Abstract
A mouse model of human ZNF198–fibroblast growth factor receptor-1 (FGFR1) stem cell leukemia lymphoma has been developed to investigate mechanisms of oncogenesis and progression. Using array-based comparative genomic hybridization, we followed disease progression after serial transplantation of ZNF198-FGFR1–transformed stem cells that give rise to a distinct myeloproliferative disorder and T-lymphoblastic leukemia. A consistent, frequently homozygous, chr14:53880459-55011545 deletion, containing the T-cell receptor α and δ genes, was identified in the bone marrow, spleen, and lymph nodes in all cases. The absence of cell-surface T-cell receptor α in tumor cells precludes CD3 recruitment, resulting in loss of a functional T-cell receptor complex, supporting the idea that prevention of maturation of CD4+/CD8+ double-positive immature T cells is important in ZNF198-FGFR1 disease development. Up-regulation of the B-cell line 2, interleukin 7 receptor α and interleuking 2 receptor α prosurvival genes in these undifferentiated tumor precursor cells suggests one mechanism that allows them to escape apoptosis in the thymus. Thus, we have defined an important event in the process of ZNF198-FGFR1–induced T-cell leukemia.
Introduction
Human stem cell leukemia-lymphoma syndrome (SCLL), also known as 8p11 myeloproliferative syndrome, is a rare atypical myeloproliferative disorder. SCLL constitutes a clinical phenotype with features of both lymphoma and eosinophilic myeloproliferative disorders and is characterized by a reciprocal chromosome translocation, resulting in a chimeric protein that activates the kinase domain of the fibroblast growth factor receptor-1 (FGFR1).1 To date, at least 8 gene partners have been shown to fuse to FGFR1, including ZNF198 on 13q12,2,3 CEP110 on 9q33,4 and FOP on 6q275 (see Tefferi and Gilliland1 for review). The most commonly observed translocation is the t(8;13) (p11;q12), in which the zinc finger domain of ZNF198 is fused to the intracellular kinase domain of FGFR1.2,3,6 Hepatosplenomegaly is characteristic of these myeloproliferative disorder patients, and almost all show T-lymphoblastic lymphoma, except for one case with B-cell acute lymphoblastic lymphoma.7 The clinical course of SCLL is aggressive, with rapid transformation to acute myeloid leukemia and lymphoblastic lymphoma of common T-cell origin.7–11 Treatment with conventional chemotherapy is often not effective,12,13 and allogeneic bone marrow (BM) transplantation seems to be the only potentially curative therapeutic option.7
In SCLL, both myeloid and lymphoid lineage cells exhibited the 8p11 translocation, suggesting a stem cell origin. The FGFR1 fusion protein in SCLL results in the constitutive and ligand-independent activitation of the FGFR1 signal transduction pathway, and is believed to be essential for disease pathogenesis, as evidenced in the clinical cases and murine models.14,15 How the ZNF198-FGFR1 fusion kinase results in disease progression, however, remains unclear, as does its relationship with the development of concurrent myeloproliferative and lymphoid malignancy.
Previously, it was reported that ZNF198-FGFR1–induced T lymphoma in mice is oligo- or monoclonal,14 which suggested that either the expression of ZNF198-FGFR1 alone is not sufficient for leukemogenesis, or that a restricted population of hematopoietic stem/progenitor target cells is sensitive to ZNF198-FGFR1 transformation. Because most cancers probably develop through the accumulation of multiple genetic and epigenetic changes,16,17 a more detailed analysis of the events that take place during ZNF198-FGFR1–initiated cancer development may provide a better understanding of the underlying molecular basis of the disease. To address this question, we used a BM transduction and transplantation mouse model for SCLL, combined with array-based comparative genomic hybridization (aCGH) to systematically characterize the occurrence and frequency of genomic changes during tumor initiation and progression. A consistent homozygous deletion occurs in the 14C2 region, which contains the T-cell receptor α (Tcra) and delta (Tcrd) genes, in transplantable T-cell lymphomas. Loss of Tcra/d is accompanied by loss of cell surface CD3, demonstrating a nonfunctional T-cell receptor (TCR) complex. These T-cell lymphomas are also CD4+ and CD8+, demonstrating a block in normal T-cell maturation, and survival of these cells is accompanied by up-regulation of the B-cell line (BCL)2, interleukin (IL)–7 receptor (R)α, and IL2Rα prosurvival genes, which provides mechanistic insights into the development of ZNF198-FGFR1–induced lymphomas. Immunophenotyping suggests that the stem cell origin of SCLL is lineage (Lin)−Sca1+Kit+.
Methods
Retroviral constructs and production
The original 3.9-kilobase ZNF198-FGFR1 cDNA2,18 was subcloned into the BamHI/NotI-digested murine stem cell virus–internal ribosome entry site–green fluorescent protein (GFP) retroviral vector19 to generate monokine induced by interferon-γ (MIG)–ZNF198-FGFR1 expressing both the fusion kinase and GFP. High-titer, helper-free retrovirus was generated using the “ping-pong.”20 Phoenix-Ampho and Phoenix-Eco packaging cell lines (ATCC) were cocultured (1:1), and plasmid DNAs were introduced using Effectene (QIAGEN), according to the manufacturer's instructions. Supernatants were collected in fresh media at 36, 48, 60, and 72 hours after transfection. Pooled supernatants were filtered through 0.45-μm membranes and stored at −80°C. Viral titers were determined by infecting NIH-3T3 cells (ATCC) with serial dilutions of virus and analyzing the percentage of GFP+ cells by flow cytometry. Viral stocks (≥ 5 × 106 colony-forming unit/μL supernatant) were used to infect primary murine BM cells.
BM retroviral transduction and transplantation
BALB/cAnNTac, 6- to 8-week-old mice (Taconic Farms) were used throughout. For BM cell transduction, male donor mice were first treated with 5-fluorouracil (200 mg/kg) intraperitoneal injection, 2 to 3 days before the BM being flushed from the femurs and tibiae. Recovered cells were then incubated in IL2, IL3, and stem cell factor (final concentration 10 ng/mL each) for 24 hours before infection. After prestimulation, 4 rounds of infection at 12-hour intervals were performed with the retroviral supernatant (25% vol/vol) in the presence of 10 μg/mL polybrene, using the same media and cytokines as described above. Female recipient mice were lethally irradiated (900 cGy), as described previously.21 Transduced cells were then transplanted by injection of 1.5 × 106 cells into the lateral tail vein of these mice. For second and third transplantations, 0.5-1.0 × 106 BM cells from primary and secondary diseased mice, respectively, were again transferred by tail vein injection into irradiated (600 cGy) female BALB/c recipient mice.
