Abstract
The base excision repair (BER) pathway recognizes and repairs most non-bulky lesions, uracil and abasic (AP) sites in DNA. Several participants are embryonic lethals in knockout mice. Since the pathway has never been investigated during embryogenesis, we characterized the first three steps of BER in zebrafish extracts from unfertilized eggs, embryos at different developmental stages and adults. Using a 45-mer double stranded substrate with a U/G mispair at position 21, we showed that extracts from all stages are capable of performing BER. Before 3 dpf aphidicolin-sensitive polymerases perform most nucleotide insertion. In fact, eggs and early stage embryos lack DNA polymerase-β protein. After hatching at 3 dpf, an aphidicolin-resistant polymerase, probably DNA polymerase β, becomes the primary polymerase. Previously we showed that when zebrafish AP endonuclease protein (ZAP1) level is knocked down, embryos cease dividing after the initial phase of rapid proliferation and die without apoptosis shortly thereafter. Nevertheless, extracts from embryos in which ZAP1 has been largely depleted process substrate equally as well as extracts from control embryos. Since apex1 and apex2 are both strongly expressed in early embryos relative to adults, these data indicate that both may play important roles in DNA repair in early development. In brief, the major differences in BER performed by early stage embryos and adults are the absence of DNA polymerase-β, leading to predominance of replicative polymerases, and the presence of backup Mg2+-dependent endonuclease activity in early stage embryos. The switch to normal, adult BER occurs fully when the embryos hatch from the chorionic membrane and encounter normal oxidative stress.
Keywords: base excision repair, DNA repair, uracil DNA glycosylase. AP endonuclease 1, AP endonuclease 2, DNA polymerase-β, aphidicolin, zebrafish
All cells make reactive oxygen species (ROS) as a by-product of ATP synthesis via the electron transport chain (1-3). ROS damage proteins, lipids, carbohydrates and DNA. Although the first three can be discarded and resynthesized, the cell cannot simply discard its DNA without losing vital genetic information. Therefore, all organisms have a pathway known as base excision repair (BER) to repair oxidatively damaged DNA (Figure 1). Four members of this pathway, AP endonuclease 1, XRCC1, flap endonuclease 1 and ligase I are required for embryonic development in mice (4-8). Another member of the pathway, DNA polymerase-β (pol-β), when deleted, results in either an embryonic lethal phenotype (9) or in abnormal neurogenesis and lung function, leading to neonatal death (10). Cell lines that are PolB-/- are viable but hypersensitive to methylating agents such as methylmethanesulfonate(11-13). Apex1 -/- murine embryos survive no longer than embryonic day 9 (4, 5) and no homozygous null cell lines have been cultured to date (14).
Figure 1. The DNA base excision repair pathway.
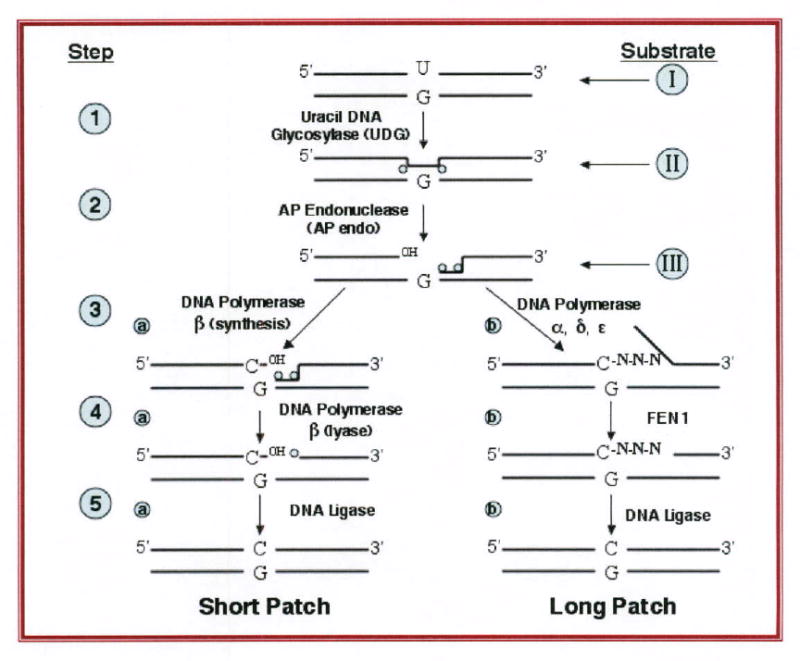
Step number is indicated on the left in Arabic numerals, while substrate used in different reactions shown in the text is indicated in Roman numerals on the right. Steps 3a. 4a and 5a occur in short patch (single nucleotide) repair. Steps 3b, 4b and 5b occur in long patch repair (insertion of 2-6 additional nucleotides). Step 3b uses the replicative DNA polymerases α, δ and ε, in the presence of PCNA (not shown).
Despite the importance of DNA repair in cellular physiology(15), relatively little is understood about the role of DNA repair in embryonic development. In an effort to understand the role of AP endonuclease 1 in early development, we recently explored the effects of knocking down expression of the gene in zebrafish embryos (16). Using morpholino oligonucleotides directed against the translation start site (TS-MO) to prevent translation of apex1, we demonstrated that full knockdown results in death after the rapid phase of cell division is complete, following the midblastula transition (MBT). Although zebrafish zygotic transcription begins at the MBT, failure to initiate zygotic transcription is unlikely to be the cause of death, because mouse embryos initiate zygotic transcription at the two-cell stage but Apex -/- mouse embryos do not fail until E7-E9.
Zebrafish embryos in which translation of apex1 mRNA has been blocked by means of TS-MO (full knockdown) can be rescued through the MBT by co-injection of the mRNA for WT human APEX1, although development is still not normal and the embryos die at ∼7 days after fertilization (dpf). The same phenotype arises in partial knockdowns (hypomorphs) if the protein is knocked down by less than 60% by means of lower concentrations of TS-MO or if the MO targets the splice sites of apex1 (16). In an effort to examine whether full or partial knockdown of apex1 results in loss of BER, we have examined the BER pathway (Figure 1) in extracts of eggs, early embryos and adult zebrafish and in full knockdowns and hypomorphic embryos at the appropriate stage of development. We find that eggs, embryos and adult zebrafish are capable of BER. Even when ZAP1 (zebrafish AP endonuclease protein) has been reduced by >60%, embryonic extracts are still capable of repairing a U/G mispair to the same extent as extracts from controls. While adult mammalian cells have a single major AP endonuclease, encoded by Apex1, here we provide functional evidence for the presence of additional Mg2+-dependent AP endonuclease activities in zebrafish embryos, most likely AP endonuclease 2. Furthermore, the lack of DNA polymerase β in unfertilized eggs and very early stage embryos necessitates the use of other, aphidicolin-sensitive and insensitive polymerases in the pathway.
