SUMMARY
Visceral leishmaniasis (VL) in northeast Brazil is a disease caused by infection with the protozoan Leishmania chagasi. Infection leads to variable clinical outcomes ranging from asymptomatic infection to potentially fatal disease. Prior studies suggest the genetic background of the host contributes to the development of different outcomes after infection, although it is not known if ancestral background itself influences outcomes. VL is endemic in peri-urban areas about the city of Natal in northeast Brazil. The population of northeast Brazil is a mixture of distinct racial and ethnic groups. We hypothesized that some sub-populations may be more susceptible than others to develop different clinical outcomes after L. chagasi infection. Using microsatellite markers, we examined whether admixture of the population as a whole, or markers likely inherited from a distinct ethnic background, differed between individuals with VL, individuals with an asymptomatic infection, or individuals with no infection. There was no apparent significant difference in overall population admixture proportions among the three clinical phenotype groups. However, one marker on Chr. 22 displayed evidence of excess ancestry from putative ancestral populations among different clinical phenotypes, suggesting this region may contains genes determining the course of L. chagasi infection.
Keywords: leishmaniasis, Brazil, admixture, STRUCTURE, susceptibility, variation
INTRODUCTION
Visceral leishmaniasis (VL) is a potentially fatal disease caused by infection with the protozoan parasite Leishmania chagasi in the New World, or L. donovani or L. infantum in the Old World.(Wilson, et al. 2005) Infection leads to a spectrum of clinical outcomes ranging from asymptomatic infection, documented by a positive delayed type hypersensitivity (DTH) skin test to Leishmania antigen in the absence of symptomatic disease, to active disease characterized by fevers, cachexia, hepatosplenomegaly and immunosuppression. Without treatment, most symptomatic patients die. Even with appropriate chemotherapy the mortality rate is approximately 10%.(Jeronimo, et al. 2000) VL has become endemic in peri-urban areas about the city of Natal in northeast Brazil since the mid-1980s.(Jeronimo, et al. 1994; Jeronimo, et al. 2004)
Genetic admixture reflects migration and mating between individuals from genetically distinct parental populations.(Griffiths, et al. 1999) Brazil’s complex history of colonialism and the trans-Atlantic slave trade contribute to a highly admixed population. Overall, it is estimated 58% of immigrants who arrived in Brazil between 1500 and 1972 were European-derived, 40% were African-derived, and 2% were Asian-derived.(Callegari-Jacques & Salzano 1999) Natal, the capital city of the northeastern state of Rio Grande do Norte, was founded in 1599 by the Portuguese, although other Europeans including Dutch and French also settled in this area. Africans brought to northeast Brazil as slaves were primarily from the Congo, Angola and Guinea.(Salas, et al. 2004) Native South American influences are primarily from the Potiguara (Tupi) and Cariri tribes.(Krieger, et al. 1965; Da Silva, et al. 1981) Although other ethnic populations have played a role in the history of other regions of Brazil (e.g. Japanese, Germans and Italians in the south of Brazil), in the northeast, the primary contributors to the current population have been European, African and Native Brazilian.(Instituto Brasileiro de Geografia Estatistica [IBGE] 2007)
Our previously published analyses of this population(Jeronimo, et al. 1994; Jeronimo, et al. 2004; Jeronimo, et al. 2007a; Jeronimo, et al. 2007b) and other studies of VL in Brazil(Cabello, et al. 1995; Blackwell, et al. 1997; Jamieson, et al. 2007) and Sudan(Mohamed, et al. 2003; Bucheton, et al. 2003; Mohamed, et al. 2004; Miller, et al. 2007) suggest a strong genetic contribution to the outcome of infection with visceralizing Leishmania species. We hypothesized sub-populations of individuals with different ethnic contributions, when exposed to Leishmania, are likely to develop different clinical outcomes of infection. We tested this hypothesis by examining whether admixture proportions of the population as a whole and whether admixture contributions at specific genomic regions differed between defined clinical phenotypes. Our approach to test this hypothesis employed microsatellite marker data from a genome-wide linkage scan,(Jeronimo, et al. 2007a) compared with microsatellite marker data from the CEPH Human Genome Diversity Project.(Cann, et al. 2002; Rosenberg, et al. 2002) With these genetic markers, we determined the ethnic contributions to our population as a whole and found regions where contributions from different ancestral backgrounds differed between phenotypic groups. Genes in these regions now become candidates for further studies of susceptibility to visceral leishmaniasis.