Analysis of diseased mice
Mice that received transplants were evaluated daily for signs of morbidity, weight loss, failure to thrive, and splenomegaly. Peripheral blood was obtained from the retroorbital venous plexus. Premorbid animals were killed by CO2 asphyxiation, and then hematopoietic tissues were removed for subsequent analysis. All animal experiments were carried out under protocols approved by the Institutional Animal Care and Use Committee of the Medical College of Georgia.
Flow cytometric analysis and stem cell sorting
Details of the specific conjugated antibodies used for flow cytometric analysis are provided in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). After staining, cells were washed once in staining medium and then analyzed using an LSR II cytometer (BD Biosciences) and FCS Express software (De Novo Software).
Mouse hematopoietic/leukemic stem cell analysis was performed using immunomagnetic-based pre-enrichment, followed by multicolor flow cytometric sorting.22 Briefly, to deplete lineage-positive cells, BM cells were reacted with a mixture of biotinylated antibodies against a panel of so-called Lin antigens (CD5, B220, CD11b, Gr-1, 7-4, and Ter-119 antibodies), and then the cells were subsequently labeled with antibiotin microbeads (Miltenyi Biotec). Lin+ cells were depleted using the Midi-MACS system (Miltenyi Biotec). Lin− cells were further reacted with phycoerythrin–Sca-1 and allophycocyanin–c-Kit antibodies (BD Pharmingen). Hematopoietic stem cells (Lin− Sca-1+ c-Kit+) or leukemic stem cells (Lin− Sca-1+ c-Kit+ GFP+) were sorted using the FACSAria.
Histopathology and immunohistochemistry
Peripheral blood smears were stained with May-Grünwald-Giemsa using standard procedures. Tissues were fixed in 10% neutral-buffered formalin for at least 48 hours. Tibiae or sternum bones were decalcified in 6% formic acid for 2 to 3 days, and then embedded in paraffin. Hematoxylin and eosin staining was performed using 3-μm sections using standard methodology.
Molecular analyses
Genomic DNA was obtained using standard phenol/chloroform methods, and the polymerase chain reaction (PCR) primers23 and conditions used in the molecular analyses are given in supplemental Table 2.
Total RNA was obtained using TRIzol (Invitrogen), retrotranscribed with a SuperScript first-strand synthesis system (Invitrogen), and amplified by PCR using conditions described in supplemental Table 2.
Western blot analysis of ZNF198-FGFR1 fusion, TCRα, BCL2, and glyceraldehyde-3-phosphate dehydrogenase protein expression was performed using standard protocols with respective primary antibodies (supplemental Table 1).
aCGH array and data analysis
The construction and analysis of the Roswell Park Cancer Institute (RPCI) custom comparative genomic hybridization (CGH) arrays have been described previously.24–26 The mouse array contains approximately 6400 RPCI-23 and RPCI-24 mouse bacterial artificial chromosome (BAC) clones that provide an average resolution across the genome of 420 kilobases. The corrected log2 ratio for each BAC was then used to segment the array using Circular Binary Segmentation (CBS) using DNAcopy software,27 and interpretation of the data was performed as previously described.28
Statistical analysis
Survival accumulation was estimated by Kaplan-Meier analysis and log-rank (Mantel-Cox) test. The t test was used to estimate the differences in spleen weights, liver weights, and white blood cell (WBC) counts. Statistical analysis was performed with the SPSS software (SPSS).
Results
Expression of the ZNF198-FGFR1 chimeric protein was achieved by cloning into the MIG retroviral expression vector, which results in the bicistronic expression of the fusion kinase and the GFP gene that was used as a marker for tumor cells carrying the chimeric kinase.
ZNF198-FGFR1 induced both myeloproliferative disorder and T-lymphoblast lymphoma
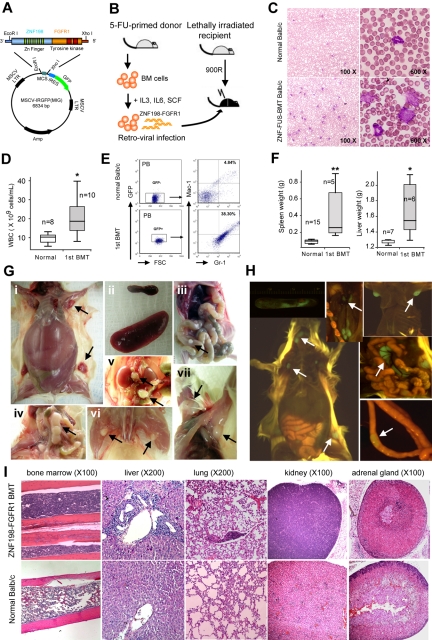
BM cells, harvested from normal male BALB/c mice, were infected with the MIG-ZNF198-FGFR-GFP retroviral construct (Figure 1A). The infection efficiency, as estimated by flow cytometry for GFP-positive cells, was approximately 16%, and the expression of the fusion protein in these transduced BM cells was confirmed either by Western blotting using the FGFR1 antibody described by Kasyapa et al29 or PCR (data not shown). After 5-day culture, these infected BM cells were transplanted into recipient female mice (Figure 1B). Analysis of peripheral blood samples from transplanted animals showed that the WBC count began to reconstitute in peripheral blood smears approximately 12 days after BM transplantation (BMT), suggesting successful engraftment. Peripheral blood smear and hematocytometer analysis showed that the WBC count was significantly increased (P < .05) 3 to 4 weeks after BMT in all mice (Figure 1C-D), indicative of the development of leukemia. These analyses conveniently monitored the temporal development of the disease in individual mice. Flow cytometric analysis of peripheral blood cells demonstrated a right shift for Mac-1+ Gr-1+ (myeloid) cells for mice receiving fusion kinase-infected BM (38.3% vs 4.84% in normal mice; Figure 1E). Nine of 10 recipient mice developed both myeloid leukemia and T lymphoma, as evidenced by leukocytosis and hepatosplenomegaly (Figure 1F-G). The median survival time in this cohort was 14 weeks (range, 5-17 weeks) posttransplantation. In 7 of 9 mice, distended abdomens were observed before the mice were euthanized. Enlarged inguinal lymph nodes (LNs) were palpable in 50% of primary recipients.
Figure 1.