Methods
Zebrafish culture
Zebrafish were grown and embryos were spawned as described (16, 17). To prepare full knockdown and hypomorphic embryos, embryos were microinjected with TS-MO as described (16).
Preparation of extracts
Extracts were prepared from eggs or embryos at different stages by lysis in 50 mM Tris-HCl, pH 7.8, containing 200 mM KCl, 2 mM EDTA, 2 mM dithiothreitol, 0.2% NP40, 40% glycerol and protease inhibitors (Complete Mini, EDTA-free protease inhibitors, Roche Diagnostics, Indianapolis, IN)(12) followed by incubation with rotation at 4 °C for 3 h. Extracts from adult fish ground in liquid nitrogen were prepared using the same protocol. Extracts were then dialyzed overnight against 50 mM HEPES, pH 7.4 containing 0.1 mM EDTA.
BER assay
The BER assay was performed as described by Singhal et al.(18) in a 50 μl volume. The reaction mix included 50 mM Hepes, pH 7.4; 5 mM MgCl2; 1 mM dithiothreitol; 0.1 mM EDTA; 2 mM ATP; 0.5 mM NAD; dATP, dTTP and dGTP at 20 μM each; 5 mM sodium phosphocreatine; 10 units of creatine phosphokinase; 120 nM of duplex oligonucleotide; 40 μM of the fourth dNTP; and 10 μg (protein) of crude cell extract unless indicated otherwise. Reactions were incubated for 0.5 – 60 min at 22 °C and stopped by addition of EDTA to a final concentration of 83 mM. DNA was extracted with phenol-chloroform and precipitated with three volumes of chilled ethanol. Substrates and products were then resolved by means of denaturing polyacrylamide gel electrophoresis employing a 15% gel in the presence of 7 M urea and observed using phosphorImager analysis (19, 20).
In all cases we used a 45-mer double stranded oligonucleotide with a U/G mismatch at position 21 (19, 20). When uracil DNA glycosylase activity (Reaction 1) alone was measured, Substrate I, end-labeled at the 5′ end by means of polynucleotide kinase (New England Biolabs, Beverly MA) and [γ32P]ATP, was used, Mg2+ was replaced with 4 mM EDTA, and the reaction was stopped by phenol extraction. The UDG inhibitor (Ugi) was the kind gift of Dr. S Bennett (U. Oregon). When Reactions 1 and 2 were measured, Substrate I was end-labeled at the 5′ end and dNTPs were omitted from the reaction mix. When AP endonuclease activity (Reaction 2) was measured individually, substrate was prepared as described (19-21). Briefly, the 5′ end-labeled ds 45-mer was treated with Ung (1 unit/100 pmole U residues) for 30 min at 37 °C after which the Ung was heat inactivated at 70 °C for 5 min and the oligonucleotide was allowed to reanneal by slowly cooling to room temperature. When Reactions 1-3 were measured, Substrate I, end-labeled at the 5′ end, was used and either dCTP alone or all 4 dNTPs were included in the reaction mix. When Reactions 3a or b were measured separately, Substrate 1 was not end-labeled. Instead, insertion of [α32P]dCTP was measured directly by using unlabeled double stranded 45-mer and dCTP (Reaction 3a) or all 4 dNTPs (Reactions 3a and 3b). Preliminary experiments determined the amount of extract protein and time required for measuring each step in the pathway.
Quantitative real-time polymerase chain reaction (qRTPCR)
Total RNA was isolated from 60 Danio rerio embryos at 3.5 hpf or from 100 mg of freeze-dried adult fish with TRIzol Reagent from Invitrogen (Carlsbad, California) according to vendor's instructions. The RNA precipitate was resuspended in 150 μL RNase-free water and stored at −20°C. The first cDNA strand was synthesized from 1 μg total RNA in a 20-μL reaction using the High Capacity cDNA Reverse Transcription Kit from Applied Biosystems (Warrington UK) according to the manufacturer's manual. The cDNA was stored at −20°C.
For qRTPCR, the synthesized cDNA above was diluted 1:100 in RNase-free water and 4 μL was added into a 20-μL qRTPCR reaction mix containing SYBR Green PCR master mix (Applied Biosystems), 2 pmol each forward and reverse DNA primers, and water, then quantified in real time with an ABI PRISM® 7000 Sequence Detection System (Applied Biosystems) programmed thus: 1 cycle of 50°C for 2 min; 1 cycle of 95°C for 10 min; 40 cycles of (94°C for 15 s; 55°C for 30 s; 68°C for 60 s); 1 cycle of 68°C for 7 min, and 1 cycle of 4°C for 2 min. Dissociation curves were generated to ensure only one PCR product was obtained. Additionally, twelve PCR products were verified independently to contain only one product by agarose gel electrophoresis and ethidium bromide staining.
The messenger RNAs of the following genes were quantified by means of primers listed in Table 1: apex1, apex2, bactin2, aprt, ef1a, and gapdh. All measures of mRNA quantity were performed in triplicate and the data were normalized to gapdh as the other genes gave ΔCT > 4 indicating a greater than 16-fold differential in the level of expression of those genes in the 3.5 hpf embryo compared to the adult fish. Gapdh is preferred over bactin2 for normalization of gene expression in zebrafish (22).
Table 1. Selected primers for quantitative real-time PCR.
| apex1for | CTATGGCATTGGTAAAGAGGA |
| apex2_1for | TTGGTCAAGATGGACTTCAG |
| apex2_2for | CCAGCATCACTTCATTGGTC |
| bactin2for | ATGAAGATCAAGATCATTGCCC |
| gapdh1for | GTTGGTATTAACGGATTCGGTC |
| aprt1for | AGAACTTTCCCTGATTTCCCT |
| ef1a1for | TACTTCTCAGGCTGACTGTG |
| apex1rev | GGAAATCCACATCCCAAGTC |
| apex2_1rev | CAAAGTTATCCACATCATCTGG |
| apex2_2rev | GATGATCACATGACTCCCTG |
| bactin2rev | TAACAGTCCGTTTAGAAGCA |
| gapdh1rev | ACCTCACCCTTGTACTTTCC |
| aprt1rev | AGCATCAAGTCCCACTATGAG |
| ef1a1rev | TCTTCTTGATGTATGCGCTG |
Western blots
Western blots were performed as described using rabbit polyclonal antibody prepared against ZAP1 in this laboratory (16), mouse monoclonal anti Arabidopsis β-actin (GeneTex, Inc., San Antonio TX), mouse monoclonal anti rat pol-β (Thermo Fisher Scientific, Freemont CA) or rabbit polyclonal antibody prepared by 21rst Century Biochemicals, (Marlboro MA) against a peptide comprising amino acids 324 - 339 (FEYIQWKYREPKDRSE) of zebrafish pol-β.
