Abstract
BH3 domains were originally discovered in the context of apoptosis regulators and they the mediate binding of proapoptotic Bcl-2 family members to antiapoptotic Bcl-2 family members. Yet, recent studies indicate that BH3 domains do not function uniquely in apoptosis regulation; they also function in the regulation of another critical pathway involved in cellular and tissue homeostasis called autophagy. Antiapoptotic Bcl-2 homologs downregulate autophagy through interactions with the essential autophagy effector and haploinsufficient tumor suppressor, Beclin 1. Beclin 1 contains a BH3 domain, similar to that of Bcl-2 proteins, which is necessary and sufficient for binding to antiapoptotic Bcl-2 homologs and required for Bcl-2-mediated inhibition of autophagy. This review will summarize the evidence that the BH3 domain of Beclin 1 serves as a key structural motif that enables Bcl-2 to function not only as an antiapoptotic protein, but also as an antiautophagy protein.
Keywords: Beclin 1, autophagy, BH3-only, apoptosis, Bcl-2, tumor suppressor
Introduction
Autophagy is the primary cellular pathway by which long-lived proteins, cytoplasmic organelles and intracellular pathogens undergo degradation. The pathway involves sequestration of these cellular constituents in double- or multimembrane cytoplasmic vesicles called autophagosomes, with subsequent delivery to the lysosome, where they are degraded and recycled (Klionsky and Emr, 2000; Levine and Klionsky, 2004). Autophagy promotes cellular survival by enabling cells to maintain macromolecular synthesis and energy homeostasis during nutrient deprivation and other forms of cellular stress; it also functions in differentiation and development, antiaging, innate and adaptive immunity, and tumor suppression (Levine and Klionsky, 2004; Shintani and Klionsky, 2004; Levine and Kroemer, 2008; Mizushima et al., 2008). The disruption of autophagy has been implicated in a wide variety of diseases including cancer, neurodegenerative disorders, skeletal and cardiac myopathies, cancer, inflammatory bowel disease and infectious diseases (Levine and Kroemer, 2008).
The autophagy pathway is conserved among all eukaryotes, and in the last decade many autophagy effectors (called Atg proteins) as well as major protein regulators have been identified (Levine and Klionsky, 2004; Xie and Klionsky, 2007). Regulators that induce autophagy include tumor suppressors, such as PTEN, TSC1 and TSC2 complexes, and DAPk; stress-activated signaling molecules, such as c-Jun N-terminal kinase 1 (JNK1), and those that respond to low energy (for example, AMP kinase) or endoplasmic reticulum (ER) stress (for example, PERK, eIF2α-kinase and IRE1), and molecules involved in innate immune signaling, such as toll-like receptors and immunity-related GTPases (Escalatine et al., 2009). Proteins that inhibit autophagy include oncogenes, such as class I phosphatidylinositol 3-OH kinase (PI3K), Akt, Ras, TOR and Bcl-2 (Pattingre and Levine, 2006; Maiuri et al., 2008). The tumor suppressor gene, p53, has been reported to play a dual role in autophagy (Levine and Abrams, 2008; Tasdemir et al., 2008b), with some studies suggesting transcription-dependent and transcription-independent positive regulation of autophagy (Feng et al., 2005; Crighton et al., 2006) and other studies suggesting the negative regulation of autophagy by wild type and mutant forms of p53 in the cytoplasm (Morselli et al., 2008; Tasdemir et al., 2008a, 2008c).
The molecular structures and mechanism(s) of action of the autophagy effectors have not been completely elucidated, but most function by participating in multi-protein complexes responsible for vesicle induction, nucleation, elongation, docking and fusion with the lysosome and degradation (Levine and Klionsky, 2004; Levine and Kroemer, 2008; Mizushima et al., 2008). Some autophagy regulators induce autophagy through the phosphorylation of key components of the induction complex, such as Atg1 and Atg13. The process by which targets are selected for autophagy is not understood, and may be different for different targets. Autophagy induction triggers the conversion of phosphatidylinositol to phosphatidylinositol 3-phosphate (PI3P) by the vesicle nucleation or class III PI3K complex, and induces the recruitment of Atg9 and associated mitochondrial lipids to the growing vesicle (He et al., 2008). PI3P serves as a signal to recruit proteins required for vesicle elongation (Obara and Ohsumi, 2008), which involves two ubiquitin-like conjugation pathways. One pathway results in the conjugation of Atg12 to Atg5 and the formation of the Atg12–Atg5–Atg16 complex, which then assists in the second pathway, resulting in the conjugation of phosphatidylethanolamine to Atg8/LC3, and the incorporation of this lipidated form of Atg8/LC3 into the growing phagophore. The docking and fusion of the completed autophagosome with the lysosome is directed by proteins, such as LAMP2 and Rab7, which are also involved in other vesicle trafficking-pathways. Ultimately, the autophagosome, along with its contents, is degraded by lysosomal hydrolases. The degraded contents are recycled by the cell to maintain macromolecular synthesis and energy homeostasis, which is important for survival during nutrient deprivation and other forms of cellular stress.
The essential autophagy effector, Beclin 1
Beclin 1, initially isolated as a Bcl-2-interacting protein (Liang et al., 1998), shares 30% sequence identity with yeast, Atg6/Vps30, which participates in a protein complex essential for autophagy in yeast (Kametaka et al., 1998). Beclin 1 was among the first mammalian autophagy effectors to be identified and along with the yeast Vps34 homolog, class III PI3K (Kihara et al., 2001), and the yeast Vps15 homolog, p150 (Panaretou et al., 1997), forms a complex responsible for autophagic vesicle nucleation in mammals (Aita et al., 1999; Liang et al., 1999; Kihara et al., 2001). The precise mechanism by which the Beclin 1/class III PI3K complex mediates vesicle nucleation is unclear.
The Beclin 1 yeast ortholog, Atg6/Vps30, was also independently discovered in a genetic screen for proteins involved in vacuolar protein sorting (Seaman et al., 1997), suggesting that Beclin 1 may also function in other class III PI3K-dependent membrane trafficking events. In a manner somewhat parallel to yeast, two distinct complexes, each of which contains class III PI3 K, p150 and Beclin 1, have recently been identified in mammalian cells (Itakura et al., 2008). One complex, considered the equivalent of the yeast complex I that is essential for autophagy, includes a human ortholog of yeast Atg14 that is required for autophagy. The other complex, considered the equivalent of the yeast complex II that functions in the vacuolar protein sorting pathway, includes ultraviolet irradiation resistance-associated gene (UVRAG), which is proposed to be the human ortholog of yeast, Vps38. Although Mizushima and colleagues (Itakura et al., 2008) found that the UVRAG-containing Beclin 1/class III PI3K complex is not required for autophagosome formation, these findings contradict an earlier report that UVRAG is required for autophagy (Liang et al., 2007). Further, Bif-1, which binds to UVRAG and regulates membrane curvature (Takahashi et al., 2007), and Ambra1, which binds to Beclin 1 in a complex containing UVRAG (Fimia et al., 2007), have each been shown to upregulate the autophagy function of the Beclin 1/class III PI3K complex. Thus, additional studies are needed to clarify the role of distinct human Atg14- and UVRAG-containing complexes in autophagy and other membrane trafficking processes.
Definitive evidence for autophagy-independent functions of Beclin 1 in mammalian cells is still lacking, and two studies showed that Vps34/class III PI3K-dependent trafficking events , such as the proteolytic processing of procathepsin D in route from the trans-Golgi network to lysosomes or the postendocytic sorting of the epidermal growth factor receptor were intact in cells without functional Beclin 1 (Furuya et al., 2005b; Zeng et al., 2006). However, in plants, Ohsumi and colleagues (Fujiki et al., 2007) postulated an autophagy-independent function of ATG6/VPS30, as ATG6/VPS30 mutants, unlike other Arabidopsis thaliana ATG gene mutants were defective in pollen germination. Furthermore, the embryonic phenotype of beclin 1 null mice is more severe than that of other autophagy gene-deficient mice (for example, Atg5−/−, Atg7−/− mice; Levine and Kroemer, 2008), suggesting that beclin 1-regulated processes other than autophagy may be essential in early mammalian development. Net, given the presence of distinct Beclin 1/class III PI3K complexes in mammalian cells, the pleiotropic effects of Beclin 1 orthologs in lower eukaryotes, and the differences between phenotypes of ATG6 or beclin 1 versus other mutant ATG genes in plants and mice, it seems likely that mammalian Beclin 1 also functions in other membrane-trafficking processes besides autophagy.