Development of SCLL in ZNF198-FGFR1–transduced BM stem cells. (A) The ZNF198-FGFR1 fusion gene was cloned into the MIG vector containing the murine stem cell virus promoter and GFP cDNA linked by the internal ribosome entry site (IRES). (B) Schematic representation of the experimental approach of retrovirus transduction and transplantation of ZNF198-FGFR1. (C) May-Grünwald-Giemsa staining and (D) hematometric analysis of peripheral blood smears show a significant (P < .05) increase in WBC counts with a population of immature or blast cells in leukemia mice (C bottom right panel). (E) Flow cytometric analysis shows a right shift for both Gr-1–positive and Mac-1–positive myeloid cells in ZNF198-FGFR1 primary recipients expressing GFP. (F) The spleens and livers in leukemia mice are significantly (P < .05, P < .01, respectively) enlarged compared with normal mice. (G) ZNF198-FGFR1 induced distinct SCLL in mice, as evidenced by hepatosplenomegaly (i-ii), and enlarged LNs, Peyer patch (iii), mesenteric (iv), renal and lumbar (v), popliteal (vi), and superficial cervical and axillary (vii) LNs. (H) Molecular imaging shows all LNs displayed GFP fluorescence (see arrows). (I) Histologic analysis shows frequent hypercellularity in BM and infiltration of lymphocytes into various organs of ZNF198-FGFR1 recipients.
Spleens and livers of the diseased mice were 397.7% and 28.6% larger than those of normal mice (P < .05; Figure 1F-G), respectively. Importantly, and in contrast with a previous report,14 all primary recipients showed significantly enlarged peripheral LNs (eg, inguinal, mesenteric, renal) and nodules in the Peyer patches of the intestines (Figure 1G). Six of 9 primary recipients presented with enlarged auxiliary, superficial cervical, and popliteal LNs (Figure 1G). Overexpression of the fusion kinase in these cells was suggested by the GFP fluorescence seen in these spleens and LNs using whole organ imaging (Figure 1H). These observations demonstrate that these leukocytes were derived from transduced donor BM cells.
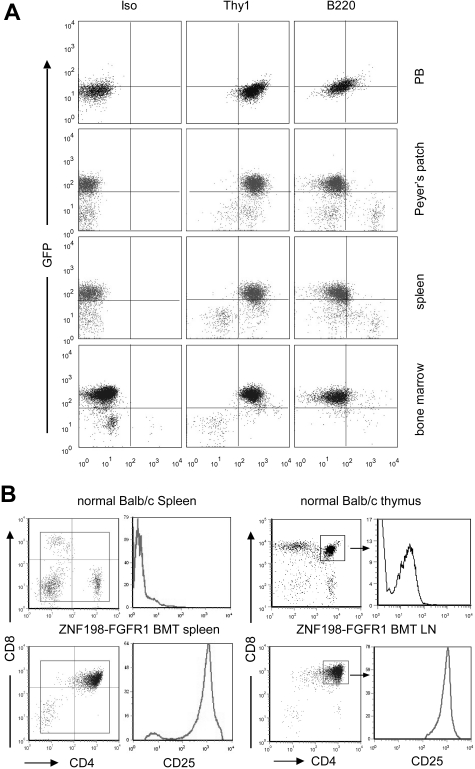
Detailed analysis of these leukemic mice indicated hypercellularity in the sternum and/or tibia marrows compared with normal mice (Figure 1I). Leukocyte infiltration was often seen in organs such as the spleens, livers, lungs, kidneys, and adrenal glands. Enlarged kidneys were seen in 20% of the primary recipients. Specifically, infiltrated leukocytes had destroyed the normal cytoarchitecture of the spleen (data not shown). Moreover, the preponderant infiltration around the central veins and portal triads in the liver was perivascular (Figure 1I). Flow cytometric analyses demonstrated that the majority of cells were Thy1+ GFP+ cells in both the BM and the peripheral LNs (Figure 2A). Further flow analysis showed that the lymphomas were CD4+CD8+, with significantly up-regulated CD25 expression (Figure 2B). These phenotypes indicate an early T-cell phenotype in both enlarged spleens and LNs. The increased numbers of GFP+ Thy1+ cells in the BM are consistent with the presence of circulating leukemia cells in this organ.
Figure 2.
Immunophenotype of hematopoietic cells of the diseased mice. (A) Flow cytometric analysis of different tissue samples, showing high levels of GFP+ Thy1+ cells in all tissues analyzed. (B) The majority of splenocytes and lymphocytes in leukemic mice are CD4 and CD8 DP, with high expression of CD25, compared with CD4+/CD8+ cells isolated from normal thymus.
The ZNF198-FGFR1 SCLL is transplantable and originates from oligo- or monoclonal hematopoietic stem/progenitor cells
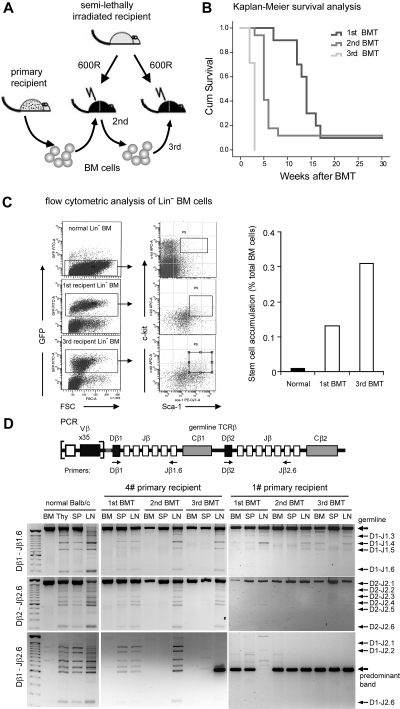
The ZNF198-FGFR1 kinase induced both myeloid leukemia and T lymphoma, suggesting a stem/progenitor cell origin, similar to human 8p11 myeloproliferative syndrome patients.7–9,11 To investigate the stem cell origin further, and explore disease progression, serial BMT experiments were performed (Figure 3A). T-lymphoma development in all second and third BMT cohorts showed a significantly shorter median survival time compared with the primary recipients, with a heavy tumor burden after only 5 and 3 weeks, respectively, versus 14 weeks for primary recipients (P < .05; Figure 3B). To quantitate the accumulation of leukemia stem cells during the serial BM transplantation, we used flow cytometery analysis to measure Lin− Sca-1+ c-Kit+ (LSK+) cell numbers. In normal BALB/c mice, approximately 0.01% of total BM cells was LSK+, whereas in primary recipients 0.13% of BM was GFP+ LSK+. By the third transplantation, 0.31% of recipient BM cells was GFP+ LSK+ cells (Figure 3C). This observation suggested the basis of the shorter survival times in the second- or third-generation transplantation recipients, even though fewer BM cells were transplanted on each successive occasion (see “Methods”). These combined data suggest that GFP+ LSK+ cells are probably the SCLL-initiated, or leukemia stem cells.
Figure 3.