Results
Extracts from eggs and adult fish are capable of performing BER
The individual steps in the BER pathway are outlined in Figure 1, which also details the substrates used to measure the entire pathway or individual steps. The choice of a substrate with a U/G mispair restricts the results to a uracil glycosylase-initiated BER cascade and minimizes the likelihood of confounding the data with pathways involving polynucleotide kinase and NEIL1 (23). In the first series of experiments we investigated whether extracts from eggs and adults could cleave a 45-mer ds substrate containing a U/G mispair at position 21 (Substrate I) and then insert the correct base opposite the orphan G residue. Loss of the 45-mer substrate and appearance of the 20-mer product are shown in Figure 2, Panels A and C. Cleavage represents both removal of the uracil residue (Reaction 1) and also cleavage by ZAP1 (Reaction 2). The same experiment was also performed in the presence of dCTP (Panels B and D), which is incorporated into the cleaved upstream strand (Reaction 3a). Appearance of the 21-mer in the presence of dCTP represents the first two reactions followed by insertion of deoxycytidine (21-mer, Reaction 3a). Extracts from both eggs and adults (10 μg protein from extract) were capable of removing uracil from a U-containing double stranded 45-mer, cleaving the resulting AP-site and inserting a deoxycytidine opposite the orphan G. However, the rate of cleavage followed by insertion was less in eggs than in adults. Since the level of exonuclease activity increased markedly after 15 min, subsequent assays were performed over 15 min intervals or less. Examination of individual reactions provided the rationale for the differences, as shown in the following sections.
Figure 2. BER in extracts from adult fish is more efficient than in extracts from eggs.
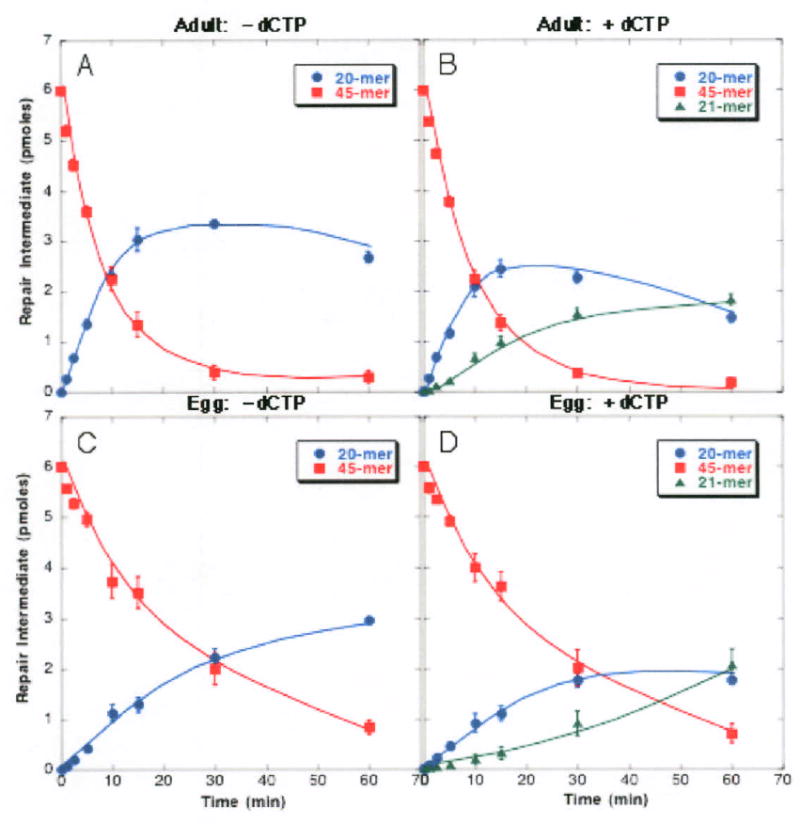
Extract (10 μg protein) from either adult fish (panels A and B) or unfertilized eggs (panels C and D) was mixed with Substrate I (45-mer containing a U/G mispair at position 21) in the absence (panels A and C) or presence of dCTP (panels B and D) for the length of time indicated. Substrate and products were resolved by denaturing polyacrylamide gel electrophoresis in the presence of urea and visualized by phosphorImager analysis. These data represent an average of three experiments +/- SE.
Extracts from eggs, embryos at different stages of early development and adults recognize and remove a U residue in DNA by means of uracil DNA glycosylase (UDG)
Cleavage of the initial 45-mer requires successive activities of a uracil DNA glycosylase (UDG) and an AP endonuclcase. To understand the slower cleavage rate on the part of egg extracts compared to extracts from adult fish, we examined each step individually. To observe the appearance of the AP-site in the substrate, we added EDTA to the reaction mix to a final concentration of 4 mM to inhibit divalent cation-dependent reactions that might cleave the AP-site, and NaBH4, which stabilizes the AP site as it forms but does not inhibit UDG or interfere with subsequent endonuclease cleavage (21, 24). In order to confirm that the uracil-removal was due to UDG, we examined to what degree Ugi (25) was able to inhibit the reaction and also the degree of enhancement on a single stranded substrate, which is a characteristic of UDG. We then subjected the purified products to cleavage with recombinant human AP endonuclease, which does not cleave the substrate unless an AP site is present. Figure 3 illustrates that extracts from all stages of development were able to remove uracil from the 45-mer and that the activity was entirely inhibited by Ugi. Uracil removal in embryos and eggs was ∼60% more efficient on a single-stranded substrate than on a ds substrate, which is characteristic of UDG (26). Extracts from adult fish, however, were not more active on a single-stranded substrate, which could imply the presence of additional Ugi-sensitive glycosylases. Alternatively, there may exist additional nucleases active on a single-stranded oligonucleotide in extracts from adult fish that are not present in eggs and early embryos. Since the rate of removal of uracil from a ds substrate in egg extracts was about half that in adult extracts, it was likely that part of the diminished activity in BER steps 1 and 2 seen in egg extracts was due to diminished processing of substrate at Step 1 in egg extracts.