Despite these possible autophagy-independent functions of Beclin 1, the best-characterized function of Beclin 1 is its role in autophagy. The autophagy function of Atg6/Beclin 1 is highly conserved throughout eukaryotic evolution and is presumed to be important in mediating many of its biological effects. Genetic knockdown or knockout studies of beclin 1, or its orthologs in lower eukaryotes have revealed several important phenotypes, many of which are likely related to autophagy, as the loss of-function mutations of ATG6/beclin 1 are phenocopied by null mutations in other ATG genes. Like all yeast ATG genes, ATG6 is essential for survival during starvation and yeast sporulation (Levine and Klionsky, 2004). Like other plant ATG genes, A. thaliana or Nicotiana benthamiana beclin 1 is essential for the prevention of premature chlorosis and the restriction of programed cell death during the innate immune response (Liu et al., 2005; Patel and Dinesh-Kumar, 2008). Like other Caenorhabditis elegans ATG genes, bec-1 is essential for dauer development (Melendez et al., 2003); survival during starvation (Hars et al., 2007); lifespan extension because of caloric restriction, insulin- signaling pathway mutations or p53 mutations (Melendez et al., 2003; Hars et al., 2007; Jia and Levine, 2007; Hansen et al., 2008; Tavernarakis et al., 2008; Tόth et al., 2008); and embryonic development (Takacs-Vellai et al., 2005).
Studies in targeted mutant mice have revealed several important functions of beclin 1. Mice with biallelic loss of beclin 1 are early embryonically lethal (Qu et al., 2003; Yue et al., 2003), and mice with monoallelic loss of beclin 1 have an increased incidence of spontaneous tumorigenesis (Qu et al., 2003; Yue et al., 2003), display abnormal proliferation of mammary epithelial cells and germinal center B lymphocytes (Qu et al., 2003), and have increased susceptibility to neurodegeneration (Pickford et al., 2008) and desmin-related cardiomyopathy (Tannous et al., 2008). In humans, monoallelic deletions of beclin 1 are frequently observed in sporadic breast, ovarian and prostate carcinoma (Aita et al., 1999), and decreased Beclin 1 expression in the central nervous system is linked to susceptibility to Alzheimer’s disease (Pickford et al., 2008). Thus, taken together, evidence is emerging that Beclin 1 plays an essential role in tumor suppression, development, aging prevention, innate immunity, neuroprotection and cardioprotection.
An antiautophagic function for antiapoptotic Bcl-2 proteins
As noted, Beclin 1 was originally isolated as a Bcl-2-interacting protein (Liang et al., 1998), and several antiapoptotic members of the Bcl-2 family, such as Bcl-2 and Bcl-XL, as well as viral Bcl-2 homologs encoded by oncogenic γ-herpesviruses, such as Kaposi’s sarcoma-associated herpesvirus (KSHV) vBcl-2 and γHV68 M11, bind to Beclin 1 and are negative regulators of its autophagy function (Liang et al., 1998; Pattingre et al., 2005; Maiuri et al., 2007; Sinha et al., 2008; Ku et al., 2008a). For example, human MCF7 breast carcinoma cells that typically express low levels of Beclin 1 do not show starvation-induced upregulation of autophagy unless Beclin 1 is ectopically expressed. This Beclin 1-mediated rescue of starvation-induced autophagy in MCF7 cells is inhibited by the exogenous expression of human Bcl-2 and Bcl-XL (Liang et al., 1998; Pattingre et al., 2005), as well as viral Bcl-2 homologs, KSHV Bcl-2 (Liang et al., 1998; Pattingre et al., 2005) and γHV68 M11 (Sinha et al., 2008; Ku et al., 2008a). These results have now been extended to numerous cell lines in numerous studies, and thus, it appears that the over-expression of cellular and viral Bcl-2 homologs is a potent inhibitor of autophagy in diverse settings (Sinha et al., 2008; Ku et al., 2008a). Furthermore, endogenous Bcl-2 also likely regulates autophagy, as autophagy-competent Hela cells undergo a twofold increase in levels of starvation-induced autophagy upon Bcl-2 siRNA knockdown (Liang et al., 1998; Pattingre et al., 2005). Finally, enforced Bcl-2 expression can also inhibit autophagy in vivo, because transgenic mice that express Bcl-2 under the control of a cardiac-specific promoter exhibit decreased levels of starvation-induced autophagy in cardiomyocytes (Liang et al., 1998; Pattingre et al., 2005).
The antiautophagy function of Bcl-2 appears to be exerted at the ER. Although an initial report suggested that endogenous Beclin 1 localized entirely to the trans-Golgi network (Kihara et al., 2001), subsequent studies showed that endogenous Beclin 1, similar to ectopically expressed Beclin 1, also localizes to mitochondria and ER (Liang et al., 1998; Pattingre et al., 2005). Only ER-targeted Bcl-2, and not mitochondrial-targeted Bcl-2, can inhibit starvation-induced autophagy, suggesting that only the interaction of Bcl-2 and Beclin 1 at the ER negatively regulates autophagosome formation. At least in certain cell types, such as HT-29 human colon carcinoma cells, enforced Bcl-2 expression decreases the amount of class III PI3K/Vps34 that coimmuno-precipitates with Bcl-2 and decreases Beclin 1-associated class III PI3K activity (Liang et al., 1998; Pattingre et al., 2005). It is not yet known how the ER-localized Bcl-2–Beclin 1 complex regulates the formation and/or activity of the Beclin 1/class III PI3K complex.
The binding of endogenous Bcl-2 antiapoptotic homologs to Beclin 1 may regulate basal autophagy levels and cell survival. In multiple different cell lines, including MCF7 cells that lack a functional caspase 3, the enforced expression of wild-type Beclin 1 does not increase basal levels of autophagy or induce cell death (Liang et al., 1998; Pattingre et al., 2005). Yet, the enforced expression of Bcl-2-bindingdefective mutants of Beclin 1 induces increased levels of autophagy, as well as cell death that is blocked by siRNA against a downstream autophagy gene, atg5, but not by caspase inhibitors. Thus, Bcl-2 homologs appear to function as a rheostat, which maintain homeostatic levels of autophagy that promote cell survival while preventing excessive levels of autophagy that promote cell death. It is not yet clear whether this antiautophagy function of Bcl-2 homologs contributes to their antideath role in physiological settings. Although the role of autophagy as a cell death pathway is controversial (Kroemer and Levine, 2008), it is nonetheless tempting to speculate that Bcl-2 homologs may serve as inhibitors of multiple death pathways, including not only apoptosis, but also autophagy.
Beclin 1 contains a BH3 domain that is sufficient and essential for binding to Bcl-2 homologs
Beclin 1 is predicted to consist of at least three domains within the evolutionarily conserved C-terminal two-thirds of the protein. The first of these is a coiled-coil domain, which shares ~25% of its sequence identity with coiled-coil domains from proteins such as myosin, and includes a poorly conserved leucine zipper. This domain is involved in heterodimerization with UVRAG, another coiled-coil domain-containing protein involved in vesicle trafficking (Liang et al., 2006). The first 10 residues of the leucine zipper also constitute a nuclear export signal essential for the cytoplasmic localization of Beclin 1, as well as its function in autophagy and tumor suppression (Liang et al., 2001). The structure and molecular function of the remaining domains in this evolutionarily conserved region have not been identified, but these domains are required for interactions with Vps34/class III PI3K, Beclin 1-dependent PI3K activity, autophagy (Furuya et al., 2005b), and peripheral membrane association (Liang et al., 1998). Although this evolutionarily conserved region of Beclin 1 is essential for the process of autophagy, the molecular mechanisms by which it mediates autophagy are not understood.