Leukemic stem cell accumulation and clonal expansion of T-lymphoma cells in serial ZNF198-FGFR1 transplantation recipients. (A) Schematic illustration of the experimental approach of serial transplantation of BM cells from primary leukemic mice. (B) Kaplan-Meier survival analysis of serially transplanted ZNF198-FGFR1 mice. The difference in survival between primary (1st BMT, n = 10), secondary (2nd BMT, n = 17), and tertiary (3rd BMT, n = 14) recipients is significantly different (P < .05, log-rank test). (C) Flow cytometric analysis of lineage-positive depleted BM cells shows hematopoietic stem cells (Lin− Sca-1+ c-Kit+) from normal BALB/c mice and leukemic stem cells (GFP+ Lin− Sca-1+ c-Kit+) from ZNF198-FGR1 recipients (left panel). The remarkable accumulation of leukemic stem cells was seen in primary and tertiary ZNF198-FGFR1 recipients (right panel). (D) Schematic representation showing the relative location of the PCR primers used to analyze the TCR locus (top panel). Gel electrophoresis of PCR products with 3 different sets of primers shows DJ arrangement of TCRβ in 2 representative serially transplanted ZNF198-FGFR1 mice (bottom panel). DNA from BM displays only one large band using either D1–Jβ1.6 or Dβ2–Jβ2.6 primer sets, reflecting no rearrangement. DNA from normal thymus (Thy), spleen (SP), and LN shows several smaller bands resulting from rearrangements between different Dβ1 and Jβ2.6 positions. In contrast, DNA from leukemic mouse BM, Thy, and SP shows only one or a few bands. Specifically, when using the Dβ1–Jβ2.6 primer sets, DNA from enlarged LNs of tertiary recipients shows only one predominant band (see arrow), indicating that lymphoma cells were monoclonal.
To determine whether ZNF198-FGFR1–induced T lymphomas were polyclonal or monoclonal, we used multiple primer sets (Figure 3D top panel) to perform PCR analysis of the arrangement of the TCRβ gene. Consistent with previous observations,14 ZNF198-FGFR1–induced T-cell lymphomas were shown to be derived from oligoclonal precursor cells in primary recipients. However, in the second and third recipients, the lymphoma became monoclonal (Figure 3D), revealing a strong biologic selection for specific, highly malignant clones. In this particular model, because the tumors/cells in the spleen and LNs were predominantly Thy1+ cells, we focused on characterizing the genetic events associated with T-cell leukemia/lymphoma in subsequent genetic analyses.
aCGH analysis of ZNF198-FGFR1–induced mouse lymphomas
To investigate the genetic changes accompanying SCLL development in this mouse model, we characterized chromosome copy number aberrations (CNAs), during serial transplantation of individually transduced BM cells in 5 different experiments using CGH and the RPCI mouse BAC arrays. In all, 43 different CGH assays were performed to study development of these lymphomas. A complete summary of the specific changes seen in these experiments is given in supplemental Table 3, and examples of some of the aCGH karyotypes are shown in supplemental Figure 1. In the following discussion, specific and consistent abnormalities seen in the different experiments are described in more detail.
In each of the 5 serially BMT mice, all of the animals showed evidence of disease at the time of sacrifice, and specific organ involvement was highly consistent across the different mice studied. All aCGH data were processed using CBS to reveal CNAs, as we described previously.28 The evolution of CNAs during tumor development is described below for each individual mouse series.
aCGH analysis of tumors in mouse no. 1
In the BM cells from primary transplanted mouse no. 1, the only detectable abnormalities involved chr12:114629205-115579903gain in addition to chr3:146946701-151460672lloss and chr14:53880459-55011545loss. Because the progression of ZNF198-FGFR1 SCLL in humans is accompanied by adenopathy and splenomegaly,30 we also determined the aCGH fingerprints for tumor cells from these sites in the same diseased animals. Interestingly, in addition to the gain involving chromosome 12 and loss involving chromosome 14, chr5:146128176-146945310gain was specific to cells from the spleen. Gains of chr10:41775968-42366767 and a whole copy gain of chr15, and losses of chr7:51149642-53437992, chr8:71805148-73890916, and chr10:78823018-81538837 were specific to cells from the mesenteric LNs (supplemental Table 3). The presence of the same Chr:14 and Chr:12 CNAs in all 3 organs suggested a common origin for these cells. The additional CNAs may have evolved during the development of the abnormal T cells and myeloid cells or may have already been present in rare clonal variants within the original transduced BM cells. Mixtures of cell clones in the BM, and to a lesser extent the spleen, would almost certainly be “masked out” in aCGH analysis by any dominant karyotypes seen in the majority of cells (see “Discussion”). Detection of CNAs therefore depends on the relative contribution of the tumor cells in the samples used for aCGH.
To examine whether karyotype evolution accompanies progression, we injected BM isolated from the first transplantation recipients into secondary transplanted mice. In these mice, the aCGH profile for BM cells showed the same chr14:53880459-55011545loss and chr12:114629205-115579903gain profile seen in the primary transplant recipient, suggesting an apparently stable karyotype. The spleen cells in this mouse, however, demonstrated numerous additional CNAs, as did the mesenteric LNs (supplemental Table 3). The CNAs in this case were the same in spleen and LNs, suggesting a common clonal origin. Thus, either the tumor cells in the spleens/LNs have a selective advantage due to the genetic contribution from their additional CNAs that are related to differentiation and proliferation of myeloid and T cells, or there are rare clones in the circulating tumor T cells carrying these abnormalities already, which have preferentially populated and expanded in the different organ sites. To examine this phenomenon more closely, organs from a third transplantation of the BM cells were also analyzed.
Consistent with the previous BM aCGH profiles, chr14:53880459-55011545loss and chr12:114736103-115579903gain were again present in the BM of the third transplantation recipient, although chr9:105719034-109334452 loss was now observed representing the first evolutionary change seen specifically in the transplanted BM cells. This ∼3.5-Mbp region is very gene dense (see http://genome.ucsc.edu/) and includes genes such as Rassf1 and Tlr9, which have been associated with tumorigenesis. This CNA was also seen in the spleen and LNs in these animals, demonstrating that it was present in the tumor stem cells. Only minor other changes were seen in the spleen cells (supplemental Table 3). In contrast, the mesenteric LNs showed more extensive CNAs, and the majority of these changes were also seen in the LNs of the second recipient mouse, with only a slight evolution of the karyotype (supplemental Table 3).
We also transplanted BM from this experiment into a fourth recipient and analyzed the BM only. In this case, a gain of a single copy of chromosome 10 was now seen in addition to the chr14:53880459-55011545loss.
aCGH analysis of tumors in mouse no. 4
To determine whether a similar evolution of genetic events took place in independently generated diseased mice, we performed the same analysis in tissue samples from a primary no. 4 recipient mouse that had received BM cells derived from a second independent in vitro infection experiment. Chr14:51787550-63581976loss was again seen in the BM from this mouse, although in this case the deletion was larger than that observed in mouse no. 1 and included the Kpna3-GucyB2 interval that corresponds to the smallest region of deletion seen in human BCLL.31 The deletion seen in mouse no. 1, however, was encompassed within the deletion seen in mouse no. 4. In addition, gain of whole chromosomes 10 and 15 was also seen in this BM as well as chr12:114736103-115579903gain. An almost identical profile was seen in the spleen and the mesenteric LNs from this mouse. Because this mouse developed prominent tumors in several other sites in the abdomen, we also profiled a pancreatic LN. Numerous additional CNAs were seen in this LN, and these are summarized in supplemental Table 3.