Figure 3. Uracil removal is performed by uracil DNA glycosylase, as shown by complete sensitivity to the UDG inhibitor Ugi.
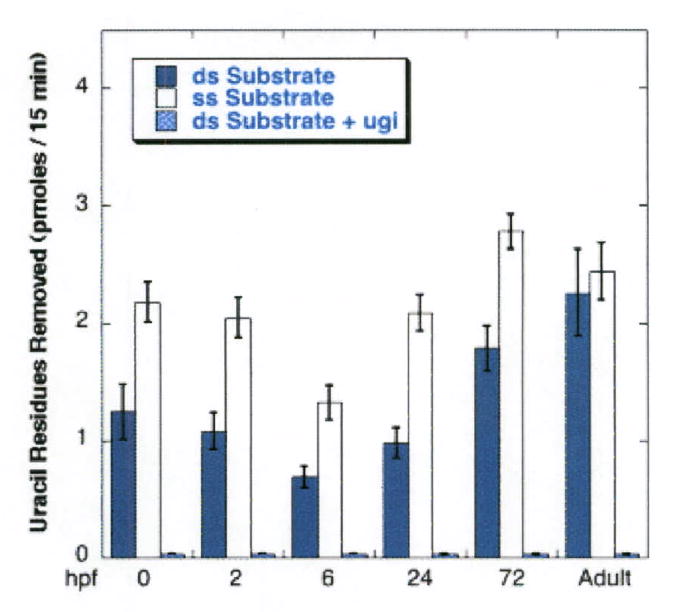
Extracts of eggs (0) and developing embryos at the indicated stages were incubated with 5′end-labeled 45-mer substrate in the absence of added Mg 2+ and the presence of 4 mM EDTA for 15 min. Substrate was either double stranded (blue bars) or single stranded, lacking its complement (white bars). Double stranded substrate was also incubated in the presence of the UDG inhibitor, Ugi. The reaction was stopped by phenol extraction. After ethanol precipitation and resuspension in TE buffer, the samples were treated with human AP endonuclease to cleave AP sites formed by the removal of uracil. There was no degradation of control substrate incubated in the absence of extract and processed in the same fashion.
Extracts from eggs, embryos at different stages of early development and adults cleave an AP-site efficiently
ZAP1 is present in eggs and throughout development (16). To avoid complications that might arise from reactions preceding the cleavage of the AP-site, we used a substrate with the same reduced AP site employed for examining the kinetics of human AP endonuclease (19, 20). This substrate allowed us to answer questions about the efficiency of AP-site cleavage by extracts from eggs, embryos and adults. The time course was linear for ∼ 2 min when 2 μg protein from either adult or egg extracts was added to the mix, as shown in Figure 4A. Using the 1 min time interval, all extracts were able to cleave an AP-site containing substrate (Figure 4B). However, extracts from unfertilized eggs had about 1/4 the activity of extracts from adults, while embryos less than 72 hours after fertilization (hpf) had about half the activity of adult extracts. We showed earlier that the protein level of ZAP1 was constant throughout early development even as cellular proliferation continued (16). Nevertheless, the diminution in efficiency of the first three steps in the BER pathway in extracts from eggs as opposed to adults was due to diminished entry into the pathway via UDG and less efficient AP endonuclease activity in extracts from the former.
Figure 4. AP site cleavage is inefficient in eggs and less efficient in pre-hatching embryos than at later times.
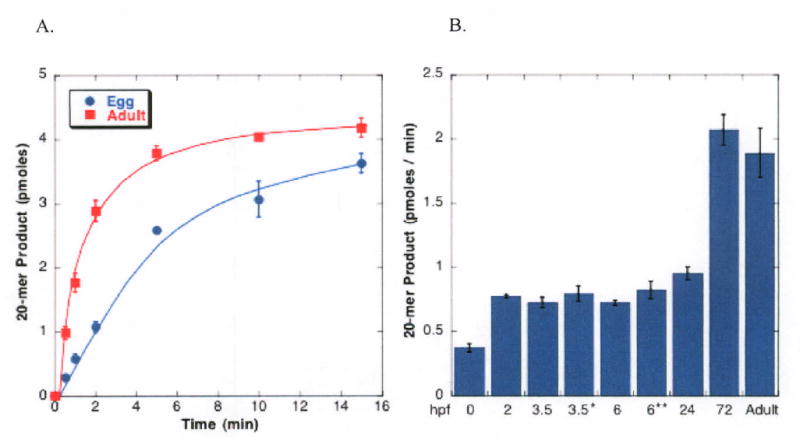
A. Extracts (2 μg protein) from eggs or adult fish were incubated for the indicated time in the presence of 5 mM Mg2+ with 5′-end labeled substrate that had been treated with Ung to remove uracil for varying time intervals (21). Reactions were stopped by addition of EDTA, and phenol extracted. Substrate and product were resolved by gel electrophoresis. AP site cleavage was linear for ∼2 min under these conditions. The rate of AP site cleavage was then examined in extracts (2 μg protein) obtained from eggs (●) or adult fish (■). B. AP site cleavage by eggs, embryos and adult fish (2 μg protein) over a 1 min interval. *: 3.5 hpf embryos in which ZAP1 has been reduced by 74-90 % by microinjection of 0.5 mM TS-MO at the 2-4 cell stage; **: extracts from 6.5 hpf embryos in which ZAP1 has been reduced by 40-56 % by microinjection of 0.2 mM TS-MO. These data are the average of three experiments +/- SE.
Knockdown of ZAP1 levels does not alter BER in early embryogenesis
Microinjection of MO into early embryos is a standard way of knocking down protein levels in zebrafish (17). The degree of knockdown varies with the amount of microinjected MO. In this series of experiments we knocked down ZAP1 levels by 74% or 54%, as shown by Western blot analysis (data not shown) and examined both the ability to cleave an AP-site (Figure 4, 3.5* and 6**) and the time course of the first three steps in BER (Figure 5). Extracts were prepared from control and full knockdown embryos just after the MBT at 3.5 hpf (3.5*); extracts were also prepared from control and hypomorphic embryos at 6.5 hpf (6**) several hours after the MBT. Despite loss of much of ZAP1 protein in full knockdowns and somewhat less in hypomorphs, we saw no difference in the first three steps of BER in comparison with controls nor was there any change in the ability to cleave an AP site.
Figure 5. Time dependent processing of 45-mer substrate is not altered by diminution of ZAP1.

These experiments were performed as described in the legend to Figure 2 except that extracts were obtained from 3.5 hpf control or full knockdown (FKD) embryos.