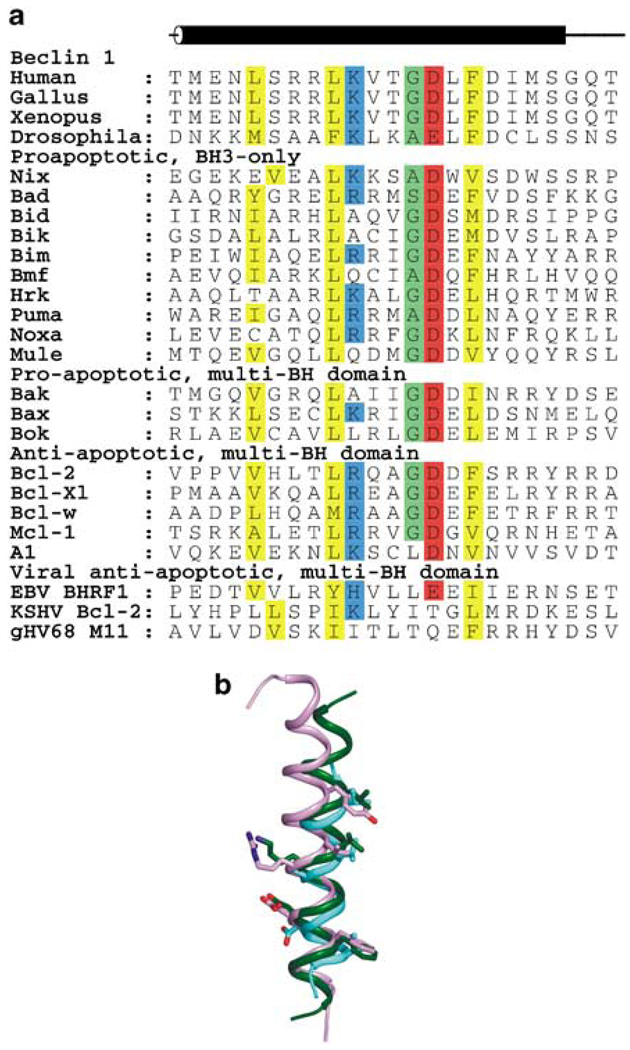
The N-terminal one-third of Beclin 1 is the most variable in sequence, with equivalent regions from evolutionarily distant orthologs sharing less than 15% sequence identity. This variable N-terminal region of Beclin 1 may serve to regulate protein interactions and/or the autophagy function of the evolutionarily conserved C-terminal region. Truncation experiments showed that residues 88–140 of human Beclin 1 are sufficient for binding to antiapoptotic Bcl-2 homologs (Liang et al., 1998; Pattingre et al., 2005). Within this region, recent structural, mutagenic and biochemical analyses show that residues 108–127 constitute a BH3 domain, similar to that required for the binding of proapoptotic proteins to antiapoptotic Bcl-2 homologs (Figure 1; Feng et al., 2007; Maiuri et al., 2007; Oberstein et al., 2007). Consistent with the variable sequence of the N-terminal regions of Beclin 1, the molecular mechanisms of the regulation of Beclin 1 may also vary among different eukaryotes; for example, yeast lack Bcl-2 homologs suggesting that Bcl-2 binding to the N-terminal region of Atg6/Beclin 1 is not conserved.
Figure 1.
Comparison of BH3 domains from Beclin 1 and different proapoptotic proteins. (a) Sequence alignment. Backgrounds of conserved residues are colored by residue type: yellow hydrophobic, green small, blue basic and red acidic. (b) Structural superposition. BH3 domains are represented by different colored ribbons; Beclin 1 (dark green,); Bad (violet) and Bak (teal). Residues conserved among BH3 domains that are involved in binding to the antiapoptotic Bcl-2 homologs are shown in atomic detail. All molecular figures were prepared with the program PYMOL (http://www.pymol.org).
Typically, a BH3 domain is defined as a four-turn amphipatic α-helix, bearing the sequence motif: Hy-X-X-X-Hy-K/R-X-X-Sm-D/E-X-Hy, in which Hy are hydrophobic residues and Sm represents small residues, typically glycine (Figure 1a). However, this domain is often difficult to identify from sequence alignments alone, as there are no invariant residues and even the pattern of residues is poorly conserved. Thus, combined analyses of sequence, structure, and a common molecular mechanism of binding to Bcl-2 homologs, may be required to identify these domains. Residues of human Beclin 1 that correspond to the residues conserved among BH3 domains are L112, L116, K117, G120, D121 and F123 (Figure 1a). Structural evidence obtained from crystal structures of peptides corresponding to the human Beclin 1 residues 108–127 bound to Bcl-XL (Oberstein et al., 2007), or from standard heteronuclear multidimensional NMR spectroscopy of variously labeled Bcl-XL-Beclin 1 (104–131) fusion protein. Feng et al. (2007) showed that like BH3 domains from proapoptotic proteins, these Beclin 1 residues fold into a four-turn α-helix when bound to Bcl-2 homologs (Figure 1b).
Proapoptotic Bcl-2 proteins are grouped into two categories: (1) the multidomain proapoptotic proteins that contain three BH domains, BH4, BH3 and BH1; and (2) the BH3-only proapoptotic proteins that contain only the BH3 domain. In both these groups, the BH3 domain is required for interaction with antiapoptotic Bcl-2 proteins. Antiapoptotic viral and cellular Bcl-2 homologs have very divergent sequences, but share similar three-dimensional structures consisting of a central hydrophobic α-helix surrounded by six amphipathic helices, delineated into four BH domains: BH4, BH3, BH1 and BH2 (Muchmore et al., 1996; Loh et al., 2005). A hydrophobic groove on the molecular surface of the antiapoptotic cellular Bcl-2 homologs (Sattler et al., 1997; Petros et al., 2000; Liu et al., 2003), as well as viral Bcl-2 homologs, such as KSHV Bcl-2 (Huang et al., 2002) and γHV68 M11 (Loh et al., 2005), is responsible for binding BH3 domains of proapoptotic proteins (Figure 2a) (Sattler et al., 1997; Petros et al., 2000; Huang et al., 2002, 2003; Liu et al., 2003; Loh et al., 2005). However, the structurally analogous surface groove of EBV BHRF1 is occluded and cannot bind BH3 domains (Huang et al., 2003). Recent structural data have shown that the BH3 domain of Beclin 1 also binds in the BH3-binding groove of Bcl-XL (Figure 2b; Feng et al., 2007; Oberstein et al., 2007) and γHV68 M11 (Figure 2c) (Sinha et al., 2008; Ku et al., 2008a).
Figure 2.
Bindingof BH3 domains to Bcl-2 homologs. Molecular surfaces of M11 and Bcl- XL are colored by atom type—oxygen red, nitrogen blue, sulfur green and carbon gray. BH3 domains are displayed in dark green ribbon with conserved residues involved in binding to antiapoptotic Bcl-2 homologs shown in atomic detail (a) Bad bound to Bcl-XL. (b) Beclin 1 bound to Bcl-XL. (c) Beclin 1 bound to M11.
Residues conserved among BH3 domains participate in the binding interface with the antiapoptotic Bcl-2 homologs (Figures 1b and Figure 2). The three hydrophobic residues conserved among the BH3 domains are packed against hydrophobic residues that line the BH3-binding groove of the antiapoptotic Bcl-2 homologs. The main chain atoms of the remaining two conserved residues, usually a glycine–aspartate pair, pack in an antiparallel manner against the main chain atoms of a glycine–arginine pair that is highly conserved among the BH1 domains of antiapoptotic Bcl-2 homologs. Further, the conserved BH3 domain aspartate ion pairs with the BH1 domain arginine conserved among the antiapoptotic Bcl-2 homologs. These interactions are preserved in the structures of the Beclin 1 BH3 domain bound to either Bcl-XL (Figure 2b; Feng et al., 2007; Oberstein et al., 2007) or γHV68 M11 (Figure 2c; Sinha et al., 2008; Ku et al., 2008a).
Several studies have identified residues within the BH3 domain of Beclin 1 required for binding to Bcl-2 homologs as well as residues within the BH3-binding groove of the Bcl-2 homologs required for binding to Beclin 1. Mutational analyses of Beclin 1 residues conserved among the BH3 domains show that the Beclin 1 BH3 domain is necessary for binding to Bcl-2. Selected substitution of key Beclin 1 residues, such as L112A, L116A, L116E, L116Q, G120E, D121A and F123A, either reduce or abrogate the interaction between Beclin 1 and either Bcl-2 or Bcl-XL, as tested by qualitative binding assays , such as analytical size exclusion chromatography, affinity pull-downs and coimmunoprecipitation assays (Feng et al., 2007; Maiuri et al., 2007; Oberstein et al., 2007). Similarly, alanine substitution of Beclin 1 residues L112, L116 and F123 reduce or prevent coimmunoprecipitation with γHV68 M11 (Sinha et al., 2008). Quantitative binding assays, such as isothermal calorimetry or fluorescence anisotropy measurements further confirm that the Beclin 1 BH3 domain residue mutants such as L116A and F123A, have unmeasurable affinity for Bcl-XL or Bcl-2 (Feng et al., 2007; Oberstein et al., 2007).