Loss of chr14:51787550-63581976 (14C2) and gain of chr:10 were detected in the BM of the second transplantation recipient as well as chr12:114736103-115579903gain (supplemental Table 3). These CNAs were more pronounced in the aCGH profile. The 14C2 loss and the chromosome 10 gain were also observed in the mesenteric LN tumor, a popliteal LN tumor, and Peyer patch tumor. The aCGH fingerprint in spleen cells, however, displayed considerably more instability (supplemental Table 3). Interestingly, the spleen profile contained many of the same aberrations seen in the second and third transplantation spleen profiles observed in mouse no. 1.
Loss of chr14:51787550-63581976 and whole chromosomal gain of 10 were again the only CNAs detected in the BM and mesenteric LNs of the third transplant recipient. Numerous additional changes were now seen in the Peyer patch and spleen. Similar to the spleen profile of the previous transplantation, many of the aberrations detected in tumors from the Peyer patch and spleen were seen in the second and third transplantation spleen profiles of mouse no. 1.
aCGH analysis of tumors in mouse no. 9
The BM sample from a primary no. 9 transplanted mouse, which was a primary recipient from the third independent in vitro BM transduction experiment, carried the same Chr14:51787550-63581976loss chr12:114736103-115579903gain seen in the previous 2 mice as the only CNAs detected using CBS. Interestingly, the cells in the spleen from this mouse did not appear to show these abnormalities, although they were present in the mesenteric LNs, which also showed an extra copy of chromosome 15. It is possible that a relatively smaller number of circulating GFP+Thy1+ cells were present in the BM, preventing detection of a discrete deletion in the presence of cells that did not carry the deletion (see “Discussion”). After the second serial transplantation, however, the BM and LN cells clearly showed the same 14C2 loss and 12F1 gain, whereas the spleen cells did not show clear evidence of the 12F1 gain. In the third serial transplanted mouse, the spleen karyotype was the same as before, but the LNs had now acquired a gain of region chr18;10974417-22424420 (supplemental Table 3). Overall, the tumors in this mouse showed the most stable karyotype within the series after BMT.
Additional primary transplants demonstrate 14C2 deletions
Because the 14C2 and 12F1 CNAs were highly consistent findings in this model, we performed additional primary transplantations from 2 independent in vitro retroviral infections. In these cases, we only undertook aCGH analysis at this primary passage to investigate the consistency of the CNAs. In mice no. 11 and no. 12, the 14D2 and 12F1 abnormalities were again seen, although they were more pronounced in the LNs and spleen as seen before. The only other abnormality involved a gain of region Chr10:73528561-81538837 in spleen and LNs of mouse no. 11, which represents a minimal region of overlap of gain within chromosome 10 compared with those tumors showing gain of a whole copy of chromosome 10. Thus, from the overall study, Thy1+ GFP+ double-positive (DP) cells showed a consistent deletion involving the Tcra locus in 5 different experiments.
ZNF198-FGFR1–transformed T cells show loss of the TCR cell- surface complex and up-regulation of prosurvival genes
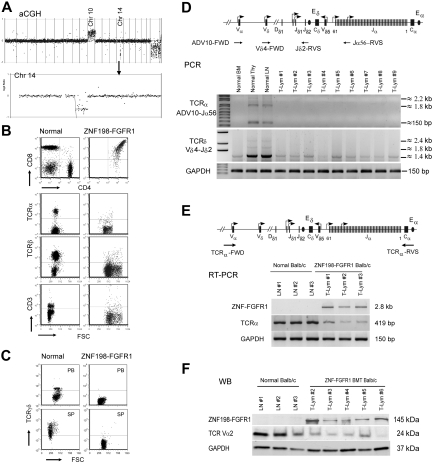
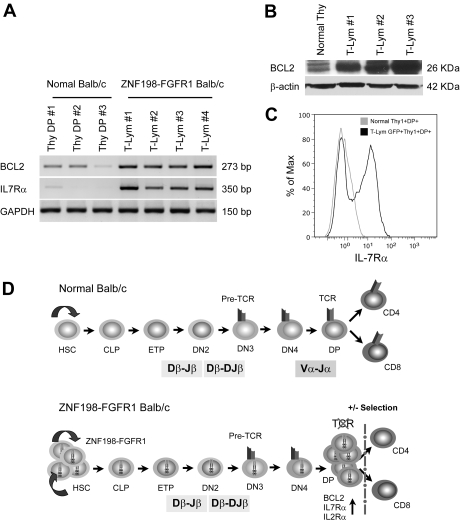
The consistent 14C2 deletion (Figure 4A) contains the TCR α and δ variable (V) and joint (J) regions. In many cases, the log2 ratios suggested homozygous deletions, which were confirmed by the absence of expression of the Tcra gene and demonstrated loss of this locus using conventional PCR. Consistent with this observation, flow cytometric analysis showed that the majority of Thy1+ tumor cells from second and third transplantation mice were CD4+ and CD8+ DP and TCRα negative on the cell surface, compared with normal lymphocytes (Figure 4B). Significantly, surface TCRδγ was also absent in lymphoma cells from these diseased mice (Figure 4C). Because loss of cell-membrane TCRα can significantly impair stability of the TCR-CD3 complex,32 we also analyzed cell-surface levels of TCRβ and CD3 in lymphocytes from second and third transplanted mice. Flow cytometric analysis using anti-TCRβ or anti-CD3ϵ demonstrated vastly reduced levels of these proteins in the tumor cells (Figure 4B). To further confirm the deletion of 14C2, we performed PCR analysis of genomic DNA with primers that recognize the rearrangement of Tcra and Tcrd (Figure 4D top panel), because the Tcrd gene is embedded within the Tcra locus in 14C2 and also deleted from the lymphoma cells in this mouse model. None of the rearrangements were detectable for both Tcra and Tcrd in lymphoma tissues (Figure 4D bottom panel). In addition, we also perfomed reverse transcription (RT)–PCR and Western blot analyses for lymphoma tissues. Both mRNA and protein levels of Tcra were significantly decreased in the lymphoma samples compared with that in the normal thymi and LNs (Figure 4E-F). Homozygous deletion of Tcrd is consistent with the flow cytometry observation (Figure 4C). The TCRδ protein is used preferentially as the TCR in cells in the Peyer patches and so may be related to the development of the lymphomas in these organs.
