During meiosis, cells deliberately form numerous DNA double-strand breaks (DSBs) in order to initiate homologous recombination, which exchanges genetic information between homologous chromosomes and promotes accurate chromosome segregation. DSB formation is restricted to a narrow window of time in meiosis, both for proper execution of the functions of recombination and to prevent production of toxic DNA lesions at inappropriate times. Two studies in this issue of Genes & Development provide important insight into the poorly understood mechanisms that ensure proper timing of DSB formation.
Function and regulation of recombination during meiosis
Meiosis is a modified version of the mitotic cell division cycle used by sexually reproducing organisms to generate haploid gametes. Meiosis comprises one round of DNA replication and two successive rounds of cell division, which reduce the genome complement by half. During the period in between replication and division in most organisms, homologous paternal and maternal chromosomes pair and align with one another along their lengths and also exchange genetic information through homologous recombination (Fig. 1). One critical function of this exchange is that it helps provide physical connections between the homologous chromosomes. These connections (called chiasmata) work together with sister chromatid cohesion to allow chromosomes to align properly on the spindle and to separate accurately at the first meiotic division. If recombination fails, chromosome segregation also frequently fails, with disastrous consequences for gamete formation (Petronczki et al. 2003).
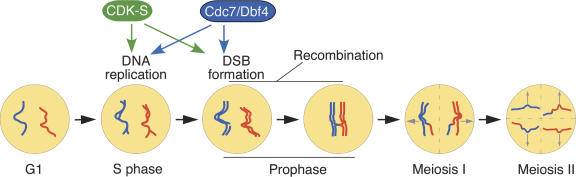
Figure 1.
Timeline of events during meiosis. Meiosis begins from the G1 stage; a diploid cell with one pair of homologous chromosomes (red and blue) is shown. (S phase) DNA is replicated, forming a pair of sister chromatids for each chromosome. Initiation of replication requires CDK-S and is probably at least partially dependent on Cdc7/Dbf4 activity. (Prophase) After completion of replication, DSBs are introduced, dependent on both CDK-S and Cdc7/Dbf4. DSBs are then repaired by homologous recombination. The resulting chiasmata physically connect homologous chromosomes and facilitate proper segregation of homologs at the first meiotic division (Meiosis I). Sister chromatids are then segregated at Meiosis II.
Meiotic recombination involves the formation and repair of DSBs—deliberately self-inflicted DNA damage—generated by the evolutionarily conserved Spo11 protein (Keeney 2007). The recombination pathway is best understood in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, but genetic and cytological data support the conclusion that Spo11-dependent DSBs are required for recombination in many other organisms, including mammals (Gerton and Hawley 2005; Keeney 2007). Thus, DSBs appear to be a universal feature of meiotic recombination.
In yeasts, where direct physical assays for DSBs are available, it is clear that DSB formation is restricted to a fairly narrow window of time during the prophase that separates replication and chromosome segregation (Fig. 1; Padmore et al. 1991; Cervantes et al. 2000). In organisms where direct DSB assays are not available (e.g., flies, worms, and mice), immunostaining of meiotic chromosomes to follow cytological DSB markers shows that restricting the timing of DSB formation is a general phenomenon (Mahadevaiah et al. 2001; Colaiacovo et al. 2003; Jang et al. 2003).
Such restrictions make sense, considering both the essential roles of meiotic DSBs and their potentially negative consequences if improperly repaired. First, as mentioned above, DSBs are necessary to generate chiasmata that physically connect homologous chromosomes. Each chiasma is a combination of a crossover generated by the repair of a DSB through recombination, plus higher-order chromosome structure changes at the site of crossing over; namely, exchange of chromosome axes and local separation of sister chromatids (Kleckner 2006). Thus, to build a chiasma, different processes must be coordinated with one another in space and time. For example, recombination must initiate only after the DNA has been replicated and sister chromatid cohesion has been established.
Second, recombination involves the search for an intact homologous DNA sequence to serve as a template to restore the integrity of the broken chromatid. In many (but not all) organisms, the homology search that occurs during recombination is a key part of the mechanism by which homologous chromosomes “find” one another (Peoples-Holst and Burgess 2005). The timing of recombination initiation must be controlled in order to effectively promote this pairing.