As predicted, mutations of residues within the BH3-binding groove of the Bcl-2 homologs also block interactions with Beclin 1. For example, mutations of conserved glycines in the BH1 domain of the Bcl-2 homologs, G145A for Bcl-2 (Pattingre et al., 2005) and G138A for Bcl-XL (Feng et al., 2007; Oberstein et al., 2007), abolish binding to wild-type Beclin 1 in coimmunoprecipitation assays. Similarly, double mutations of conserved γHV68 M11 residues G86A+R87A (Sinha et al., 2008; Ku et al., 2008a) in the BH1 domain, or of residues Y60A+L74A (Sinha et al., 2008) lining the hydrophobic groove, prevent γHV68 M11 binding to wild-type Beclin 1. Thus, the combined structural, biochemical and mutagenic analyses show that Beclin 1 contains a BH3 domain that is sufficient and essential for binding to Bcl-2 homologs. Further, the BH3 domains from Beclin 1 and various proapoptotic proteins bind to the Bcl-2 homologs by very similar mechanisms.
Although interactions of the BH3 domain residues appear to be conserved in each interacting BH3 domain-Bcl-2 homolog pair, the contribution of each of these residues to the total affinity of binding varies significantly between each pair of interactions. For instance, the conserved Beclin 1 BH3 domain residues, G120 and D121, are not essential for binding to γHV68 M11 (Sinha et al., 2008). As suggested by NMR studies examining residues of γHV68 M11 affected by binding to the BH3 domain of Beclin 1 versus BH3 domains from the proapoptotic proteins, Bad or Bax (Sinha et al. 2008), the variation in amino acids constituting the BH3 domains, as well as in the residues lining the BH3-domain-binding groove of the interacting Bcl-2 homologs, probably result in a wide range of binding affinities for each pair of interactions, enforcing a degree of specificity in these interactions.
Mechanisms of Bcl-2-mediated inhibition of Beclin 1-dependent autophagy
The BH3 domain of Beclin 1, and interactions between the BH3 domain of Beclin 1 and Bcl-2 homologs, are essential for the antiautophagy activity of Bcl-2 proteins. Two major lines of evidence suggest that Bcl-2 proteins inhibit autophagy by binding to the BH3 domain of Beclin 1. First, mutation of residues in either Beclin 1 or Bcl-2 homologs that block Beclin 1–Bcl-2 homolog interactions, without exception, also block the ability of Bcl-2 to inhibit autophagy. Bcl-2, Bcl-XL and viral Bcl-2 s inhibit autophagy when wild-type Beclin 1 is expressed, but not when Bcl-2 binding-defective Beclin 1 mutants are expressed. (Liang et al., 1998; Pattingre et al., 2005). Conversely, Bcl-2 homolog mutants that cannot bind to Beclin 1, such as Bcl-2 G145A (Pattingre et al., 2005), Bcl-XL G138A (Maiuri et al., 2007) and γHV68 M11 G86A+R87A or Y60A+L74A (Sinha et al., 2008; Ku et al., 2008a), are defective in autophagy inhibition. Second, studies using the peptidomimetic inhibitor, ABT-737, a small molecule designed to bind to the BH3-binding groove of Bcl-2 or Bcl-XL (Oltersdorf et al., 2005), show that ABT-737 competitively inhibits the binding of peptides corresponding to the Beclin 1 BH3 domain to Bcl-XL, and abrogates the inhibition of autophagy by Bcl-XL (Maiuri et al., 2007), providing further evidence that Bcl-2-mediated down-regulation of autophagy depends on binding through the Bcl-XL BH3 binding groove.
Although the research reviewed above conclusively demonstrates that the binding of the Beclin 1 BH3 domain to Bcl-2 homologs is essential for the Bcl-2-mediated downregulation of autophagy, as yet, we do not understand how this interaction results in autophagy inhibition. In mammals, Beclin 1, class III PI3K and p150 (Kihara et al., 2001; Furuya et al., 2005b), and either Atg14 (Itakura et al., 2008) or UVRAG (Liang et al., 2007), Bif-1/Endophilin 1 (Takahashi et al., 2007) and Ambra1 (Fimia et al., 2007) have been shown to be essential components of the multiprotein, membrane-associated, autophagosome nucleation complex. The binding of Beclin 1 to Bcl-2 may inhibit the interaction of Beclin 1 with class III PI3K, although class III PI3K does not appear to interact directly with the Beclin 1 BH3 domain (Pattingre et al., 2005; Ku et al., 2008b). Furthermore, both Beclin 1 and Bif-1 can form homo-oligomers (Takahashi et al., 2007; Ku et al., 2008b), and the binding of Bcl-2 homologs may also prevent higher-order oligomerization of Beclin 1 (Liang et al., 2008). Therefore, it appears likely that the binding of Bcl-2 homologs to Beclin 1 sterically hinders its interactions with other essential components of the autophagy nucleation complex, inhibiting the autophagy-inducing function of Beclin 1 (Figure 3). However, the molecular mechanism(s) by which Beclin 1 functions in autophagy and consequently, the exact mechanism(s) by which the Beclin 1–Bcl-2 interaction disrupts the autophagy function of Beclin 1 remain to be understood.
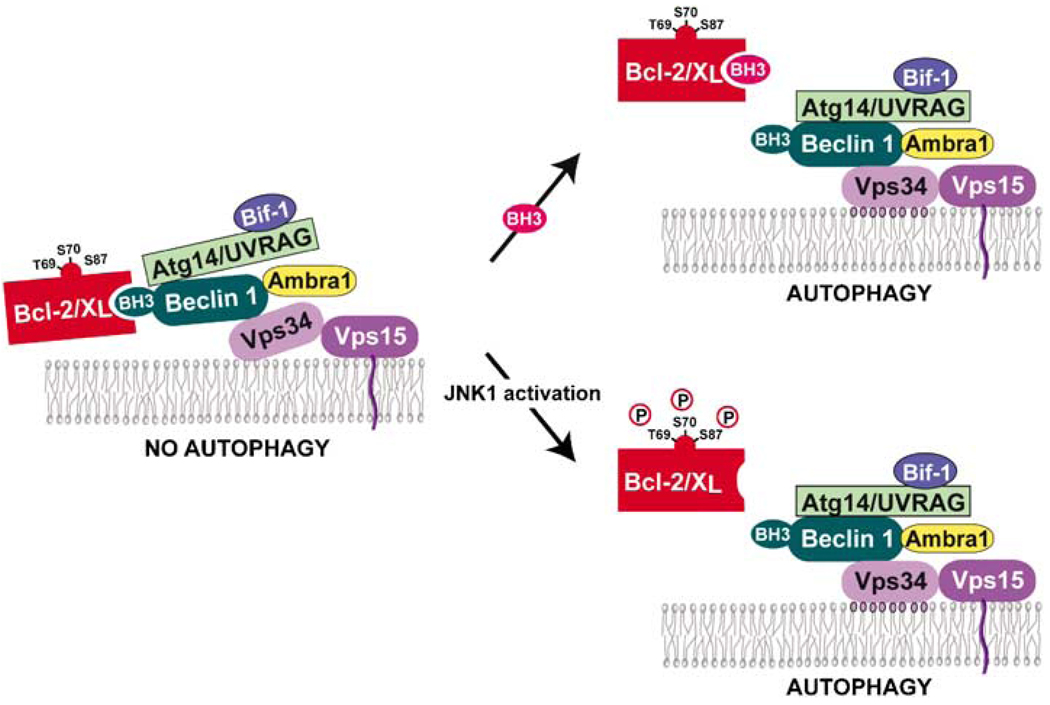
Figure 3.
Schematic representation of the role of the Beclin 1 BH3 domain in the regulation of autophagy. The vesicle nucleation complex consists of Vps34 (also known as class III PI3K) and activators, such as Beclin 1, either Atg14 or UVRAG, Ambra1 and Bif-1. The binding of Bcl-2 homologs to the BH3 domain of Beclin 1 prevents the assembly of an autophagy-competent complex (left). Disruption of the Beclin 1–Bcl-2 homolog interaction , either by competitive displacement of the Beclin 1 BH3 domain by other BH3 domain-containing proteins (upper right) or due to JNK1-mediated phosphorylation of Bcl-2 (lower right) results in the assembly of an autophagy-competent complex. The lipid kinase activity of this complex then catalyses the conversion of PI to PI3P (shown with violet circles) resulting in autophagy.