Figure 4.
Frequent loss of TCRα and TCRδ in lymph tissues in ZNF198-FGFR1 mice. (A) A representative whole genome aCGH profile from a mouse lymphoma showed 14qC2 segmental loss as well as gain of Chr:10. (B) Flow cytometric analysis revealed that lymphocytes from a serial transplanted mouse are DP with negative TCRα, TCRβ, and CD3e. (C) Both peripheral WBCs and splenocytes from leukemic mice are TCRγδ negative using the anti-TCRγδ. (D) Genomic DNA PCR analysis, using primers located within the TCRα ADV10 variable segment (forward) and joint region 56 (reverse, top panel), was used to analyze the TCRα rearrangement. A forward primer located in the TCRδ Vδ4 segment and a reverse primer located in the Jδ2 region (top panel) to amplify the TCRδ rearrangement were used. Both PCR analyses suggested that TCRα and TCRδ rearrangements were absent in the peripheral lymph tissues from ZNF198-FGFR1 mice. (E) Semiquantitative RT-PCR, using a forward primer located in TCRα variable region and a reverse primer located in the constant, C, region (top panel), showed that TCRα transcription levels were significantly reduced in T lymphomas compared with normal mesenteric LNs. (F) Western blot analysis showed positive expression of the ZNF198-FGFR1 fusion protein, but low expression of TCR Vα2 in lymphoid tissues of disease mice.
More than 98% of CD4+/CD8+ DP cells normally undergo apoptosis in the thymus during maturation to single-positive thymocytes.33 In our analysis, the majority of the T-cell population in the tumors showed the DP phenotype, suggesting they had escaped thymic apoptosis. To determine whether this is due to up-regulation of prosurvival genes, we analyzed transcription levels of the Bcl2 and Il7r prosurvival genes and found remarkable up-regulation of both in T-lymphoma samples versus normal, DP thymocytes (Figure 5A). Western blot analysis of BCL2 levels and flow analysis of IL2Rα (CD25) and IL7Rα (CD127) in the T-lymphoma cells from the diseased mice confirmed the up-regulation of these genes (Figures 2B and 5B-C).
Figure 5.
Up-regulation of prosurvival genes protected immature T cells from ongoing apoptosis. (A) RT-PCR shows that transcription levels of Bcl2 and IL7R were remarkably up-regulated in T lymphomas compared with normal Thy1+CD4+CD8+ thymocytes. (B) High expression of BCL2 protein was also seen in T lymphoma versus that in normal BALB/c thymus by immunoblotting. (C) Flow cytometric analysis with anti-IL7Rα (CD127-PE) showing cell membrane IL7Rα expression levels in Thy1+DP lymphoma cells is approximately 10 times higher than that seen in normal Thy1+DP thymocytes. (D) A proposed mechanism for T-lineage lymphoblastic lymphoma development in the ZNF198-FGFR1 mouse model. The top panel from a normal mouse shows that early thymocyte progenitors (ETP) migrate from BM common lymphoid progenitors (CLP) into the thymus, and then sequentially differentiate into CD4−CD8− double-negative and CD4+CD8+ DP cells after Tcrb and Tcra rearrangements. Cells that express a TCR-CD3 complex can pass through both positive and negative selection and differentiate into CD4+, CD8+ mature T cells, or natural killer T cells (data not shown). The bottom panel proposes that self-renewal of hematopoietic stem cell (HSC) is accelerated when the ZNF198-FGFR1 fusion kinase is expressed. Concordantly, constitutive activation of high expression of pre-Tα promotes the progenitors to differentiate into DP cells, although loss of Tcra prevents immature T cells from forming a functional TCR-CD3 cell-surface complex, which results in arrested T-cell development at the DP stage. Furthermore, due to high expression of BCL2, IL2Rα, and IL7Rα, these DP cells can escape apoptosis and maintain high rates of cell proliferation.
Discussion
We have generated a SCLL mouse model using the ZNF198-FGFR1 fusion kinase to investigate genetic events that occur during T-cell lymphomagenesis and progression. This model closely mimics the human disease phenotypes. From our data, and that of others,14 the fusion kinase, in itself, is clearly only an initiating factor for leukemogenesis, and progression requires other genetic events. The fusion kinase can clearly stimulate the self-renewal of the target stem cell (GFP+ LSK+) through unknown mechanisms. The contribution of these cells to the BM increased more than 30-fold after serial transplantation, supporting the idea that this may be the critical stem cell for this disease. As in the human disease, these stem cells give rise to proliferating myeloid cells and T cells that develop into leukemia/lymphoma, which are Mac1+ Gr1+ and CD4+CD8+Thy1+, respectively. The reduced latency period after serial transplantation is related to the expansion of the stem cell population in the BM of donor animals carrying the early transformation events. In our pilot studies, maintaining the transplant inoculum at 5 million cells resulted in rapid mortality of the animals before characteristic LN involvement was seen. When this inoculum was reduced, the more typical time course of the disease was re-established. These observations support the idea that the cells that seed to the spleen and LNs are derived from transformed clones within the BM.
This mouse model closely mimicked human 8p11 SCLL, as evidenced by increased WBCs, hepatosplenomegaly, and myeloid proliferation with lymphoma. Interestingly, the majority of human cases showed lymphadenopathy and T-lymphoblastic lymphoma,8 and the thymuses appeared normal. Unlike previous studies,14 in which ZNF198-FGFR1 recipient mice had minimal peripheral LN enlargement, we showed that all leukemic mice developed remarkably enlarged T-cell lymphomas in various organ systems, whereas the sizes of their thymuses were normal, even though they contained abundant GFP-positive cells (data not shown). This difference between these 2 studies could be due to the ZNF198-FGFR1 vector used by Roumiantsev et al,14 which was Myc tagged at the 3′ terminus, potentially attenuating the pathogenesis of ZNF198-FGFR1.
