zhang X, Neibuhr A, Albertson D, Yang H, Gray J, Niebuhr E, Bolund L, Pinkel D. High-resolution mapping of genotype-phenotype relationships in Cri Du Chat syndrome using array comparative genomic hybridization. Am J Hum Genet. 2005;76:312–326. doi: 10.1086/427762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.