Third, DSBs are a hazardous type of genomic damage that cells normally go to great lengths to avoid, but meiotic cells must deliberately introduce these chromosomal lesions. To minimize the risk of deleterious effects, cells must regulate Spo11 activity to occur only at the right time and right place so that DSBs are converted efficiently into appropriate end products (Hochwagen and Amon 2006; Keeney 2007).
Relationship of Spo11-dependent DSB formation to meiotic DNA replication
One manifestation of the control of DSB formation is the apparent coupling of DSBs with DNA replication. A particularly revealing set of experiments examined the consequences of delaying replication of the left arm of chromosome III in Saccharomyces cerevisiae (Borde et al. 2000; Murakami et al. 2003). Deleting the three major replication origins on this arm delays meiotic replication by ∼1 h, presumably because the arm must now be replicated by forks that originate in the right arm. Replication timing in the right arm is unaffected, however, so the effect of origin deletion is local (i.e., neither chromosome- nor genome-wide). Strikingly, DSB formation on the left arm was delayed by the same margin, while DSB timing on the right arm was unaffected. The net result was that DSBs followed local replication by the same margin (∼1.5 h) irrespective of whether the DNA replicated early or late.
How DSB timing might be regulated relative to replication is not understood. One hypothesis has been that there is a strict dependence of DSBs on replication (Borde et al. 2000; Smith et al. 2001). This idea was based on the fact that DSBs do not form in many conditions where meiotic replication is blocked, such as in the presence of the deoxynucleotide synthesis inhibitor hydroxyurea (HU), or when the cell cycle regulator cyclin-dependent kinase (CDK) is not active (Borde et al. 2000; Smith et al. 2001; Benjamin et al. 2003). However, HU also blocks induction of early meiotic genes, including SPO11 (Lamb and Mitchell 2001), and CDK directly regulates DSB formation separately from regulation of replication (Henderson et al. 2006; see below).
These issues undermine the argument that DSB formation depends strictly on replication, and other experiments provide direct evidence against this hypothesis. For example, when meiotic expression of the replication initiation factor Cdc6 is eliminated, DSB formation occurs even though replication apparently does not (Hochwagen et al. 2005). Similarly in Schizosaccharomyces pombe, when completion of replication is inhibited by reduced levels of core replication proteins, DSB formation still occurs (Murakami and Nurse 2001). Moreover, elimination of replication checkpoint factors allows recombination to occur even in the presence of HU (which does not block early meiotic gene expression in these S. pombe cultures) (Tonami et al. 2005; Ogino and Masai 2006). These findings suggest that checkpoint responses to defects in ongoing replication can block DSB formation, but also suggest that replication is not a prerequisite for DSBs per se (Hochwagen and Amon 2006).
If replication is not a prerequisite for DSB formation, then how can we account for the coordinate timing of the two processes in the origin-deletion experiments and, more importantly, how do cells normally ensure that DSBs form only on DNA that has already replicated? Clues to answer these questions are provided by several recent studies, including two in this issue of Genes & Development, that uncover important new molecular details of the regulation of DSB formation (Henderson et al. 2006; Ogino et al. 2006; Valentin et al. 2006; Wan et al. 2006, 2008; Sasanuma et al. 2008).
Control of DSB formation by CDK-S
Spo11 is the catalytic subunit of the meiotic DSB-forming activity, but it does not work alone. In S. cerevisiae, at least nine other proteins are also required (for review, see Keeney 2007). Four of these (Mei4, Rec102, Rec104, and Rec114) are meiosis-specific and, like Spo11, their expression is controlled primarily at the level of transcription. Four other DSB proteins also have roles in vegetative cells. Ski8 (a.k.a. Rec103) functions in RNA metabolism in vegetatively growing cells, and the evolutionarily conserved Mre11–Rad50–Xrs2 complex has multiple functions in meiotic and nonmeiotic cells, including DSB repair, telomere maintenance, and DNA damage checkpoint activation. The identities of these proteins have been known for some time, but we still understand relatively little about their biochemical functions during DSB formation. However, recent studies have traced out functional interactions that connect these proteins to one another and implicate all of them as direct players along with Spo11 at the sites on chromosomes where DSBs are formed (Arora et al. 2004; Kee et al. 2004; Prieler et al. 2005; Li et al. 2006; Maleki et al. 2007; Sasanuma et al. 2007).