Our genetic analysis of T-cell leukemias suggests that loss of Tcra and Tcrd is a critical event that influences the final maturation stages into CD4+ or CD8+ T cells. Tcra loss in primary transplants suggests that this is an early event, and the presence of this deletion in the BM probably reflects high levels of circulating T cells. The absence of thymic tumors in most animals suggests that precursor T cells are not transformed. Because the majority of cells in the spleen and LNs are DP, a block in the thymus-dependent maturation of these T cells has occurred. During normal T-cell maturation, more than 98% of the DP cells undergo apoptosis in the thymus, with only a small fraction becoming mature lymphocytes. The up-regulation of prosurvival genes in ZNF198-FGFR1–transformed Thy1+ cells provides one explanation why these immature T cells can escape apoptosis. Thus, loss of the TCR complex, which includes CD3, prevents maturation, and a combination of oncogene-driven proliferation in these cells with prevention of apoptosis provides an expanded pool of immature precursor T cells for malignant transformation (Figure 5). The absence of the TCR complex also accounts for the absence of mature CD4+ or CD8+ cells that normally develop in the thymus (Figure 5). Interestingly, the DP phenotype with high expression of CD25, and lack of surface TCR, was also reported for T-cell lymphoblastic lymphomas that developed in ataxia telangiectasia mutated (ATM)–deficient mice.34,35 The ATM deficiency apparently impairs Tcra locus integrity, which results in biallelic loss of the distal part of the Tcra gene, thus also preventing DP cells from maturing into CD4+ or CD8+ T cells.34,35 Tumors from the ZNF198-FGFR1–transduced and ZNF198-FGFR1–transplanted mice, however, showed the same levels of ATM expression seen in T cells from normal mice (data not shown), suggesting a different mechanism for Tcra deletion in cells expressing the fusion kinase.
One focus of our genetic analysis was to identify potentially important secondary events that occur during progression of the disease. Despite some tumors showing extensive genetic changes, the only consistent change other than chr14:53880459-55011545loss was chr12:114736103-115579903gain containing the IgH and ADAM6 genes, although the significance of a single copy gain of this region is not clear. The deletion that was seen adjacent to this region in mouse no. 4 encompasses the Tcrb gene, but also carries Jag2, Akt1, Hsp90, and Rage, which also have potentially important implications in leukemogenesis. Other frequently observed CNAs included whole copy gains of chromosomes 10 and 15. It has been noted, however, that these chromosomes are also frequently involved in several other studies of mouse leukemias,36 as well as in some solid tumors.37
Given the shorter tumor latency period in the third and fourth transplants, it is very unlikely that the multiple CNAs seen in the LNs develop de novo each time, in exactly the same way in each passage. It is more likely that a specific clone gives rise to the lymphomas in the different organs. The determination of which specific clone will establish at specific sites, however, is not immediately obvious, although this in vivo model provides an opportunity to study trafficking of these cancer cells. It is possible that the genetic events that have occurred in these cells are required for growth at these other sites, or that the consequences of these events facilitate their tissue-specific relocation.
In summary, the model described in this study closely resembles the human SCLL disease and provides an opportunity to study the genetic events that occur downstream of initiation by the ZNF198-FGFR1 fusion kinase. The 14C2 deletion that demonstrates that correct assembly of the TCR complex, and induction of prosurvival genes in DP cells, is important for T-cell lymphoma development. In addition, the ability to track the movement of cells, using CNAs, provides an opportunity to study tumor cell trafficking. The increased levels, specifically of the LSK stem cell during transplantation, suggest that this is potentially the target stem cell, which provides important information for identifying changes in gene expression studies.
Supplementary Material
Acknowledgments
We are grateful to Dr Ken Lo for assistance with the aCGH analysis, K. Sossey-Alaoui for cloning help, and to Dr Bonnie Hylander for helping in immunohistochemistry study. We thank Drs George Deeb and Karoly Toth for assistance with pathologic analyses, and Dr James Clements for his useful discussions and suggestions.
This work was supported by grant CA076167 from the National Institutes of Health.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.R. performed the animal experiments and molecular analyses of the tumors; X.L. performed cell and molecular studies; and J.K.C. analyzed the aCGH data, coordinated the project, and wrote the manuscript with M.R.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John K. Cowell, Medical College of Georgia Cancer Center, CN4112, 1120 15th St, Augusta, GA 30912; e-mail: jcowell@mcg.edu.
References
- 1.Tefferi A, Gilliland DG. Oncogenes in myeloproliferative disorders. Cell Cycle. 2007;6:550–566. doi: 10.4161/cc.6.5.3919. [DOI] [PubMed] [Google Scholar]
- 2.Still IH, Chernova O, Hurd D, Stone RM, Cowell JK. Molecular characterization of the t(8;13)(p11;q12) translocation associated with an atypical myeloproliferative disorder: evidence for three discrete loci involved in myeloid leukemias on 8p11. Blood. 1997;90:3136–3141. [PubMed] [Google Scholar]
- 3.Xiao S, Nalabolu SR, Aster JC, et al. FGFR1 is fused with a novel zinc-finger gene, ZNF198, in the t(8;13) leukemia/lymphoma syndrome. Nat Genet. 1998;18:84–87. doi: 10.1038/ng0198-84. [DOI] [PubMed] [Google Scholar]
- 4.Guasch G, Mack GJ, Popovici C, et al. FGFR1 is fused to the centrosome-associated protein CEP110 in the 8p12 stem cell myeloproliferative disorder with t(8;9)(p12;q33). Blood. 2000;95:1788–1796. [PubMed] [Google Scholar]
- 5.Popovici C, Zhang B, Gregoire MJ, et al. The t(6;8)(q27;p11) translocation in a stem cell myeloproliferative disorder fuses a novel gene, FOP, to fibroblast growth factor receptor 1. Blood. 1999;93:1381–1389. [PubMed] [Google Scholar]
- 6.Still IH, Cowell JK. The t(8;13) atypical myeloproliferative disorder: further analysis of the ZNF198 gene and lack of evidence for multiple genes disrupted on chromosome 13. Blood. 1998;92:1456–1458. [PubMed] [Google Scholar]
- 7.Suzan F, Guasch G, Terre C, et al. Long-term complete haematologic and molecular remission after allogeneic bone marrow transplantation in a patient with a stem cell myeloproliferative disorder associated with t(8;13)(p12;q12). Br J Haematol. 2003;121:312–314. doi: 10.1046/j.1365-2141.2003.04269.x. [DOI] [PubMed] [Google Scholar]