Regulation of Bcl-2-mediated inhibition of Beclin 1-dependent autophagy
Two major mechanisms have been identified that regulate Bcl-2/Bcl-XL binding to Beclin 1 and Bcl-2/Bcl-XL inhibition of Beclin 1-dependent autophagy (Figure 3), including JNK1-mediated Bcl-2 phoshorylation and competitive disruption of Bcl-2/Bcl-XL binding to Beclin 1 by other BH3 domains. Earlier, we showed that the Bcl-2–Beclin 1 interaction is regulated by nutrient status, with the highest levels of interaction in nutrient-rich conditions (when autophagy is suppressed) and the lowest levels of interaction in starvation conditions (when autophagy is maximally stimulated; Liang et al., 1998; Pattingre et al., 2005). More recently, we showed that the starvation-induced activation of JNK1 causes multisite phosphorylation at residues T69, S70 and S87 in the unstructured loop between the BH4 and BH3 domains of Bcl-2, which leads to the dissociation of Bcl-2 and Beclin 1 (Wei et al., 2008a). Bcl-2 is not phosphorylated in cells lacking JNK1, or in which JNK1 is inhibited. In these cells, starvation-induced dissociation of Bcl-2 and Beclin 1 and starvation-induced autophagy are abrogated. Similarly, in cells that express a T69A+S70A+S87A Bcl-2 mutant that cannot be phosphorylated instead of wild-type Bcl-2, there is no starvation-induced dissociation of Bcl-2 and Beclin 1, and no starvation-induced autophagy. Conversely, the expression of constitutively activated JNK1 leads to Bcl-2 phosphorylation, the lack of Bcl-2 and Beclin 1 binding during normal growth conditions, and increased basal levels of autophagy. Further, a multisite T69E+S70E+S87E Bcl-2 phosphomimetic mutant does not bind to Beclin 1 and does not inhibit autophagy (Wei et al., 2008a). Thus, multisite phosphorylation of the Bcl-2 unstructured loop by JNK1 is responsible for the disruption of Bcl-2–Beclin 1-binding during starvation, and subsequent autophagy activation. Interestingly, viral Bcl-2s lack this unstructured loop with analogous phosphorylation sites and constitutively inhibit the autophagy function of Beclin 1 (Wei et al., 2008a), suggesting that they have evolved mechanisms to escape normal physiological regulation of Bcl-2–Beclin 1 interactions.
The competitive binding of BH3 domains to Bcl-2 homologs represents another mechanism by which Bcl-2/Bcl-XL-mediated inhibition of Beclin-1-dependent autophagy can be modulated. This was shown experimentally by the competitive displacement of Beclin 1 bound to Bcl-2 or Bcl-XL by Bad, a BH3-only proapoptotic protein. As discussed above, nutrient deprivation reduces the amount of Beclin 1 that coimmunoprecipitates with Bcl-2/Bcl-XL, resulting in increased levels of starvation-induced autophagy. This correlates with an increase in the amount of Bad that coimmunoprecipitates with Bcl-2/Bcl-XL (Maiuri et al., 2007). Increased levels of autophagy in response to starvation are not observed upon siRNA-mediated knock-down of bad in human HeLa cells or in bad knockout mouse embryonic fibroblasts (MEFs), but starvation-induced autophagy is restored in these cells upon treatment with ABT-737, a BH3 domain peptidomimetic. Finally, enforced expression of Bad, but not a Bcl-2-binding defective Bad mutant, is sufficient to induce autophagy, both in normal conditions and upon caspase inhibition.
Therefore, various BH3 domain-containing proteins that either bind with higher affinity to Bcl-2 homologs than does the BH3 domain of Beclin 1, or that are present in substantially higher concentrations, may competitively displace the Beclin 1 BH3 domain bound to Bcl-2, leading to abrogation of Bcl-2/Bcl-XL-mediated inhibition of Beclin 1-dependent autophagy. Indeed, in addition to Bad, other BH3-only proteins, such as Nix/Bnip3 (Daido et al., 2004; Chang et al., 2007; Hamacher-Brady et al., 2007; Schweers et al., 2007; Zhang et al., 2008), Bik (Rashmi et al., 2008), Noxa, Puma and BimEL (Abedin et al., 2007) have been shown to induce autophagy and may, in a similar manner, function as competitive inhibitors of Beclin 1–Bcl-2/Bcl-XL interactions. Thus, the BH3 domain not only functions directly in autophagy regulation by mediating interactions between Beclin 1 and antiapoptotic Bcl-2 homologs, it may also function, in the context of the BH3 domain-containing proapoptotic Bcl-2 proteins, in the indirect modulation of autophagy by influencing the interaction of Beclin 1 with Bcl-2/Bcl-XL.
In addition to the two regulatory mechanisms discussed above, it is likely that other mechanisms, such as regulation of Beclin 1 and Bcl-2 expression and changes in the subcellular localization of either protein and/or post-translational modifications of Beclin 1, also modulate the interaction of Beclin 1 with Bcl-2 homologs as well as Bcl-2 homolog-mediated inhibition of Beclin 1-dependent autophagy. These potential regulatory mechanisms are important areas of future research.
The BH3 domain: a structural motif integrating the regulation of autophagy and apoptosis
BH3 domain-containing proteins bind with differential affinity to various Bcl-2 homologs, and may competitively affect the binding of other BH3 domain-containing proteins to Bcl-2. The competitive binding of the BH3 domain-containing proteins may be further modulated by the combined effect of post-translational modifications as well as spatial and temporal variations in the concentrations of each of the BH3 domain-containing proteins. Thus, Beclin 1 and proapoptotic BH3 domain-containing proteins may each indirectly regulate apoptosis and autophagy, respectively. As noted, proapoptotic BH3-only proteins induce autophagy by competitively disrupting the low-affinity Beclin 1-Bcl-2/Bcl-XL interaction (Maiuri et al., 2007). An important question is whether, through this mechanism, autophagy is invariably activated in cells exposed to apoptotic stimuli. A corollary question is whether the proapoptotic BH3-only proteins can also serve a prosurvival function by activating an adaptive autophagy response, and/or an apoptosis-independent pro-death function by activating toxic levels of autophagy that lead to self-cannibalization.
Bcl-2 phosphorylation and its differential effect on the binding of different BH3 domain-containing proteins, provide a mechanism to further fine-tune the regulation of these cellular events. This concerted regulation has been investigated only in the context of cellular response to nutrient starvation (Wei et al., 2008b), although similar parallel mechanisms may well regulate the response to other stress-inducing stimuli . Nutrient deprivation results in increasing levels of JNK1-mediated Bcl-2 multisite phosphorylation, with levels of phosphorylated Bcl-2 dependent upon the duration of starvation. This may lead to the differential Bcl-2 phosphorylation level-dependent dissociation of different BH3 domain-containing proteins bound to Bcl-2 (Wei et al., 2008b). For example, Wei et al. (2008b) showed that early after starvation, low levels of multisite Bcl-2 phosphorylation result in increased dissociation of the Bcl-2–Beclin 1 complex and increased Beclin 1-dependent autophagy, which may be an attempt to promote cell survival. However, after prolonged starvation, when autophagy can no longer keep cells alive, higher levels of multisite phosphorylated Bcl-2 lead to its dissociation from the BH3 domain containing proapoptotic proteins, such as Bax, concurrent with caspase 3 activation and apoptosis (Wei et al., 2008b). Thus, the differential affinities of Bcl-2 for the BH3 domain of Beclin 1 versus that of proapoptotic Bcl-2 family members may provide a mechanism for post-translational modifications of Bcl-2 to temporally coordinate the dual regulation of autophagy and apoptosis by Bcl-2.
Beclin 1 as a novel BH3-only protein
The first structure of the complex of Bcl-XL bound to the minimal Beclin 1 region required for binding to Bcl-2 homologs, indicated that the Bcl-2-binding region constituted a BH3 domain, which bound to Bcl-XL in a manner similar to BH3 domains from proapoptotic proteins (Oberstein et al., 2007). On the basis of this structural evidence, the authors postulated that Beclin 1 may function as a proapoptotic BH3-only protein. Further, it was suggested that the role of Beclin 1 in tumor suppression may be explained by this proposed proapoptotic role, rather than by its proautophagic function. However, to date, there is no physiologically relevant evidence of a proapoptotic function for Beclin 1 or that such a proposed function contributes to its tumor suppressor action.