Another AP endonuclease is expressed in very early stage embryos
Since suppression of translation of ZAP1 did not result in complete loss of the protein, there may still have been sufficient ZAP1 in hypomorphs and full knockdown embryos to provide endonuclease activity for BER. Nevertheless, the possibility remained that another AP endonuclease might be expressed during development. A potential candidate is AP endonuclease 2, whose enzymatic activity in humans is 1% that of its paralogue (27, 28) but which is involved in processing of AP sites during immunoglobulin class switching (29, 30). Therefore, we chose to examine whether AP endonuclease 2 might be expressed duing early embryological developmental stages. Using gapdh as the standard of normalization, qRT-PCR revealed that both AP endonucleasel and 2 were expressed in early stage embryos at a level 13 and 20-fold that of adults respectively (Figure 6). Therefore, cleavage of an AP-site by extracts of unfertilized eggs and early embryos could be the result of both proteins.
Figure 6. AP endonuclease 2 may serve a redundant function for AP endonuclease 1 during very early development.
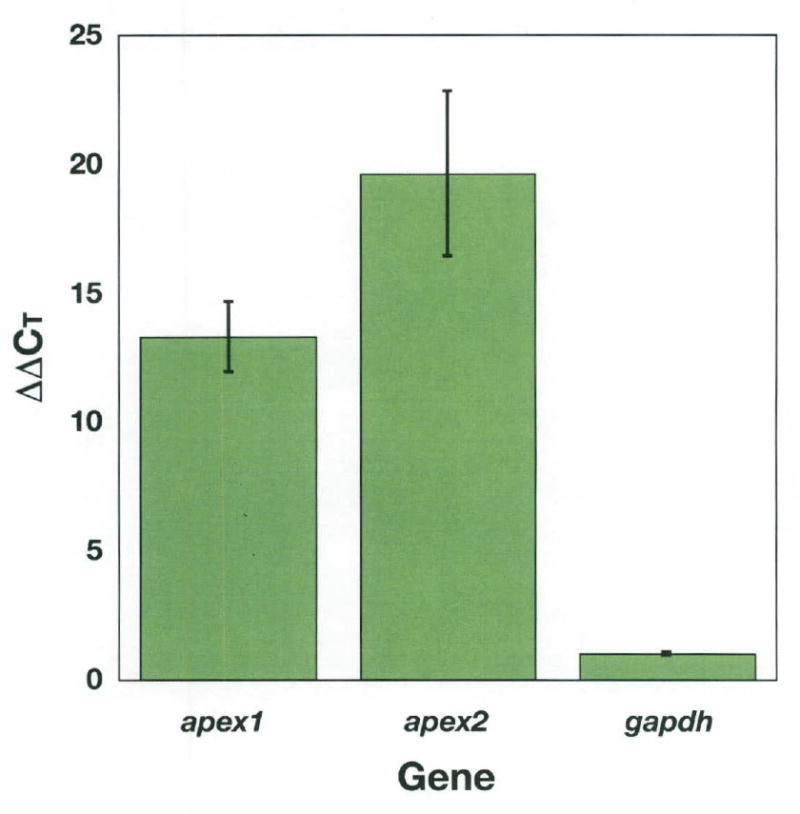
qRTPCR was performed to examine expression levels of apex 1, apex 2 and gapdh in early stage, untreated embryos (3.5 hpf). ΔCT values were calculated by normalizing to gapdh for the embryo and adult fish. Relative gene expression (plotted), ΔΔCT, was calculated by subtracting the ΔCT from the the ΔCT of the embryo for apex1, apex 2, and gapdh. Data are means of triplicate values calculated from at least one primer pair of each gene over two independent experimental runs. Error bars indicate standard error of the mean.
Before hatching at 72 hpf, replicative polymerases predominate in BER
Po1-β is the preferred polymerase for insertion of a single nucleotide during BER of cultured cells and tissues from adult animals (12, 13, 18). During long patch repair, po1-β, the replicative polymerases α, and δ/ε together with PCNA or one of several bypass polymerases may participate (31-35). The presence in the reaction mix of the single nucleotide dCTP limited the reaction to short patch repair, whereas the presence of all four dNTPs enabled progression of the alternative long patch pathway. In order to examine the preferred polymerases during short patch (single nucleotide insertion) and long patch (insertion of 2-6 nucleotides) BER during development, we examined whether aphidicolin or dideoxyCTP could inhibit nucleotide insertion. Inhibition by aphidicolin is diagnostic for participation of the replicative polymerases α, δ and ε (36), while inhibition by ddCTP is considered diagnostic for po1-β (18) (Figures 7 and 8). Because po1-β has no proofreading ability, it readily inserts but cannot remove ddCTP, which then prevents further chain elongation or ligation.
Figure 7. The bulk of short patch repair before hatching (72 hpf) is mediated by replicative polymerases.

Extracts from eggs, embryos at various stages and adult fish were incubated in the presence or absence of aphidicolin (40 μM) with 45-mer substrate which had been treated with Ung and human AP endonuclease to generate the free 3′ hydroxyl group capable of accepting a new incoming nucleotide. The incubation medium included [α32P]dCTP. Insertion of dCTP was measured after phenol extraction and resolution of substrate and product by denaturing gel electrophoresis in the presence of 7 M urea. Average of two experiments +/- range. Solid bars: - aphidicolin; open bars: + aphidicolin.
Figure 8. Unlike synthesis in adult extract, both short patch and long patch insertion in egg extracts is mediated by replicative polymerases.
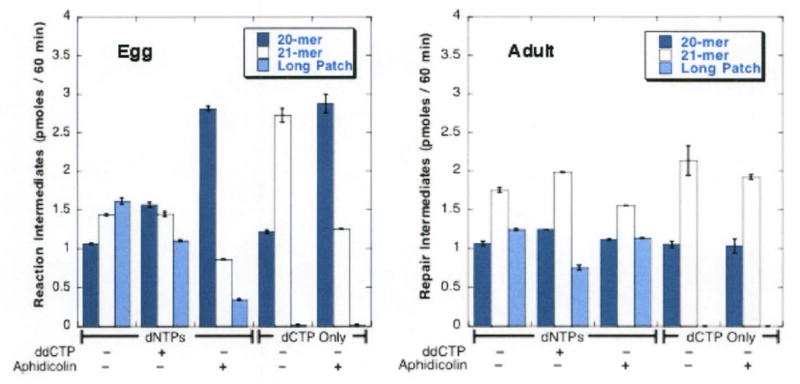
For this series of experiments, substrate was 45-mer oligonucleotide end-labeled at the 5′ end of the U-containing strand. Extracts from unfertilized eggs or adult fish were assayed in the presence or absence of 40 μM aphidicolin or 400 μM dideoxyCTP for products formed over a 60-min interval in the presence of all 4 dNTPs or dCTP only in the presence or absence of aphidicolin or dideoxyCTP. Dark blue bars: 20-mer; clear bars: 21-mer; light blue bars: total 22-45-mer.