- 8.Wong WS, Cheng KC, Lau KM, et al. Clonal evolution of 8p11 stem cell syndrome in a 14-year-old Chinese boy: a review of literature of t(8;13) associated myeloproliferative diseases. Leuk Res. 2007;31:235–238. doi: 10.1016/j.leukres.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 9.Kuskonmaz B, Kafali C, Akcoren Z, Karabulut HG, Akalin I, Tuncer MA. The 8p11 myeloproliferative syndrome in a 3-year-old child. Leuk Res. 2008;32:198–199. doi: 10.1016/j.leukres.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Sahin F, Sercan Z, Ertan Y, et al. Rapid transformation of atypical myeloproliferative disorder with consistent t(8;13) to B-cell acute lymphoblastic leukemia: a case report. Hematology. 2007;12:489–492. doi: 10.1080/10245330701562204. [DOI] [PubMed] [Google Scholar]
- 11.Etienne A, Gelsi-Boyer V, Carbuccia N, et al. Combined translocation with ZNF198-FGFR1 gene fusion and deletion of potential tumor suppressors in a myeloproliferative disorder. Cancer Genet Cytogenet. 2007;173:154–158. doi: 10.1016/j.cancergencyto.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Krause DS, Van Etten RA. Right on target: eradicating leukemic stem cells. Trends Mol Med. 2007;13:470–481. doi: 10.1016/j.molmed.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mozziconacci MJ, Carbuccia N, Prebet T, et al. Common features of myeloproliferative disorders with t(8;9)(p12;q33) and CEP110-FGFR1 fusion: report of a new case and review of the literature. Leuk Res. 2008;32:1304–1308. doi: 10.1016/j.leukres.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 14.Roumiantsev S, Krause DS, Neumann CA, et al. Distinct stem cell myeloproliferative/T lymphoma syndromes induced by ZNF198-FGFR1 and BCR-FGFR1 fusion genes from 8p11 translocations. Cancer Cell. 2004;5:287–298. doi: 10.1016/s1535-6108(04)00053-4. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Deangelo DJ, Kutok JL, et al. PKC412 inhibits the zinc finger 198-fibroblast growth factor receptor 1 fusion tyrosine kinase and is active in treatment of stem cell myeloproliferative disorder. Proc Natl Acad Sci U S A. 2004;101:14479–14484. doi: 10.1073/pnas.0404438101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 17.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baumann H, Kunapuli P, Tracy E, Cowell JK. The oncogenic fusion protein-tyrosine kinase ZNF198/fibroblast growth factor receptor-1 has signaling function comparable with interleukin-6 cytokine receptors. J Biol Chem. 2003;278:16198–16208. doi: 10.1074/jbc.M300018200. [DOI] [PubMed] [Google Scholar]
- 19.Schwaller J, Frantsve J, Aster J, et al. Transformation of hematopoietic cell lines to growth-factor independence and induction of a fatal myelo- and lymphoproliferative disease in mice by retrovirally transduced TEL/JAK2 fusion genes. EMBO J. 1998;17:5321–5333. doi: 10.1093/emboj/17.18.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozak SL, Kabat D. Ping-pong amplification of a retroviral vector achieves high-level gene expression: human growth hormone production. J Virol. 1990;64:3500–3508. doi: 10.1128/jvi.64.7.3500-3508.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S, Ilaria RL, Jr, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189:1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ema H, Morita Y, Yamazaki S, et al. Adult mouse hematopoietic stem cells: purification and single-cell assays. Nat Protoc. 2006;1:2979–2987. doi: 10.1038/nprot.2006.447. [DOI] [PubMed] [Google Scholar]
- 23.Kawamoto H, Ohmura K, Fujimoto S, Lu M, Ikawa T, Katsura Y. Extensive proliferation of T cell lineage-restricted progenitors in the thymus: an essential process for clonal expression of diverse T cell receptor β chains. Eur J Immunol. 2003;33:606–615. doi: 10.1002/eji.200323461. [DOI] [PubMed] [Google Scholar]
- 24.Cowell JK, Nowak NJ. High-resolution analysis of genetic events in cancer cells using bacterial artificial chromosome arrays and comparative genome hybridization. Adv Cancer Res. 2003;90:91–125. doi: 10.1016/s0065-230x(03)90003-0. [DOI] [PubMed] [Google Scholar]
- 25.Cowell JK, Matsui S, Wang YD, et al. Application of bacterial artificial chromosome array-based comparative genomic hybridization and spectral karyotyping to the analysis of glioblastoma multiforme. Cancer Genet Cytogenet. 2004;151:36–51. doi: 10.1016/j.cancergencyto.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Rossi MR, La Duca J, Matsui S, Nowak NJ, Hawthorn L, Cowell JK. Novel amplicons on the short arm of chromosome 7 identified using high resolution array CGH contain overexpressed genes in addition to EGFR in glioblastoma multiforme. Genes Chromosomes Cancer. 2005;44:392–404. doi: 10.1002/gcc.20256. [DOI] [PubMed] [Google Scholar]
- 27.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 28.Lo KC, Rossi MR, Eberhart CG, Cowell JK. Genome wide copy number abnormalities in pediatric medulloblastomas as assessed by array comparative genome hybridization. Brain Pathol. 2007;17:282–296. doi: 10.1111/j.1750-3639.2007.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasyapa CS, Kunapuli P, Hawthorn L, Cowell JK. Induction of the plasminogen activator inhibitor-2 in cells expressing the ZNF198/FGFR1 fusion kinase that is involved in atypical myeloproliferative disease. Blood. 2006;107:3693–3699. doi: 10.1182/blood-2005-04-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inhorn RC, Aster JC, Roach SA, et al. A syndrome of lymphoblastic lymphoma, eosinophilia, and myeloid hyperplasia/malignancy associated with t(8;13)(p11;q11): description of a distinctive clinicopathologic entity. Blood. 1995;85:1881–1887. [PubMed] [Google Scholar]
- 31.Hawthorn LA, Chapman R, Oscier D, Cowell JK. The consistent 13q14 translocation breakpoint seen in chronic B cell leukemia (BCLL) involves deletion of the D13S25 locus which lies distal to the retinoblastoma predisposition gene. Oncogene. 1993;8:1415–1419. [PubMed] [Google Scholar]
- 32.Kuhns MS, Davis MM. The safety on the TCR trigger. Cell. 2008;135:594–596. doi: 10.1016/j.cell.2008.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson G, Moore NC, Owen JJ, Jenkinson EJ. Cellular interactions in thymocyte development. Annu Rev Immunol. 1996;14:73–99. doi: 10.1146/annurev.immunol.14.1.73. [DOI] [PubMed] [Google Scholar]
- 34.Matei IR, Gladdy RA, Nutter LM, Canty A, Guidos CJ, Danska JS. ATM deficiency disrupts Tcra locus integrity and the maturation of CD4+CD8+ thymocytes. Blood. 2007;109:1887–1896. doi: 10.1182/blood-2006-05-020917. [DOI] [PubMed] [Google Scholar]
- 35.Vacchio MS, Olaru A, Livak F, Hodes RJ. ATM deficiency impairs thymocyte maturation because of defective resolution of T cell receptor α locus coding end breaks. Proc Natl Acad Sci U S A. 2007;104:6323–6328. doi: 10.1073/pnas.0611222104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiener F, Ohno S, Spira J, Haran-Ghera N, Klein G. Chromosome changes (trisomies #15 and 17) associated with tumor progression in leukemias induced by radiation leukemia virus. J Natl Cancer Inst. 1978;61:227–237. doi: 10.1093/jnci/61.1.227. [DOI] [PubMed] [Google Scholar]
- 37.Cowell JK. Consistent chromosome abnormalities associated with mouse bladder epithelial cell lines transformed in vitro. J Natl Cancer Inst. 1980;65:955–961. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.