Mauiri et al. (2007) found that, while the microinjection of high concentrations of a peptide corresponding to the BH3 domain of Beclin 1 induces Bax-dependent apoptosis, enforced overexpression of full-length Beclin 1 fails to trigger mitochondrial membrane permeability transition or apoptosis. This suggests that some region outside of the BH3 domain of Beclin 1 may exert a preventive, antiapoptotic function that ‘neutralizes’ the potential apoptogenic effects of the isolated BH3 domain. Furthermore, the reduced expression or knockdown of mammalian Beclin 1 has been shown to increase, rather than decrease, apoptosis in several different settings, such as in HeLa cells subjected to nutrient deprivation (Boya et al., 2005); endothelial cells treated with angiogenesis inhibitors (Nguyen et al., 2007; Ramakrishnan et al., 2007); chondrocytes subjected to hypoxia (Bohensky et al., 2007); conjunctival cell death upon exposure to benzalkonium chloride or ultraviolet radiation (Bohensky et al., 2007); HEPG2 cells after antiFas antibody or doxorubicin treatment (Daniel et al., 2006); MEFs treated with C(2)-ceramide (Fujiwara et a,l., 2008); human myeloid leukemia cells treated with retinoic acid or vitamin D3 (Wang, 2008); human bronchial epithelial cells exposed to cigarette smoke extract (Kim et al., 2008); cultured hepatoma cells (Harada et al., 2008); various human carcinoma cell lines upon irradiation (Apel et al., 2008); and renal tubular epithelial cells after cisplatin injury (Yang et al., 2008). In addition, increased apoptosis has been reported in nematodes with a null mutation in the C. elegans beclin 1 ortholog, bec-1 (Takacs-Vellai et al., 2005).
Similarly, there is no evidence that Beclin 1 functions as a tumor suppressor by playinga proapoptotic role. In tumor-prone epithelial cells in beclin 1+/− mice or in tumor-prone immortalized epithelial cells derived from such mice, there is no evidence of decreased apoptosis with allelic loss of beclin 1 (Karantza-Wadsworth et al., 2007; Mathew et al., 2007; Qu et al., 2007). Yet, in the setting of beclin 1 allelic loss, epithelial cells display a defect in cell growth control (Qu et al., 2003) and are more prone to DNA damage and chromsomal instability (Karantza-Wadsworth et al., 2007; Mathew et al., 2007), suggesting that apoptosis-independent functions of Beclin 1 contribute to its tumor suppressor action. Thus, it does not appear that the BH3 domain enables Beclin 1 to function physiologically as a BH3-only proapoptotic protein, or that such a proposed role contributes to its tumor suppressor activity.
Another theoretical argument for a proapoptotic function of Beclin 1, in addition to the observation that it is a BH3-only protein, is based on the observed competitive inhibition of BH3 domains from different BH3-only proteins in binding to Bcl-2 antiapoptotic homologs. As discussed, the competitive inhibition of Beclin 1 binding to Bcl-2 homologs by BH3-only proapoptotic proteins is a mechanism by which these proteins indirectly upregulate autophagy (Feng et al., 2007; Oberstein et al., 2007). Similarly, it is theoretically possible that the Beclin 1 BH3 domain might induce apoptosis by the competitive inhibition of Bcl-2 binding to BH3 domains of proapoptotic Bcl-2 proteins. However, this seems unlikely because all cellular Bcl-2 homologs bind BH3 domains of specific subsets of proapoptotic proteins with substantially higher affinity compared with their affinity for the Beclin 1 BH3 domain (Sinha et al., 2008). Instead, it seems more likely that the BH3 domain of Beclin 1 serves as a structural motif that enables the coordinate regulation of apoptosis and autophagy. Thus, although the formal possibility that Beclin 1 may function as a proapoptotic protein in certain settings remains, we propose that Beclin 1 is a novel type of BH3-only protein, in which the BH3 domain enables the antiapoptotic Bcl-2 homologs to inhibit autophagy (through direct binding) and enables the proapoptotic BH3-only homologs to stimulate autophagy (through competitive inhibition of binding to Bcl-2/Bcl-XL).
Bcl-2 inhibition of Beclin 1-dependent autophagy: a potential mechanism of oncogenesis?
Bcl-2 was the first cellular protein shown to function as an oncoprotein by blocking apoptotic cell death, rather than by increasing cellular proliferation (McDonnell et al., 1989; Vaux et al., 1998). In light of emerging evidence that Beclin 1 and the autophagy pathway function as tumor suppressors (Edinger and Thompson, 2003; Ogier-Denis and Codogno, 2003; Gozuacik and Kimchi, 2004; Shintani and Klionsky, 2004; Furuya et al., 2005a; Jin, 2006; Levine, 2006; Pattingre and Levine, 2006), a critical unanswered question is whether the antiautophagy activity of Bcl-2 also contributes to its oncogenic activity. Although in normal cells, autophagy is activated by the disruption of Bcl-2–Beclin 1 interactions; Bcl-2 overexpression, a common occurrence in human cancers, can also suppress the physiological activation of autophagy both in cultured cells and in mice (Liang et al., 1998; Pattingre et al., 2005). Furthermore, KSHV vBcl-2, and likely other viral Bcl-2s encoded by other oncogenic γ-herpesviruses, irreversibly bind to Beclin 1 and function as constitutive inhibitors of autophagy, because they lack the unstructured loop between the BH4 and BH3 domain that undergoes regulatory phosphorylation (Wei et al., 2008a). Therefore, it is plausible that cellular Bcl-2 overexpression and viral Bcl-2s disrupt the tumor suppressor activity of the autophagy pathway by blocking the Beclin 1 function.
It should be noted that not only is Beclin 1 a haploinsufficient tumor suppressor, but other proteins that positively regulate the Beclin 1-class III PI3K complex, including UVRAG, Bif-1 and Ambra1 also function in negative growth control and/or tumor suppression (Levine and Kroemer, 2008). Thus, the Beclin 1–class III PI3K vesicle nucleation complex is probably an important nexus in autophagy control and tumor suppression, including the suppression of lymphomagenesis. B-cell lymphomas are the most common spontaneous tumors that arise in mice with either monoallelic deletion of beclin 1 (Qu et al., 2003; Yue et al., 2003) or biallelic loss of bif-1 (Takahashi et al., 2007). Therefore, it is likely that cellular Bcl-2, which contributes to lymphomas in patients and mouse models, and viral Bcl-2s, may exert their oncogenic activity, at least in part, through autophagy inhibition. Specific agents that disrupt Bcl-2/Bcl-XL–Beclin 1 BH3 domain binding without disrupting Bcl-2/Bcl-XL-proapoptotic BH3 protein binding, and/or mutations in Bcl-2/Bcl-XL that selectively disrupt its interaction with Beclin 1, will be required to address this hypothesis.
A related question is whether, beyond tumor suppression, cellular and viral Bcl-2 homologs also regulate other processes involving Beclin 1 and autophagy, such as apoptotic corpse clearance, innate immunity, the prevention of neurodegenerative disease and adaptive and maladaptive responses in heart disease (Levine and Kroemer, 2008). At a minimum, given the central role of Beclin 1 and autophagy in intracellular pathogen degradation (xenophagy), and activation of innate and adaptive immunity (Schmid et al., 2006; Levine and Deretic, 2007; Orvedahl and Levine, 2008), it seems likely that viral Bcl-2 targeting of the Beclin 1 function may contribute to γ-herpesvirus pathogenesis. Indeed, autophagy inhibition by another herpesvirus-encoded protein, herpes simplex virus type I ICP34.5, which binds to a different region of Beclin 1, has been shown to be essential for herpes simplex virus type I-induced lethal encephalitis (Orvedahl et al., 2007). A high research priority will be to determine whether the cellular and viral Bcl-2 homologs also contribute to human disease through binding to the BH3 domain of Beclin 1 and subsequent autophagy inhibition.
Bcl-2–Beclin 1 interactions: a novel therapeutic target for autophagy induction?
Beclin 1 is a central player in autophagosome formation, and Bcl-2/Bcl-XL function as important regulators of levels of Beclin 1-mediated autophagy. Thus, the Bcl-2/Bcl-XL–Beclin 1 complex represents an important therapeutic target for autophagy modulation. Compounds that disrupt the complex will lead to increased autophagy and may be useful for the prevention and/or treatment of certain diseases, such as aging, certain cancers, infectious diseases and neurodegenerative diseases. Compounds that stabilize the complex may be useful in scenarios in which it may be desirable to inhibit autophagy, such as in established tumors in which autophagy is considered a prosurvival mechanism.