Mer2 is the final member of the group of proteins needed for Spo11 activity. Mer2 is also induced during meiosis, but its expression is controlled differently: MER2 is constitutively transcribed, but contains an intron that is spliced efficiently only during meiosis (Engebrecht et al. 1991). As a result, Mer2 protein is present at low levels during vegetative growth and at substantially higher levels during meiosis (Henderson et al. 2006; Li et al. 2006). Mer2 has recently emerged as a key control point through which the activity of Spo11 is influenced by two cell cycle regulatory kinases, Cdc28 and Cdc7.
Cdc28 is the principal CDK regulating cell cycle progression in S. cerevisiae. Cdc28 associates with various protein-binding partners known as cyclins, which activate the kinase and target it toward specific substrates. Cdc28 and two of its cyclin partners, Clb5 and Clb6, are required both for meiotic DNA replication and for DSB formation (Stuart and Wittenberg 1998; Smith et al. 2001; Benjamin et al. 2003). Complexes of Cdc28 with one or the other of these cyclins will be referred to below as CDK-S (for S-phase CDK).
Mer2 is an essential target of CDK-S in regulating DSB formation. Mer2 is phosphorylated during early meiosis, based on shift in electrophoretic mobility of the protein, and phosphorylation does not require Spo11 or any other DSB proteins, and is thus not a consequence of DSB formation (Henderson et al. 2006; Li et al. 2006). Several lines of evidence establish that CDK is one of the kinases responsible (Henderson et al. 2006). Inactivation of Cdc28 using a specific small-molecule inhibitor or through elimination of Clb5 and Clb6 eliminates much of the electrophoretic mobility shift, and Cdc28 can phosphorylate recombinant Mer2 protein in vitro. The consensus CDK target is the sequence T/S-P-x-K/R (Ubersax et al. 2003). Mer2 has an optimal site (SPFR) at Ser30 and a minimal site (SP) at Ser271 (Fig. 2A). Both the mobility shift in vivo and the ability to phosphorylate Mer2 in vitro were dependent on both CDK sites, indicating that both are direct, bona fide targets of CDK-S. However, only Ser30 is essential for DSB formation: Mutating Ser30 to alanine causes a DSB defect similar to deleting the MER2 gene entirely, whereas mutation of Ser271 has little effect. These findings provided insights into the molecular connections between cell cycle progression (as reflected in CDK-S activity) and DSB formation.
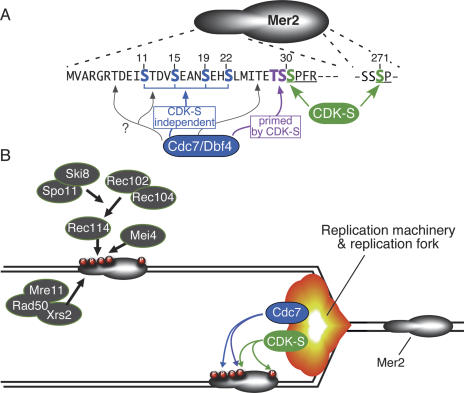
Figure 2.
CDK-S and Cdc7/Dbf4 regulate DSB formation via phosphorylation of Mer2. (A) Phosphorylation sites on Mer2. Amino acid sequence is shown for the N-terminal region (residues 1–33) and a phosphorylation site near the C terminus (residues 269–272). Green indicates residues phosphorylated by CDK-S (Ser30 and Ser271). Two types of Cdc7/Dbf4 target are shown: Blue indicates serine residues phosphorylated by Cdc7 independently of CDK-S, and magenta indicates residues phosphorylated by Cdc7 only after CDK-S has primed the sites by phosphorylating the adjacent serine. Three additional threonine residues (gray arrows) are present in this region and are also potential Cdc7 targets. See the text for further details. (B) Model for coupling of replication with DSB formation. Mer2 is proposed to be preferentially phosphorylated in the wake of replication fork passage, perhaps through physical association of CDK-S and/or Cdc7/Dbf4 with components of the replication machinery. Phosphorylated Mer2 is then able to recruit other DSB proteins, including Spo11 itself. See the text for further details.