In extracts from egg and early stages of development single nucleotide insertion was far more sensitive to inhibition by aphidocolin than in extracts from 24 hpf or older embryos and adults (Figure 7). Neither the ability to insert a single nucleotide nor the sensitivity to aphidicolin was altered by apex1 knockdown. By 24 hpf, aphidicolin's ability to reduce insertion had decreased. At this time the drug decreased incorporation by only 33% from control values, indicating a shift towards insertion by a polymerase resistant to aphidicolin. After hatching (3 dpf) the rate of insertion even exceeded levels in extracts from adult fish. Finally, in extracts from adult fish aphidicolin was able to decrease single nucleotide insertion by only ∼15%. This last level was consistent with aphidicolin's effect on BER (31, 37, 38) and with the participation of pol-β in both long and short patch repair (39) from a variety of adult tissues and from many cultured cell lines.
Comparison of repair intermediates by extracts obtained from adults and unfertilized eggs in the presence of all four dNTPs using a 5′-end labeled substrate enabled us to confirm and extend the results obtained in the presence of dCTP alone (Figure 8). Despite the presence of exonuclease activity, the incubation time was continued to 60 min in order to better measure long patch repair. Figure 7 illustrates the involvement of replicative polymerases in both short and long patch repair as shown by sensitivity to aphidicolin in extracts from unfertilized eggs but not in extracts from adult fish. Figure 7 also shows the relative insensitivity to dideoxyCTP of BER by egg extracts, as shown by a lower 21-mer to long patch product ratio. These data confirm the surprising lack of involvement of pol-β in BER performed by extracts from eggs.
Pol-β protein does not appear until after the midblastula transition
The use of aphidicolin-dependent polymerases in BER in the unfertilized egg and very early embryogenesis could be due to replicative polymerases outcompeting pol-β for the cleaved AP site or to a lack of pol-β. Therefore, we performed Western blot analysis on embryonic extracts obtained from unfertilized eggs and from embryos at different stages of development. Figure 9 represents data showing that pol-β was not detectable in unfertilized eggs and very early stage embryos. However, the amount of pol-β relative to β-actin reached adult levels by 13 hpf.
Figure 9. Pol-β is not detected in unfertilized eggs and very early stage embryos.
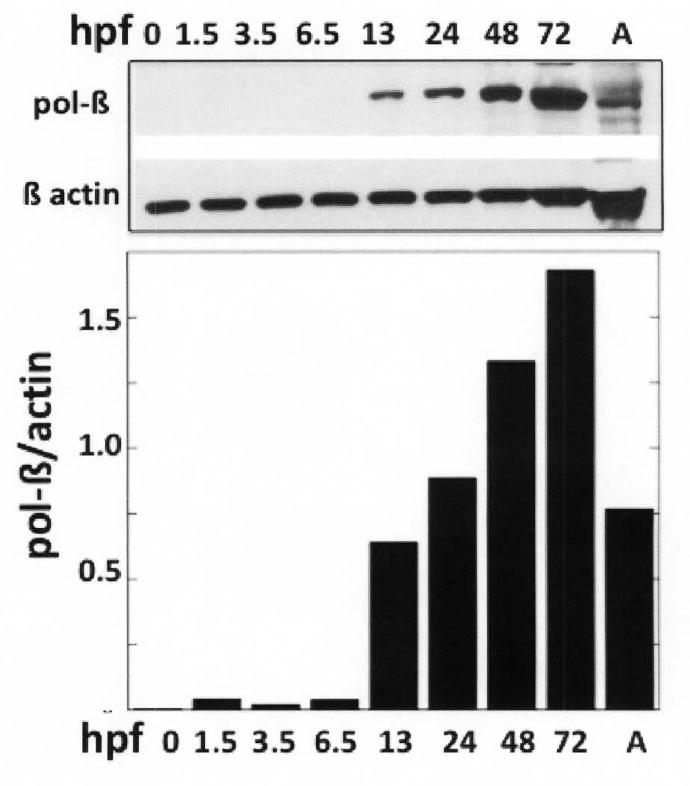
Protein (100 μg/lane) in extracts of unfertilized eggs and embryos at various stages was resolved by SDS-PAGE, transferred to membranes and probed with mouse anti-rat pol-β. The same extracts (40 μg/lane) were used for preparing membranes probed with mouse anti β actin. The ratio of pol-β/β-actin at each time is shown in the chart. Time after fertilization when embryos were harvested (hpf) is indicated above the Western blot and below each lane in the bar graph. A, extract prepared from adult fish.
Discussion
Here we have presented the first data on the first three steps of the BER pathway in early zebrafish embryogenesis. Cell-free extracts from unfertilized eggs and early stage embryos as well as from adult fish are capable of recognizing and removing uracil in a U/G mispair by uracil DNA glycosylase, cleaving the resulting AP-site, and inserting the correct cytosine residue either during short patch or long patch repair. The overall pathway is less efficient in extracts from unfertilized eggs and early stage embryos. At the glycosylase step and the endonuclease step, extracts from unfertilized eggs and very early stage embryos have 1/4-1/2 the enzymatic activity of UDG and ZAP1 found in extracts from adult fish, which contributes to the lower rate of deoxynucleotide insertion in short patch synthesis. The difference lessens at ∼3 dpf when hatching has occurred. This observation is consistent with the role that BER plays in repairing oxidatively damaged DNA. After hatching, the embryo is no longer protected by the chorionic membrane and must be prepared for exposure to whatever conditions it encounters in the environment.
Once a zebrafish egg is fertilized, it undergoes ten rapid division cycles to form the blastula in less than 3 hours (17). The preponderance of replicative polymerases in both short and long patch repair in eggs and very early stage embryos can be explained by the lack of pol-β and large amounts of replicative polymerases stored in preparation for the rapid cell division that follows fertilization. At the MBT cell division slows, zygotic transcription begins, spatial differentiation arises and the cells become motile. The egg stockpiles all the components necessary for the initial rapid cell division including DNA polymerases and accessory proteins, dNTPs and the mitochondrial systems to regenerate the required ATP. However, ATP generation is inevitably accompanied by production of ROS with the concomitant damage to DNA (1-3). Therefore, the BER pathway is an important component in early embryogenesis.