The BH3 peptidomimetics were originally developed in the context of targeted cancer therapies designed to increase apoptosis in tumor cells by disrupting interactions between antiapoptotic Bcl-2 homologs and proapoptotic BH3-only proteins. Not surprisingly, given the central role of BH3 domains in Bcl-2 regulation of both apoptosis and autophagy, they also constitute a class of drugs that can induce autophagy by disrupting the interaction between Bcl-2 homologs and the BH3 domain of Beclin 1. The first example of such an inhibitor was ABT-737 (Oltersdorf et al., 2005), which specifically targets antiapoptotic Bcl-2 homologs that bind the Bad BH3 domain with high affinity. Assays measuring the binding of synthetic peptides to purified recombinant Bcl-XL in vitro show that ABT-737 also competitively inhibits the binding of Beclin 1 BH3 peptides, with an IC50 in the micromolar range (Maiuri et al., 2007). Consistent with this finding, in cells resistant to the proapoptotic action of ABT-737, pretreatment with this inhibitor abolishes the immunoprecipitation of Beclin 1 with Bcl-2 or Bcl-XL and induces high levels of autophagy (Maiuri et al., 2007). ABT-737-induced autophagy cannot be inhibited by Bcl-2 or Bcl-XL overexpression, yet is abolished upon either transfection with Mcl-1, which does not bind ABT-737, or by the siRNA-mediated knockdown of Beclin 1. Together these results clearly show that competitive disruption of the Beclin 1 interaction with Bcl-2 or Bcl-XL by small molecule inhibitors suffices to induce autophagy (Maiuri et al., 2007). Further refinements of BH3 peptidomimetics, based on structural analyses of Bcl-2 homolog/Beclin 1 BH3 domain complexes, may enable an increase in the specificity and/or potency of autophagy induction by this class of agents.
At present, it is controversial whether the proautophagy action of BH3 peptidomimetics should be enhanced or reduced in developing agents for cancer therapy. Autophagy induction may promote the survival of tumor cells, thereby counteracting or limiting the efficacy of apoptosis induction by these compounds. However, excessive autophagy may also promote cell death through self-cannibalization and help kill tumor cells. Indeed, recent evidence has shown that an experimental BH3 mimetic, obatoclax, kills gluococorticoid-resistant leukemic cells in a manner that is independent of Bax and Bak, but dependent on Atg5, Atg7 and Beclin 1 (Bornhauser et al., 2008). Therefore, different BH3 mimetics may induce either autophagy-dependent cell survival or autophagy-dependent cell death, depending on the magnitude of autophagy induction. It will be important to determine the optimal levels of autophagy induction by BH3 peptidomimetics not only for the treatment of specific cancers, but also in the potential treatment of other diseases in which autophagy stimulation may be beneficial.
Conclusion
In conclusion, the research we have reviewed here shows that the essential autophagy effector, Beclin 1, contains a BH3 domain with a conserved molecular mechanism of binding to antiapoptotic Bcl-2 homologs. However, despite this conserved mechanism and unlike the canonical BH3 domain-containing proteins, Beclin 1 does not appear to play a significant role in apoptosis. Rather, studies with the Beclin 1 BH3 domain point to a new function of BH3 domains in enabling the antiapoptotic Bcl-2 homologs to inhibit autophagy, another fundamental cellular pathway involved in tissue homeostasis. Thus, the Beclin 1 BH3 domain facilitates the coordinate regulation of autophagy, primarily a cell survival pathway, and apoptosis, a cell death pathway, by Bcl-2 homologs. Perhaps, through the common molecular mechanism of binding to ‘antiapoptotic’ Bcl-2 homologs, the simple BH3 domain has, in fact, complex cellular functions.
Acknowledgements
The work in the authors’ laboratory was funded by NIH Grants R21 AI78108-01 to SS and RO1 CA109618 and R01 CA084254 to BL.
Footnotes
Conflict of interest
The authors have declared no conflict of interest.
Contributor Information
S Sinha, Email: sangita.sinha@ndsu.edu.
B Levine, Email: Beth.Levine@UTSouthwestern.edu.
References
- Abedin M, Wang D, McDonnell M, Lehmann U, Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14:500–510. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- Aita VM, Liang XH, Murty VVVS, Pincus DL, Yu W, Cayanis E, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- Apel A, Herr I, Schwarz H, Rodemann H, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- Bohensky J, Shapiro I, Leshinsky S, Terkhorn S, Adams C, Srinivas V. HIF-1 regulation of chondrocyte apoptosis: induction of the autophagic pathway. Autophagy. 2007;3:207–214. doi: 10.4161/auto.3708. [DOI] [PubMed] [Google Scholar]
- Bornhauser BC, Bonapace L, Schmitz M, Schrappe M, Walensky LD, Bourquin J-P. The BH3 mimetic obatoclax overcomes glucocorticoid-resistance by activating autophagic cell death. In: McDonnell TJ, Hickman JA, Debatin K-M, editors. Seventh International Euroconference on Mechanisms of cell death and disease: Advances in therapeutic intervention and drug development. Portugal: Cascais; 2008. p. Poster 28. [Google Scholar]
- Boya P, Gonzalez-Polo R-A, Casares N, Perfettini J-L, Dessen P, Larochette N, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Yang M, Liu H, Lin Y, Lei H. Concanavalin A induces autophagy in hepatoma cells and has a therapeutic effect in a murine in situ hepatoma model. Hepatology. 2007;45:286–296. doi: 10.1002/hep.21509. [DOI] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison P, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–124. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 2004;64:4286–4293. doi: 10.1158/0008-5472.CAN-03-3084. [DOI] [PubMed] [Google Scholar]
- Daniel F, Legrand A, Pessayre D, Vadrot N, Descatoire V, Bernuau D. Partial Beclin 1 silencing aggravates doxorubicin-and Fas-induced apoptosis in HepG2 cells. World J Gastroenterol. 2006;12:2895–2900. doi: 10.3748/wjg.v12.i18.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger AL, Thompson CB. Defective autophagy leads to cancer. Cancer Cell. 2003;4:422–424. doi: 10.1016/s1535-6108(03)00306-4. [DOI] [PubMed] [Google Scholar]
- Escalatine A, Chaumorcel M, Codogno P. Levine B, editor. Macroautophagy signaling and regulation. Autophagy in Infection and Immunity. 2009 doi: 10.1007/978-3-642-00302-8_2. (in press) [DOI] [PubMed] [Google Scholar]
- Feng W, Huang S, Wu H, Zhang M. Molecular basis of Bcl-XL′s target recognition versatility revealed by the structure of Bcl-XL in complex with the BH3 domain of Beclin-1. J Mol Biol. 2007;372:223–235. doi: 10.1016/j.jmb.2007.06.069. [DOI] [PubMed] [Google Scholar]
- Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia G, Stoykova A, Romagnoli A, Giunta L, Bartolomeo S, Nardacci R, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Yoshimoto K, Ohsumi Y. An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 2007;143:1132–1139. doi: 10.1104/pp.106.093864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K, Daido S, Yamamoto A, Kobayashi R, Yokoyama T, Aoki H, et al. Pivotal role of the cyclin-dependent kinase inhibitor p21WAF1/CIP1 in apoptosis and autophagy. J Biol Chem. 2008;283:388–397. doi: 10.1074/jbc.M611043200. [DOI] [PubMed] [Google Scholar]
- Furuya N, Liang X, Levine B. Klionsky DJ, editor. Autophagy and cancer. Autophagy. 2005a:242–255. [Google Scholar]
- Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of beclin 1 is required for Vps34 binding, autophagy, and tumor supppressor function. Autophagy. 2005b;1:41–47. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- Hamacher-Brady A, Brady N, Logue S, Sayen M, Jinno M, Kirshenbaum L, et al. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Diff. 2007;14:146–157. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic L, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada M, Strnad P, Toivola D, Omary M. Autophagy modulates keratin-containing inclusion formation and apoptosis in cell culture in a context-dependent fashion. Exp Cell Res. 2008;314:1753–1764. doi: 10.1016/j.yexcr.2008.01.035. [DOI] [PubMed] [Google Scholar]
- Hars E, Qi H, Ryazanov A, Jin S, Cai L, Hu C, et al. Autophagy regulates ageing in C. elegans. Autophagy. 2007;3:93–95. doi: 10.4161/auto.3636. [DOI] [PubMed] [Google Scholar]
- He C, Baba M, Cao Y, Klionsky D. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol Biol Cell. 2008;19:5506–5516. doi: 10.1091/mbc.E08-05-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Petros AM, Virgin HW, Fesik SW, Olejniczak ET. Solution structure of a Bcl-2 homologfrom Kaposi sarcoma virus. Proc Nat Acad Sci. 2002;99:3428–3433. doi: 10.1073/pnas.062525799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Petros AM, Virgin HW, Fesik SW, Olejniczak ET. Solution structure of the BHRF1 protein from Epstein-Barr virus, a homologof human Bcl-2. J Mol Biol. 2003;332:1123–1130. doi: 10.1016/j.jmb.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- Jin S. Autophagy, mitochondrial quality control, and oncogenesis. Autophagy. 2006;2:80–84. doi: 10.4161/auto.2.2.2460. [DOI] [PubMed] [Google Scholar]