SUBJECTS, MATERIALS AND METHODS
Brazilian study participants
Brazilian subjects living in neighborhoods with endemic L. chagasi infection were recruited for a family study described elsewhere.(Jeronimo, et al. 2004; Jeronimo, et al. 2007a) In the current study, three mutually exclusive phenotype categories were employed: (1) VL: subjects who had either ongoing or a history of active, symptomatic VL disease. (2) DTH+: subjects with no history of documented VL who had a positive DTH skin test to Leishmania antigen.(Melo, et al. 1977) These individuals had likely had an asymptomatic infection in the past, and might be immune to reinfection with L. chagasi. (3) DTH−: subjects who lived in a household where 40% or more family members were infected (VL+ or DTH+), and either in or near a household where an active case of VL had occurred, but they themselves had a negative Montenegro test, negative serology, and no history of VL.
Ethical approvals
Approval for work with human subjects was obtained from the Ethical Research Committee at the Universidade Federal do Rio Grande do Norte, the Comissão Nacional de Ètica em Pesquisa (CONEP), the University of Iowa, Johns Hopkins University, the University of Virginia and the National Institutes of Health/National Human Genome Research Institute boards. The UFRN Institutional Review Board is registered with the NIH. All subjects or guardians of minors signed an approved informed consent in Portuguese.
Genotype Data
A total of 405 (385 autosomal, 20 sex chromosomal) multiallelic microsatellite markers across the genome were typed on 1254 individuals from 191 families at the Center for Inherited Disease Research (CIDR).(Jeronimo, et al. 2007a) Initially, we selected all persons who did not have parents in the database (irrespective of clinical phenotype) and designated this group as our ‘Natal Founders’ group (n=321). This group was used to calculate overall admixture proportions of this Natal cohort and for preliminary analysis of the statistical models. Subsequent analysis of the three specific clinical phenotype groups was accomplished by again searching through all family pedigrees and choosing unrelated individuals from each phenotype group with genotype data available (VL+, n=105; DTH+, n=193; DTH−, n=110).
The microsatellite markers used for the Natal data were based on the Marshfield Genetics Version 8 marker set with modifications as outlined on the CIDR website. CEPH Human Genome Diversity Project data were downloaded from Dr. Noah Rosenberg’s website. Lymphoblastoid cell lines from 52 indigenous populations were genotyped at 377 microsatellite markers(Rosenberg, et al. 2002; Rosenberg, 2006) covering all autosomes using a marker set modified from the Marshfield Genetics Version 10 marker set. Ideally to analyze our Natal, Brazil cohort, we would have preferred to use specific genotype data from Angola, the Congo Basin and Portuguese Europeans. Because genotype data from these populations are not publicly available, ancestral populations used as training data for our study were chosen from 3 continental groups of CEPH Diversity genotype data: European [French and Basque, n=52 total], African [Mandenka and Yoruba, n=44 total] and native Brazilian [Surui and Karitiana, n=22 total] after excluding all related individuals.(Rosenberg, 2006) It is likely that Natal’s “true” ancestral populations are different from the “putative” ancestral populations used in our analyses. However, Rosenberg (Rosenberg, et al. 2002) suggested inter-continental ethnic differences are larger than most intra-continental ethnic differences. Therefore, use of these population groups as training data for the Structure program should provide a reasonable approximation to the true ethnic background.
Of the 377 autosomal markers in the CEPH Diversity marker set, 289 were common to the CIDR marker set. To accommodate slight variations in allele size due to genotyping the CEPH Diversity data and the CIDR data on different platforms at different institutions, the CEPH Diversity data were manually re-binned to match the amplified allele sizes of the CIDR data for all 289 overlapping markers, using two CEPH individuals who had been genotyped by both CIDR and the CEPH Diversity Project (CEPH individuals 1331-01 and 1331-02) as a reference.
Structure Data Analysis
STRUCTURE v.2.1(Falush, et al. 2003) uses Bayesian inference and Monte Carlo Markov Chain (MCMC) modeling to cluster multi-locus genotype data into a user-specified number of subpopulations (K). STRUCTURE has two primary outputs: estimated admixture proportions both for each experimental individual and for the experimental group as a whole, and an estimated ‘Posterior Probability’ (PP) expressed as an estimated natural logarithm for the probability of the ‘fit’ of the experimental data to the training data under the model being tested.(Pritchard, et al. 2003) Less negative PPs indicate a better ‘fit’ of the model. After initial testing, we used a burnin of 20,000 iterations and an MCMC run of 10,000 iterations. All models were run at least three times. All PPs are the mean of at least three runs. We tested three different statistical models; (i) admixture model/correlated allele frequencies, (ii) admixture model/independent allele frequencies, and (iii) linkage model/independent allele frequencies.