The reduction in ZAP1 levels by 74% in full knockdown embryos leads to little, if any, reduction in BER activity. Although the remaining protein might be sufficient to provide endonuclease cleavage, there is still the possibility that a backup activity is present in eggs and early stage embryos. To our surprise, there is 20 times the apex2 message in early stage embryos as in adults. In fact, mRNA of both apex genes is expressed at higher levels in the early stage embryos in comparison to adult fish. Thus, AP endonuclease 2 with less than 1% the activity of AP endonuclease 1 on a molar basis in vitro may play an important role in early embryological development. The former has recently been shown to participate in processing of AP sites during class switching in lymphocyte maturation (29).
Finally, although pol-β message is expressed in early development (XJ Yang, Unpublished results), pol-β protein appears in detectable amounts only after 6.5 hpf. The aphidicolin-sensitive replicative polymerase activity that provide nucleotide insertion during BER in unfertilized eggs and early stage embryos gradually diminishes to levels found in adult tissues. While the involvement of replicative polymerases in BER could be enhanced or promoted by the presence of a factor that blocks pol-β access much like the APC protein (40), the tight coordination inherent in single nucleotide BER through the interaction of pol-β, DNA and AP endonuclease seen in adult tissues (41, 42) is abrogated by the lack of pol-β in very early embryonic development.
In short, BER activity in eggs and early stage zebrafish embryos has several unexpected features consistent with rapid cellular proliferation before differentiation begins, most notably the use of aphidicolin-sensitive replicative polymerases in both short and long patch repair and the presence of standby AP endonuclease activity. Once differentiation is initiated, the pathway gradually evolves to the one found in cultured cells and adult tissues. By the time the embryo hatches from the chorionic membrane and is fully exposed to environmental conditions, the transformation is complete.
Acknowledgments
The authors are grateful to Dr. Samuel Bennett for the gift of the uracil DNA glycosylase inhibitor, Ugi.
Abbreviations
- AP
abasic site
- BER
base excision repair
- dpf
days post fertilization
- ds
double strand
- hpf
hours post fertilization
- MBT
midblastula transition
- MO
morpholino oligonucleotide
- pol-β
DNA polymerase-β
- qRTPCR
quantitative real-time polymerase chain reaction
- ROS
reactive oxygen species
- TS-MO
morpholino oligonucleotide directed against the translation start site
- UDG
eukaryotic uracil DNA glycosylase
- Ung
prokaryotic uracil DNA glycosylase
- ZAP1
zebrafish AP endonuclease 1 protein
References
- 1.Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci STKE. 2007;2007:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- 2.Semenza GL. Life with oxygen. Science. 2007;318:62–64. doi: 10.1126/science.1147949. [DOI] [PubMed] [Google Scholar]
- 3.Dedon PC, Tannenbaum SR. Reactive nitrogen species in the chemical biology of inflammation. Arch Biochem Biophys. 2004;423:12–22. doi: 10.1016/j.abb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 4.Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci U S A. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meira LB, Devaraj S, Kisby GE, Burns DK, Daniel RL, Hammer RE, Grundy S, Jialal I, Friedberg EC. Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Res. 2001;61:5552–5557. [PubMed] [Google Scholar]
- 6.Tebbs RS, Thompson LH, Cleaver JE. Rescue of Xrccl knockout mouse embryo lethality by transgene-complementation. DNA Repair (Amst) 2003;2:1405–1417. doi: 10.1016/j.dnarep.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Kucherlapati M, Yang K, Kuraguchi M, Zhao J, Lia M, Heyer J, Kane MF, Fan K, Russell R, Brown AM, Kneitz B, Edelmann W, Kolodner RD, Lipkin M, Kucherlapati R. Haploinsufficiency of Flap endonuclease (Fenl) leads to rapid tumor progression. Proc Natl Acad Sci U S A. 2002;99:9924–9929. doi: 10.1073/pnas.152321699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bentley D, Selfridge J, Millar JK, Samuel K, Hole N, Ansell JD, Melton DW. DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nat Genet. 1996;13:489–491. doi: 10.1038/ng0896-489. [DOI] [PubMed] [Google Scholar]
- 9.Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 10.Sugo N, Aratani Y, Nagashima Y, Kubota Y, Koyama H. Neonatal lethality with abnormal neurogenesis in mice deficient in DNA polymerase beta. Embo J. 2000;19:1397–1404. doi: 10.1093/emboj/19.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horton JK, Prasad R, Hou E, Wilson SH. Protection against methylation-induced cytotoxicity by DNA polymerase beta-dependent long patch base excision repair. J Biol Chem. 2000;275:2211–2218. doi: 10.1074/jbc.275.3.2211. [DOI] [PubMed] [Google Scholar]
- 12.Horton JK, Baker A, Berg BJ, Sobol RW, Wilson SH. Involvement of DNA polymerase beta in protection against the cytotoxicity of oxidative DNA damage. DNA Repair (Amst) 2002;1:317–333. doi: 10.1016/s1568-7864(02)00008-3. [DOI] [PubMed] [Google Scholar]
- 13.Horton JK, Joyce-Gray DF, Pachkowski BF, Swenberg JA, Wilson SH. Hypersensitivity of DNA polymerase beta null mouse fibroblasts reflects accumulation of cytotoxic repair intermediates from site-specific alkyl DNA lesions. DNA Repair (Amst) 2003;2:27–48. doi: 10.1016/s1568-7864(02)00184-2. [DOI] [PubMed] [Google Scholar]
- 14.Fung H, Demple B. A vital role for apel/refl protein in repairing spontaneous DNA damage in human cells. Mol Cell. 2005;17:463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 15.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2. ASM Press; Washington, DC: 2006. [Google Scholar]
- 16.Wang Y, Shupenko CC, Melo LF, Strauss PR. DNA repair protein involved in heart and blood development. Mol Cell Biol. 2006;26:9083–9093. doi: 10.1128/MCB.01216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westerfield M. The Zebrafish Book. 3rd. University of Oregon Press; Eugene, OR: 1995. [Google Scholar]