- Kametaka S, Okano T, Ohsumi M, Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Develop. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-golgi network. EMBO Rep. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HP, Wang X, Lee S-J, Huang M-H, Wang Y, Ryter SW, et al. Autophagic proteins regulate cigarette smoke induced apoptosis: Protective role of heme oxygenase-1. Autophagy. 2008;4:887–895. doi: 10.4161/auto.6767. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku B, Woo J-S, Liang C, Lee K-H, Hong H-S, Xiaofei E, et al. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral Bcl-2 of murine γ-herpesvirus 68. PLoS Path. 2008a;4:e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku B, Woo J-S, Liang C, Lee K-H, Jung JU, Oh B-H. An insight into the mechanistic role of Beclin 1 and its inhibition by prosurvival Bcl-2 family proteins. Autophagy. 2008b;4:519–520. doi: 10.4161/auto.5846. [DOI] [PubMed] [Google Scholar]
- Levine B. Unravelingthe role of autophagy in cancer. Autophagy. 2006;2:65–66. doi: 10.4161/auto.2.2.2457. [DOI] [PubMed] [Google Scholar]
- Levine B, Abrams J. p53: The Janus of autophagy? Nat Cell Biol. 2008;10:637–639. doi: 10.1038/ncb0608-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Deretic V. Unveilingthe roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Xiaofei E, Jung JU. Downregulation of autophagy by herpesvirus Bcl-2 homologs. Autophagy. 2008;4:268–272. doi: 10.4161/auto.5210. [DOI] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, B-H O, Jung JU. UVRAG: a new player in autophagy and tumor cell growth. Autophagy. 2007;3:69–71. doi: 10.4161/auto.3437. [DOI] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by Beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, et al. Protection against fatal Sindbis virus encephalitis by Beclin 1, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Yu J, Brown K, Levine B. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 2001;61:3443–3449. [PubMed] [Google Scholar]
- Liu X, Dai SC, Zhu Y, Marrack P, Kappler JW. The structure of a Bcl-xL/Bim fragment complex: implications for Bim function. Immunity. 2003;19:341–352. doi: 10.1016/s1074-7613(03)00234-6. [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Loh J, Huang Q, Petros AM, Nettesheim D, van Dyk LF, Labrada L, et al. A surface groove essential for viral Bcl-2 function duringchronic infection in vivo. PLoS Path. 2005;1:80–91. doi: 10.1371/journal.ppat.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri M, Le Toumelin G, Criollo A, Rain J, Gautier F, Juin P, et al. Functional and physical interaction between Bcl-XL and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri M, Tasdemir E, Criollo A, Morselli E, Vicencio J, Carnuccio R, et al. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ. 2008;16:87–93. doi: 10.1038/cdd.2008.131. [DOI] [PubMed] [Google Scholar]
- Mathew R, Kongara S, Beaudoin B, Karp C, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell T, Deane N, Platt F, Nunez G, Jaeger U, McKearn JP, et al. bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell. 1989;57:79–88. doi: 10.1016/0092-8674(89)90174-8. [DOI] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen E-L, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo A, Klionsky D. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Tasdemir E, Maiuri M, Galluzzi L, Kepp O, Criollo A, et al. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle. 2008;7:3056–3061. doi: 10.4161/cc.7.19.6751. [DOI] [PubMed] [Google Scholar]
- Muchmore SW, Sattler M, Liang H, Meadows RP, Harlan JE, Yoon HS, et al. X-ray and NMR structure of human Bcl-XL, an inhibitor of programmed cell death. Nature. 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Subramanian I, Kelekar A, Ramakrishnan S. Kringle 5 of human plasminogen, an angiogenesis inhibitor, induces both autophagy and apoptotic death in endothelial cells. Blood. 2007;109:4793–4802. doi: 10.1182/blood-2006-11-059352. [DOI] [PubMed] [Google Scholar]
- Obara K, Ohsumi Y. Dynamics and function of PtdIns(3)P in autophagy. Autophagy. 2008;4:952–954. doi: 10.4161/auto.6790. [DOI] [PubMed] [Google Scholar]
- Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3 only protein. J Biol Chem. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- Ogier-Denis E, Codogno P. Autophagy: a barrier or an adaptive response to cancer. Biochim Biophys Acta. 2003;1603:113–128. doi: 10.1016/s0304-419x(03)00004-0. [DOI] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Orvedahl A, Levine B. Eatingthe enemy within: Autophagy in infectious diseases. Cell Death Diff. 2008;16:57–69. doi: 10.1038/cdd.2008.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou C, Domin J, Cockcroft S, Waterfield MD. Characterization of p150, an adaptor protein for the human phosphatidylinositol (PtdIns) 3-kinase. Substrate presentation by phosphatidylinositol transfer protein to the p150 PtdIns 3-kinase complex. J Biol Chem. 1997;272:2477–2485. doi: 10.1074/jbc.272.4.2477. [DOI] [PubMed] [Google Scholar]
- Patel S, Dinesh-Kumar S. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy. 2008;4:20–27. doi: 10.4161/auto.5056. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Levine B. Bcl-2 inhibition of autophagy: A new route to cancer? Cancer Res. 2006;66:2885–2888. doi: 10.1158/0008-5472.CAN-05-4412. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Petros AM, Nettseheim DG, Wang Y, Olejniczak ET, Meadows RP, Mack J, et al. Rationale for Bcl-xL/BAD peptide complex formation from structure, mutagenesis, and biophysical methods. Protein Sci. 2000;9:2528–2534. doi: 10.1110/ps.9.12.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger P, et al. The autophagy-related protein Beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan R, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:833–836. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan S, Nguyen T, Subramanian I, Kelekar A. Autophagy and angiogenesis inhibition. Autophagy. 2007;3:512–515. doi: 10.4161/auto.4734. [DOI] [PubMed] [Google Scholar]
- Rashmi R, Pillai S, Vijayalingam S, Ryerse J, Chinnadurai G. BH3-only protein BIK induces caspase-independent cell death with autophagic features in Bcl-2 null cells. Oncogene. 2008;27:1366–1375. doi: 10.1038/sj.onc.1210783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, et al. Structure of the Bcl-XL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- Schmid D, Dengjel J, Schoor O, Stevanovic S, Munz C. Autophagy in innate and adaptive immunity against intracellular pathogens. J Mol Med. 2006;84:194–202. doi: 10.1007/s00109-005-0014-4. [DOI] [PubMed] [Google Scholar]
- Schweers R, Zhang J, Randall M, Loyd M, Li W, Dorsey F, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN, Marcusson EG, Cereghino JL, Emr SD. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J Cell Biol. 1997;137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Colbert CL, Becker N, Wei Y, Levine B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the γ-herpesvirus 68 Bcl-2 homolog M11. Autophagy. 2008;4:989–997. doi: 10.4161/auto.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacs-Vellai K, Vellai T, Puoti A, Passannante M, Wicky C, Streit A, et al. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr Biol. 2005;15:1513–1517. doi: 10.1016/j.cub.2005.07.035. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Coppola D, Matsushita N, Cualing H, Sun M, Sato S, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous P, Zhu H, Johnstone J, Shelton J, Rajasekaran N, Benjamin I, et al. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci USA. 2008;105:9745–9750. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri M, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008a;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri M, Morselli E, Criollo A, D’Amelio M, Djavaheri-Mergny M, et al. A dual role of p53 in the control of autophagy. Autophagy. 2008b;4:810–814. doi: 10.4161/auto.6486. [DOI] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri M, Orhon I, Kepp O, Morselli E, Criollo A, et al. p53 represses autophagy in a cell cycle-dependent fashion. Cell Cycle. 2008c;7:3006–3011. doi: 10.4161/cc.7.19.6702. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Pasparaki A, Tasdemir E, Maiuri M, Kroemer G. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy. 2008;4:870–873. doi: 10.4161/auto.6730. [DOI] [PubMed] [Google Scholar]
- Tόth M, Sigmond T, Borsos E, Barna J, Erdélyi P, Takács-Vellai K, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- Vaux D, Cory S, Adams J. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1998;335:440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- Wang J. Beclin 1 bridges autophagy, apoptosis and differentiation. Autophagy. 2008;4:947–948. doi: 10.4161/auto.6787. [DOI] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008a;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Sinha S, Levine B. Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy. 2008b;4:949–951. doi: 10.4161/auto.6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yang C, Kaushal V, Shah S, Kaushal G. Autophagy is associated with apoptosis in cisplatin injury to renal tubular epithelial cells. Am J Physiol Renal Physiol. 2008;294:F777–F787. doi: 10.1152/ajprenal.00590.2007. [DOI] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Nat Acad Sci. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Overmeyer J, Maltese W. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- Zhang H, Bosch-Marce M, Shimoda L, Tan Y, Baek J, Wesley J, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]