Z-Score Calculation and Analysis
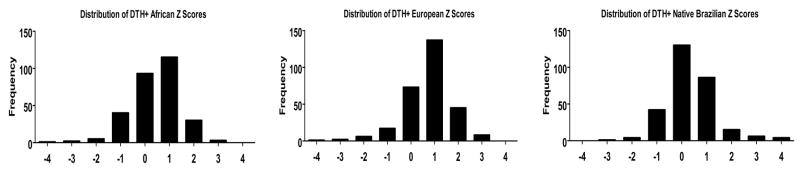
Following the method of Zhu, et al.,(Zhu, et al. 2005) excess ancestry (EA) for each population (African, European or native Brazilian) at each marker was calculated for each of the three clinical phenotype groups (VL+, DTH+, DTH−) using estimated admixture data from the primary STRUCTURE clustering assuming K=3 under the linkage/independent allele frequencies model for each individual. First, for each individual, the estimated genome-wide ancestry from each hypothetical ancestral population (Mi) and for each marker (qil) was obtained from the primary STRUCTURE results. These data were used to calculate for marker location l and for each clinical phenotype group (in comparison to each of the three ancestral populations), ΔΠl, where . We next calculated the EA as a Z-score, where . Z score distributions are plotted in Figure 4 and Supplementary Figure 1. Significant EA for each ancestral background at individual markers in a given clinical phenotype group was defined as markers yielding a |Z-score| > 2 and an empiric p value < 0.0005.
Figure 4. Distribution of DTH+ Z scores.
The distribution of African-derived, European-derived and Native Brazilian-derived Z scores for the DTH+ (asymptomatic infection) clinical phenotype group are plotted. Data for the VL+ and DTH− clinical phenotype groups was similar (Supplementary Figure 1).
Empiric p-values for each Z-score were generated by permutation. A group of individuals was randomly chosen from the total data set of all three clinical phenotype groups and then EA Z-scores were re-calculated at each marker and for each ancestral background in this randomly chosen group. A running tabulation was maintained for markers where these calculated |Z-scores| on the permuted data set were >2. This was repeated 10,000 times for all markers. These tabulations were then divided by 10,000 to generate empiric p-values at each marker relative to each ancestral background (e.g. a p-value = 0.0010 represents a marker with a Z-score > 2 only 10 times out 10,000).
Z-scores > 2 for a particular marker, relative to a particular ancestral group, should indicate the presence of alleles in that clinical phenotype group derived from one particular ancestral group more than expected. Similarly, Z-scores < −2 at a particular marker should indicate the unexpected absence of alleles derived from a putative ancestral group in that clinical phenotype group.
RESULTS
Supplementary Table I shows the distribution by gender and age for all the individuals used in our analysis (n=439). As an initial test of our method, genotypes of the six CEPH Diversity populations were analyzed by unsupervised clustering using STRUCTURE with the ‘admixture/correlated allele frequency model’ assuming three subpopulations (K=3; Figure 1). This analysis correctly identified and clustered each individual in the six CEPH Diversity populations into three continental groups. Some European individuals shared genetic similarities with native Brazilians (as evidenced by the slight red at the top of some of the European individuals) and there was apparently one native Brazilian individual with some European ancestry (see the green section contained within one of the native Brazilians). Since our CEPH Diversity populations were approximations of the known “true” ancestral populations, we did not test clustering of the CEPH Diversity project data beyond K > 3. Our interest was only to evaluate whether there were measurable differences among the “continental” groups, not differences specific to a particular ethnic group. Therefore, although genotype data from all six CEPH Diversity populations were used our subsequent analyses, we felt it simplest to conceptualize the data in reference to these three continental groups (European, African, Native Brazilian).
Figure 1. Unsupervised Clustering of CEPH Diversity Project Data.
The extracted CEPH Diversity Project data for 289 overlapping markers in six populations (French, Basque, Mandenka, Yoruba, Karitiana, Surui) were clustered using STRUCTURE under the admixture model, assuming correlated allele frequencies and with K=3. Each vertical line represents a separate individual and is a graphical representation of that individual’s admixture proportions.
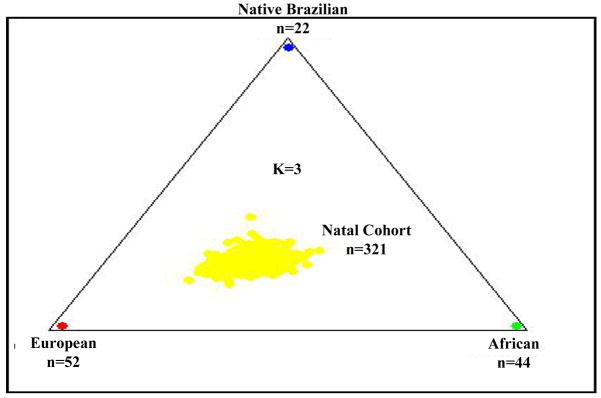
Next, using the CEPH Diversity populations as ‘training data,’ we performed a supervised clustering of genotype data from unrelated individuals in our ‘Natal Founders’ cohort (n=321) under the ‘admixture model’, testing both the correlated allele frequency and the independent allele frequency models for various values of K. Under the assumption that K=3 (Figure 2), our ‘Natal Founders’ group clustered more closely with the European and African data than the native Brazilian data. Subsequent analysis suggested the independent allele frequency model and higher values of K improved the supervised clustering (i.e. improved the ‘fit’) of our data (data not shown). We also analyzed our Natal Founders cohort to test whether removing one or more hypothetical ancestral populations would improve the fit to data from the Natal Founders. Removing either the Basque or Mandenka data only marginally improved the PPs at K=3, 4 or 5 (Supplementary Table II). Therefore, all six of the initial CEPH Diversity data sets were retained in subsequent analysis of clinical phenotypes.