- 18.Singhal RK, Prasad R, Wilson SH. DNA polymerase beta conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J Biol Chem. 1995;270:949–957. doi: 10.1074/jbc.270.2.949. [DOI] [PubMed] [Google Scholar]
- 19.Lucas JA, Masuda Y, Bennett RA, Strauss NS, Strauss PR. Single-turnover analysis of mutant human apurinic/apyrimidinic endonuclease. Biochemistry. 1999;38:4958–4964. doi: 10.1021/bi982052v. [DOI] [PubMed] [Google Scholar]
- 20.Mundle ST, Fattal M, Melo LM, Strauss PR. A novel mechanism for human AP endonuclease 1. DNA Repair (Amst) 2004;3:1447–1455. doi: 10.1016/j.dnarep.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Strauss PR, Beard WA, Patterson TA, Wilson SH. Substrate binding by human apurinic/apyrimidinic endonuclease indicates a Briggs-Haldane mechanism. J Biol Chem. 1997;272:1302–1307. doi: 10.1074/jbc.272.2.1302. [DOI] [PubMed] [Google Scholar]
- 22.Pei DS, Sun YH, Chen SP, Wang YP, Hu W, Zhu ZY. Zebrafish GAPDH can be used as a reference gene for expression analysis in cross-subfamily cloned embryos. Anal Biochem. 2007;363:291–293. doi: 10.1016/j.ab.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Mol CD, Arvai AS, Sanderson RJ, Slupphaug G, Kavli B, Krokan HE, Mosbaugh DW, Tainer JA. Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell. 1995;82:701–708. doi: 10.1016/0092-8674(95)90467-0. [DOI] [PubMed] [Google Scholar]
- 25.Bennett SE, Schimerlik MI, Mosbaugh DW. Kinetics of the uracil-DNA glycosylase/inhibitor protein association. Ung interaction with Ugi, nucleic acids, and uracil compounds. J Biol Chem. 1993;268:26879–26885. [PubMed] [Google Scholar]
- 26.Chen CY, Mosbaugh DW, Bennett SE. Mutations at Arginine 276 transform human uracil-DNA glycosylase into a single-stranded DNA-specific uracil-DNA glycosylase. DNA Repair (Amst) 2005;4:793–805. doi: 10.1016/j.dnarep.2005.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hadi MZ, Wilson DM., 3rd Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III. Environ Mol Mutagen. 2000;36:312–324. [PubMed] [Google Scholar]
- 28.Burkovics P, Szukacsov V, Unk I, Haracska L. Human Ape2 protein has a 3′-5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res. 2006;34:2508–2515. doi: 10.1093/nar/gkl259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guikema JE, Linehan EK, Tsuchimoto D, Nakabeppu Y, Strauss PR, Stavnezer J, Schrader CE. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J Exp Med. 2007;204:3017–3026. doi: 10.1084/jem.20071289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dan Y, Ohta Y, Tsuchimoto D, Ohno M, Ide Y, Sami M, Kanda T, Sakumi K, Nakabeppu Y. Altered gene expression profiles and higher frequency of spontaneous DNA strand breaks in APEX2-null thymus. DNA Repair (Amst) 2008;7:1437–1454. doi: 10.1016/j.dnarep.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Dogliotti E, Fortini P, Pascucci B, Parlanti E. The mechanism of switching among multiple BER pathways. Prog Nucleic Acid Res Mol Biol. 2001;68:3–27. doi: 10.1016/s0079-6603(01)68086-3. [DOI] [PubMed] [Google Scholar]
- 32.Fortini P, Pascucci B, Parlanti E, Sobol RW, Wilson SH, Dogliotti E. Different DNA polymerases are involved in the short- and long-patch base excision repair in mammalian cells. Biochemistry. 1998;37:3575–3580. doi: 10.1021/bi972999h. [DOI] [PubMed] [Google Scholar]
- 33.Bennett RA. The Saccharomyces cerevisiae ETH1 gene, an inducible homolog of exonuclease III that provides resistance to DNA-damaging agents and limits spontaneous mutagenesis. Mol Cell Biol. 1999;19:1800–1809. doi: 10.1128/mcb.19.3.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Auerbach P, Bennett RA, Bailey EA, Krokan HE, Demple B. Mutagenic specificity of endogenously generated abasic sites in Saccharomyces cerevisiae chromosomal DNA. Proc Natl Acad Sci U S A. 2005;102:17711–17716. doi: 10.1073/pnas.0504643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petta TB, Nakajima S, Zlatanou A, Despras E, Couve-Privat S, Ishchenko A, Sarasin A, Yasui A, Kannouche P. Human DNA polymerase iota protects cells against oxidative stress. Embo J. 2008;27:2883–2895. doi: 10.1038/emboj.2008.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamada K, Itoh R. Involvement of DNA polymerase delta and/or epsilon in joining UV-induced DNA single strand breaks in human fibroblasts (comparison of effects of butylphenyldeoxyguanosine with aphidicolin) Biochim Biophys Acta. 1994;1219:302–306. doi: 10.1016/0167-4781(94)90052-3. [DOI] [PubMed] [Google Scholar]
- 37.Stucki M, Pascucci B, Parlanti E, Fortini P, Wilson SH, Hubscher U, Dogliotti E. Mammalian base excision repair by DNA polymerases delta and epsilon. Oncogene. 1998;17:835–843. doi: 10.1038/sj.onc.1202001. [DOI] [PubMed] [Google Scholar]
- 38.Parlanti E, Pascucci B, Terrados G, Blanco L, Dogliotti E. Aphidicolin-resistant and -sensitive base excision repair in wild-type and DNA polymerase beta-defective mouse cells. DNA Repair (Amst) 2004;3:703–710. doi: 10.1016/j.dnarep.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 39.Dianov GL, Prasad R, Wilson SH, Bohr VA. Role of DNA polymerase beta in the excision step of long patch mammalian base excision repair. J Biol Chem. 1999;274:13741–13743. doi: 10.1074/jbc.274.20.13741. [DOI] [PubMed] [Google Scholar]
- 40.Kundu CN, Balusu R, Jaiswal AS, Narayan S. Adenomatous polyposis coli-mediated hypersensitivity of mouse embryonic fibroblast cell lines to methylmethane sulfonate treatment: implication of base excision repair pathways. Carcinogenesis. 2007;28:2089–2095. doi: 10.1093/carcin/bgm125. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Prasad R, Beard WA, Kedar PS, Hou EW, Shock DD, Wilson SH. Coordination of steps in single-nucleotide base excision repair mediated by apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta. J Biol Chem. 2007;282:13532–13541. doi: 10.1074/jbc.M611295200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abyzov A, Uzun A, Strauss PR, Ilyin VA. An AP endonuclease 1-DNA polymerase beta complex: theoretical prediction of interacting surfaces. PLoS Comput Biol. 2008;4:e1000066. doi: 10.1371/journal.pcbi.1000066. [DOI] [PMC free article] [PubMed] [Google Scholar]