Figure 2. Supervised Clustering Of Unrelated Individuals From Natal Brazilian Cohort.
Each dot represents one individual. The ‘Natal Founders’ cohort data consisted of unrelated individuals irrespective of phenotype. Using STRUCTURE, the data were analyzed using the CEPH Diversity data from six ethnic groups (three continental groups; European, African, Native Brazilian) as ‘training data’ in a supervised clustering under the admixture model, assuming correlated allele frequencies with K=3.
Next, we performed separate supervised clusterings for each of the three clinical phenotype groups (VL+, n=105; DTH+, n=193; DTH−, n=110,) using individuals from all six CEPH Diversity populations grouped into the 3 continental groups as training data (See Methods section for details of phenotype definitions). Table I shows the PPs for the VL+, DTH+ or DTH− phenotype groups analyzed under the Admixture model, assuming either correlated allele frequencies or independent allele frequencies for K = 2–5. (K=1 and K=6 were also tested, but these data are not shown.) Within each clinical phenotype, the trend was similar: as K increased up to 6, PP values became less negative, indicating an improved fit. Additionally, for each value of K, the independent allele frequency models fit the data better than the correlated models and the ‘linkage/independent allele frequencies’ model outperformed the ‘admixture/independent allele frequencies’ model (Table I). As described in Falush, et al.,(Falush, et al. 2003) the ‘admixture model’ allows individuals to have mixed ancestry and assumes each individual will have some fraction of their genome inherited from one of the K reference populations. The ‘linkage model’ is a generalization of this admixture model that incorporates multipoint analysis and admixture linkage disequilibrium (see Methods).
Table I.
Posterior Probabilities For Each Leishmaniasis Clinical Phenotype Group Under Several Analysis Modelsa
| Clinical Phenotype | VL+ | DTH+ | DTH− | |
|---|---|---|---|---|
| # of Individuals | 105 | 193 | 110 | |
| # of Diversity | 118 | 118 | 118 | |
| Model Choice | K | |||
| Admixture, Correlated | 2 | −3,767,149 | −12,081,099 | −4,518,476 |
| 3 | −2,159,683 | −6,877,618 | −2,492,999 | |
| 4 | −540,150 | −2,253,407 | −599,426 | |
| 5 | −373,337 | −829,615 | −449,287 | |
| Admixture, Independent | 2 | −448,861 | −1,100,109 | −469,277 |
| 3 | −383,298 | −895,070 | −425,052 | |
| 4 | −342,407 | −744,724 | −368,642 | |
| 5 | −306,673 | −616,457 | −318,084 | |
| Linkage, Independent | 3 | −356,317 | −791,652 | −372,799 |
| 4 | −324,597 | −673,970 | −339,270 | |
| 5 | −293,340 | −565,196 | −306,215 |
Analysis performed using Diversity Project data as ‘training data’ for various K under several analysis models. The ‘Linkage/independent allele frequency’ model generates the best ‘fit.’
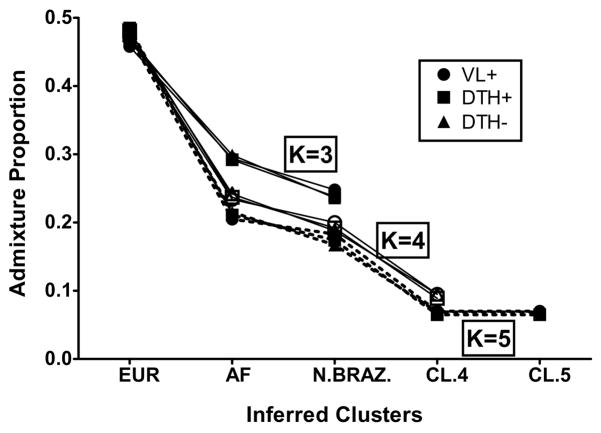
Figure 3 graphically summarizes results from supervised clusterings of each clinical phenotype group (VL+ vs. DTH+ vs. DTH−) using all six CEPH Diversity populations (represented as three continental groups) employing the ‘linkage/independent allele frequency’ model assuming K = 3, 4 or 5. Two main conclusions can be drawn. First, for each value of K, the three lines representing the three different phenotype groups essentially overlap, indicating no significant genome-wide differences in admixture proportions when comparing the VL+ group vs. the DTH+ group vs. the DTH− group; i.e. there was not a distinct ethnic group over-represented in one of these phenotype groups. Second, as K increased, even though the overall ‘fit’ of the data improved (Table I), the proportion of European ancestry for each clinical group changed very little. In contrast, when K was increased from 3 to 5 the proportion of native Brazilian and African ancestry dropped considerably.
Figure 3. Genome-Wide Admixture Proportions By Leishmaniasis Phenotype.
Genome-wide admixture proportions by clinical phenotype under the ‘Linkage’ model, assuming independent allele frequencies, for various K values. There were no differences between clinical phenotypes in the estimated admixture proportions for K=3, 4 or 5 under the model parameters. As K increased, the component of European admixture stayed relatively constant, whereas the estimated African component showed the greatest drop, suggesting STRUCTURE detected the greater genetic CEPH Diversity of African-derived ancestral genotypes.
Since we did not see evidence of genome-wide differences in admixture proportions, we next analyzed each individual marker for evidence of excess ancestry (EA) to determine whether portions of chromosomes derived from distinct ethnic backgrounds might be over-represented in one of the phenotype groups, which could contain potential susceptibility genes. We followed the methodology of Zhu, et al.,(Zhu, et al. 2005) which generates an estimated EA Z-score for each individual marker relative to the three putative continental backgrounds (K=3, representing European, African, Native Brazilian ancestral populations) for each clinical phenotype group (see the Methods section for a detailed explanation). We chose an EA Z-score cutoff of ±2, which should represent the 5% most outlying data. Z score distributions for the African-derived, European-derived and Native Brazilian-derived DTH+ clinical phenotype group are plotted in Figure 4. Distribution plots for all other clinical phenotype groups were similar (Supplementary Figure 1). Statistical significance was assessed via permutation using randomly chosen phenotype groups to re-calculate Z-scores as a way to generate empiric p-values for each marker (as explained in the Methods section). Selected allele frequency (AF) data are presented in Figure 5. Among 289 tested markers, only 51 markers had Z-scores >2, or <−2 in at least one comparison (i.e. at least one phenotypic group showed a significant Z-score when considering one reference population). Of those 51 markers, only 1 marker on chromosome 22 retained statistical significance after permutation. The data for all 51 markers with |Z-scores| > 2 and their associated p-values are listed in Supplementary Table III.
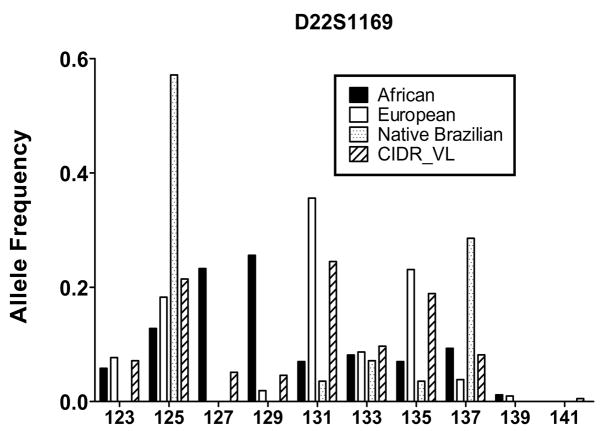
Figure 5. Allele Frequency Data At A Previously Reported Candidate Susceptibility Locus.
AF data for a marker that demonstrated significant Excess Ancestry (EA) (see Methods for further definition) and was near a previously reported VL susceptibility locus(Bucheton, et al. 2003) are displayed. The plot shows the distribution of alleles at D22S1169, which was determined to have significant European-derived EA in VL+ individuals relative to the putative ancestral population groups (European, African, Native Brazilian).
SLC11A1 (which encodes a pH-dependent divalent cation carrier previously called NRAMP1), TNFA encoding TNF-α and markers on Chromosome 22q12 have all been reported to be associated with or to be linked to genes controlling susceptibility to VL.(Blackwell, et al. 1997; Bucheton, et al. 2003; Karplus, et al. 2002) The chromosome 22q12 region has shown evidence of linkage for VL in some Sudanese populations.(Bucheton, et al. 2003) In our data, a marker near this region (D22S1169) also showed European-derived EA Z-scores >2 for VL+ individuals only (p < 0.0001) (Figure 5).
Previous work has shown different markers in TNFA, which encodes the TNF-α gene product, are associated with different phenotypic outcomes of leishmania infection, and varying levels of serum TNF-α have been associated with different clinical forms of leishmaniasis. (Karplus, et al. 2002) In addition, the gene SLC11A1 (NRAMP1) has been shown both in animal models and in some human populations to contribute to susceptibility to Leishmania infection. (Blackwell, et al. 1997) Nonetheless, after permutation we did not find evidence for significant EA at marker D2S427 near SLC11A1 or at D6S2439 near TNFA for any ancestral background across any of the three clinical phenotypes.
In contrast to a disorder like cleft lip where the dichotomous definitions of ‘affected’ and ‘unaffected’ can be clearly delineated, we have ordinal categories of clinical disease in response to infection: individuals who have symptomatic disease (VL+), individuals who have been exposed but who have controlled the infection (DTH+) and individuals who have been exposed but show no evident immune response to the parasite (DTH−). These ordered groups should be regarded as corresponding to different host innate and/or adaptive immune responses. Therefore, to make comparisons with physiological relevance, we examined markers with the highest differences in the EA Z-scores between the VL+ group versus the DTH+ group or the difference in the EA Z-scores of the DTH+ group versus the DTH− group (Supplementary Table IV). Chromosomes 1 and 4 each had several markers with large differences in Z-scores. Interestingly, some markers were found to have large EA values (i.e., |Z-scores| > 2) for all clinical phenotype and permutation groups examined (i.e. their empiric p-values were close to 1). These most likely reflect ethnic-specific markers (Supplementary Table III) but are not associated with our phenotypes.
There are several genes known to have clear allelic differences among ancestral populations, including SLC24A5 (a cation exchange channel at 15q21.1 identified as influencing skin pigmentation(Lamason, et al. 2005)), DARC (the Duffy antigen at 1q21-q22(Hamblin, et al. 2002)), LCT (the lactase gene at 2q21(Tishkoff, et al. 2007)) and ABCC11 (the earwax gene at 16q12.1(Yoshiura, et al. 2006)). In our data, there were no significant EA Z-scores for markers near any of these genes when comparing any of the thee clinical phenotypes examined here (data not shown).
DISCUSSION
Studies of human leishmaniasis have identified several genes and/or regions that may contain genes controlling susceptibility to clinical outcomes of infection in exposed populations.(Jeronimo, et al. 2007a; Blackwell, et al. 1997; Jamieson, et al. 2007; Bucheton, et al. 2003; Karplus, et al. 2002) Here, we examine leishmaniasis in a Brazilian population endemic for Leishmania chagasi, the cause of South American visceral leishmaniasis (VL). This study tested the hypothesis that individuals in each of three clinical phenotype groups might have different levels of admixture of ancestral backgrounds compared to the overall population, either overall or at specific markers. We considered three distinct clinical phenotypes: symptomatic infection (VL+), asymptomatic self-curing infection (DTH+), and living in a highly endemic situation without apparent immune response to the parasite (DTH−). We estimated genetic admixture of these exposed individuals using microsatellite marker data from a genome wide linkage scan in a family study.(Jeronimo, et al. 2007a) Although there were no detectable overall differences in admixture proportions among clinical phenotypes (Figure 3), we were able to detect one marker with significant excess ancestry (EA) for a particular clinical phenotype group compared to the other two groups (D22S1169, Figure 5). This region of EA warrants further investigation to identify candidate susceptibility genes predisposing to disease status.
Earlier studies of genetic admixture in this region of Brazil using limited numbers of genetic markers (e.g. red blood cell and serum protein polymorphisms) estimated Brazilians in Natal had approximately 58% European ancestry, 25–30% African ancestry and 11–17% Native Brazilian ancestry.(Krieger, et al. 1965; Franco, et al. 1982) We compared our Natal subjects to groups of unrelated individuals representing European [French and Basque], African [Mandenka and Yoruba] and native Brazilian [Surui and Karitiana] populations drawn from the CEPH Human Genome Diversity Project. Analysis of the 321 unrelated individuals in our ‘Natal Founders’ group exhibited approximately 47% European, 29% African and 24% Native Brazilian ancestry (Figure 2). Interestingly, when we increased the number of hypothetical sub-populations (K) in our analysis, the putative European component did not change appreciably, whereas both the African and native Brazilian components decreased (Figure 3). These results suggest as K increases, the STRUCTURE algorithm parses the African ancestral component of our Natal cohort into ever smaller groups. This is not entirely unexpected since Africans brought to Natal were not from a single region of Africa and, on average, Africans have smaller haplotype blocks and greater genetic diversity.(Tishkoff & Williams, 2002)
In contrast to some other diseases, there is no published evidence that any single ethnic group is at higher risk for developing symptomatic VL versus asymptomatic L. chagasi infection. Consistent with this, our Natal data showed no significant differences in genome-wide admixture proportions when comparing individuals from to the VL+, DTH+ or DTH− clinical phenotype groups (Figure 3). However, when we queried whether specific markers would display EA for any particular ancestral population in one of the three clinical phenotype groups, the data suggested several regions of the genome could potentially control susceptibility or resistance to L. chagasi infection. One marker in the genome showed significant EA (i.e. |Z-scores| > 2 and was near a locus previously reported as potentially linked to or associated with VL susceptibility) (Figure 5). Marker D22S1169 near the chromosome 22q12 susceptibility region identified in linkage studies of Sudanese families (Bucheton, et al. 2003) demonstrated positive European-derived EA among VL+ individuals. This observation suggests individuals who inherited alleles at these markers similar to those of the European population may be predisposed to developing symptomatic infections.
As an alternative strategy to correlating our EA Z-scores with previous publications, we searched for markers which exhibited large differences among clinical phenotype groups. For example, markers showing large EA differences for any particular ancestral background when comparing VL+ to DTH+ individuals might lie near a region containing a gene controlling development of symptomatic disease as opposed to asymptomatic infection. Similarly, markers which displayed large EA differences between DTH+ and DTH− individuals might lie near a gene whose product contributes to or protects from infection. Therefore, we took Z-scores for each ancestral background and clinical phenotype group and ordered them by their absolute values of the Z-score differences, |VL+_Zscore DTH+_Zscore| and |DTH+_Zscore DTH−_Zscore|. There were few regions with large differences between Z-scores across these three phenotype groups, consistent with a highly admixed population (Supplementary Table IV). It has already been shown that loci controlling susceptibility in one population [e.g. Sudan](Miller, et al. 2007) may not be the same as those controlling susceptibility in another population [e.g. northern Brazil].(Jeronimo, et al. 2007a) The paucity of regions yielding large differences in EA Z-scores, the lack of differences in genome-wide admixture proportions among the three clinical phenotype groups under various models and the lack of EA in genomic regions near ethnic specific markers most likely reflect in the extensive admixture of this population.
Evolutionary pressure exerted by infectious diseases has undoubtedly helped to shape our genetic makeup in many ways.(Burgner, et al. 2006) Only recently have we begun to understand how molecular mechanisms can play a role in genetic susceptibility to infectious disease. Studying admixed populations offers a novel way to investigate the pathogenesis of infectious diseases and to identify genes playing a causal role in modulating human susceptibility and/or resistance to infectious pathogens.
Supplementary Material
Distribution of VL+ and DTH− Z scores. The distribution of African-derived, European-derived and Native Brazilian-derived Z scores for the (A) VL+ (symptomatic infection) clinical phenotype group and the (B) DTH− (exposed, but uninfected) clinical phenotype group are plotted.
Acknowledgments
This work was supported in part by NIH R01 grants AI048822, AI067874, and AI045540 from the National Institutes of Health (MEW), Merit Review and Gulf War RFA grants from the Department of Veterans’ Affairs (MEW), NIH TMRC grant P50 AI-30639 (RFSB, ETN, SMBJ, RDP) and in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. The work of NAE was supported by NIH T32 AI07511.
Abbreviations used
- VL
visceral leishmaniasis
- DTH
delayed type hypersensitivity
- CIDR
Center for Inherited Diseases Research
- PP
posterior probability
- MCMC
Monte Carlo Markov Chain
- EA
excess ancestry
- AF
allele frequency
- VL+
individuals with documented current or historical symptomatic infection
- DTH+
individuals with documented asymptomatic infection
- DTH−
individuals with a documented negative DTH reaction who live in an endemic household
References
- Blackwell JM, et al. Immunogenetics of leishmanial and mycobacterial infections: the Belem Family Study. Philos Trans R Soc Lond, B, Biol Sci. 1997;352:1331–1345. doi: 10.1098/rstb.1997.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheton B, et al. A major susceptibility locus on chromosome 22q12 plays a critical role in the control of kala-azar. Am J Hum Genet. 2003;73:1052–1060. doi: 10.1086/379084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgner D, et al. Genetic susceptibility to infectious diseases: big is beautiful, but will bigger be even better? Lancet Infect Dis. 2006;6:653–663. doi: 10.1016/S1473-3099(06)70601-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello PH, et al. Familial aggregation of Leishmania chagasi infection in Northeastern Brazil. Am J Trop Med Hyg. 1995;52:364–365. doi: 10.4269/ajtmh.1995.52.364. [DOI] [PubMed] [Google Scholar]
- Callegari-Jacques SM, Salzano FM. Brazilian Indian/non-Indian interactions and their effects. Ciênc Cult. 1999;51:166–174. [Google Scholar]
- Cann HM, et al. A Human Genome Diversity Cell Line Panel. Science. 2002;296:261b–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Da Silva MIAF, et al. Migration, inbreeding, blood groups and hemoglobin types in Natal, Brazil. Stud Phys Anthropol. 1981;7:3–11. [Google Scholar]
- Falush D, et al. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco MHLP, et al. Blood polymorphisms and racial admixture in two Brazilian populations. Am J Phys Anthropol. 1982;58:127–132. doi: 10.1002/ajpa.1330580204. [DOI] [PubMed] [Google Scholar]
- Griffiths AJF, Gelbart WM, et al. Modern Genetic Analysis. 1. New York: W.H. Freeman; 1999. [Google Scholar]
- Hamblin MT, et al. Complex signatures of natural selection at the Duffy blood group locus. Am J Hum Genet. 2002;70:369–383. doi: 10.1086/338628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Instituto Brasileiro de Geografia Estatística [IBGE] Brasil: 500 Anos de Povoamento. Available: http://www.ibge.gov.br/brasil500/index2.html.
- Jamieson SE, et al. Genome-wide scan for visceral leishmaniasis susceptibility genes in Brazil. Genes Immun. 2007;8:84–90. doi: 10.1038/sj.gene.6364357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo SMB, et al. An emerging peri-urban pattern of infection with Leishmania chagasi, the protozoan causing visceral leishmaniasis in Northeast Brazil. Scand J Infect Dis. 2004;36:443–449. doi: 10.1080/00365540410020451. [DOI] [PubMed] [Google Scholar]
- Jeronimo SMB, et al. Genetic predisposition to self-curing infection with the protozoan Leishmania chagasi: a genomewide scan. J Infect Dis. 2007a;196:1261–1269. doi: 10.1086/521682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo SMB, et al. Genes at human chromosome 5q31.1 regulate delayed-type hypersensitivity responses associated with Leishmania chagasi infection. Genes Immun. 2007b;8:539–551. doi: 10.1038/sj.gene.6364422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo SMB, et al. An urban outbreak of visceral leishmaniasis in Natal, Brazil. Trans R Soc Trop Med Hyg. 1994;88:386–388. doi: 10.1016/0035-9203(94)90393-x. [DOI] [PubMed] [Google Scholar]
- Jeronimo SMB, et al. Natural history of Leishmania (Leishmania) chagasi infection in Northeastern Brazil: long-term follow-up. Clin Infect Dis. 2000;30:608–609. doi: 10.1086/313697. [DOI] [PubMed] [Google Scholar]
- Karplus TM, et al. Association between the tumor necrosis factor locus and the clinical outcome of Leishmania chagasi infection. Infect Immun. 2002;70:6919–6925. doi: 10.1128/IAI.70.12.6919-6925.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger H, et al. Racial admixture in north-eastern Brazil. Ann Hum Genet. 1965;29:113–125. doi: 10.1111/j.1469-1809.1965.tb00507.x. [DOI] [PubMed] [Google Scholar]
- Lamason RL, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- Melo MN, et al. Padronizacao do Antigeno de Montenegro [Standardization of the Montenegro antigen] Rev Inst Med Trop São Paulo. 1977;19:161–164. [PubMed] [Google Scholar]
- Miller EN, et al. Y chromosome lineage- and village-specific genes on chromosomes 1p22 and 6q27 control visceral leishmaniasis in Sudan. PLoS Genet. 2007;3:e71. doi: 10.1371/journal.pgen.0030071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed HS, et al. Genetic susceptibility to visceral leishmaniasis in The Sudan: linkage and association with IL4 and IFNGR1. Genes Immun. 2003;4:351–355. doi: 10.1038/sj.gene.6363977. [DOI] [PubMed] [Google Scholar]
- Mohamed HS, et al. SLC11A1 (formerly NRAMP1) and susceptibility to visceral leishmaniasis in The Sudan. Eur J Hum Genet. 2004;12:66–74. doi: 10.1038/sj.ejhg.5201089. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, et al. Structure. 2003:2.1. [Google Scholar]
- Rosenberg NA. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Salas A, et al. The African diaspora: mitochondrial DNA and the Atlantic slave trade. Am J Hum Genet. 2004;74:454–465. doi: 10.1086/382194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Williams SM. Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet. 2002;3:611–621. doi: 10.1038/nrg865. [DOI] [PubMed] [Google Scholar]
- Wilson ME, et al. Immunopathogenesis of infection with the visceralizing leishmania species. Microb Pathog. 2005;38:147–160. doi: 10.1016/j.micpath.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Yoshiura K, et al. A SNP in the ABCC11 gene is the determinant of human earwax type. Nat Genet. 2006;38:324–330. doi: 10.1038/ng1733. [DOI] [PubMed] [Google Scholar]
- Zhu X, et al. Admixture mapping for hypertension loci with genome-scan markers. Nat Genet. 2005;37:177–181. doi: 10.1038/ng1510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of VL+ and DTH− Z scores. The distribution of African-derived, European-derived and Native Brazilian-derived Z scores for the (A) VL+ (symptomatic infection) clinical phenotype group and the (B) DTH− (exposed, but uninfected) clinical phenotype group are plotted.