Control of DSB formation by Cdc7
The Mer2 story grows more complex with intriguing reports from the Hollingsworth and Ohta laboratories (Sasanuma et al. 2008; Wan et al. 2008). Cdc7 is a Ser/Thr kinase essential for firing of DNA replication origins (for review, see Sclafani 2000). Analogous to the Cdc28–cyclin relationship, Cdc7 protein levels are relatively constant, but kinase activity fluctuates because of changes in the level of an accessory subunit, Dbf4, which is abundant and chromatin-associated in late G1 and S phase. In meiosis, Cdc7 is required for timely and efficient replication, DSBs, and divisions (Schild and Byers 1978; Valentin et al. 2006; Wan et al. 2006).
Hollingsworth’s group (Wan et al. 2006) generated a conditional cdc7 allele by introducing a mutation in the kinase-active site to enlarge the ATP-binding pocket so that bulky analogs of ATP or kinase inhibitors can be bound. With this allele (cdc7-as3, for “analog-sensitive”), the inhibitor PP1 blocks DSB formation and meiotic divisions (Wan et al. 2006, 2008). Ohta’s group (Sasanuma et al. 2008) pursued a different strategy for inactivating Cdc7, taking advantage of the fact that the essential requirement for Cdc7 in mitotic DNA replication is bypassed by a mutation known as mcm5-bob1 (Sclafani 2000). Whereas a cdc7-null mutation is lethal, cdc7 mcm5-bob1 double mutants are viable. However, these double mutants are unable to form DSBs or to complete the meiotic divisions, again implicating Cdc7 activity in both processes (Sasanuma et al. 2008). In these and in earlier studies using temperature-sensitive cdc7 alleles, meiotic DNA replication was slowed, but not eliminated. However, DNA replication is almost completely blocked in mutants engineered to eliminate most or all expression of the regulatory subunit Dbf4 prior to meiotic entry, strongly suggesting that Cdc7 is required for initiation of DNA replication in meiosis, as in mitosis (Fig. 1; Valentin et al. 2006).
Why is Cdc7 required for DSBs? At least part of the answer is that Mer2 is an essential Cdc7 target. Mer2 phosphorylation is greatly reduced when Cdc7-as3 is inhibited and in cdc7 mcm5-bob1 mutants. Interestingly, if the Cdc7-as3 inhibitor is washed out of an arrested culture, cells are able to resume meiosis. In these circumstances, Mer2 phosphorylation occurs fairly rapidly, followed ∼80 min later by DSB formation (Wan et al. 2006, 2008). CDK-S kinase activity increases more or less normally when Cdc7-as3 is inhibited, arguing against an indirect role for Cdc7 via activation of CDK-S.
Coordinated control of DSB formation by CDK-S and Cdc7
Analysis of phosphorylation sites on Mer2 revealed a complex and intriguing pattern of partially interdependent regulation by the two kinases. Ohta’s group (Sasanuma et al. 2008) used mass spectrometry to identify phosphorylated proteolytic peptides in Mer2 protein purified from meiotic cells. Phosphopeptides were identified that likely derived from phosphorylation of Ser30 or Ser271 by CDK-S, but more importantly, phosphopeptides were also observed corresponding to up to three separate phosphate groups on the peptide containing Ser30. To identify the additional phosphorylation sites, serine residues in this region were mutated individually or in combination (Fig. 2A). Single mutations of Ser11, Ser15, Ser19, and Ser22 confer little or no DSB defects, but double, triple, and quadruple mutant combinations cause progressively more severe defects, culminating in up to 10-fold reduction in DSBs. These findings are consistent with the interpretation that phosphorylation of one or more of these residues is important for DSB formation, but that the specific residue targeted by the kinase(s) is not critical. More strikingly, mutation of Ser29 to alanine reduces phosphorylation-dependent electrophoretic shift of Mer2 protein and confers a DSB-defective phenotype similar to a mer2 null. Thus, like CDK-S-targeted Ser30, Ser29 is essential for DSBs.
Hollingsworth’s group (Wan et al. 2008) likewise identified Ser29 as an essential residue in Mer2, but from a different approach. Cdc7 prefers to phosphorylate Ser/Thr residues that are followed by an acidic amino acid (aspartate or glutamate), but for two sites in Mcm2 (a replicative helicase subunit regulated by Cdc7), requirement for a negative charge is met by CDK phosphorylating an adjacent serine (Cho et al. 2006; Montagnoli et al. 2006). Wan et al. (2008) noted that the sequence contexts of these sites in Mcm2 and Mer2 are similar, and thus proposed that Mer2 phosphorylation follows the sequence Ser30(CDK) → Ser29(Cdc7) (Fig. 2A). This hypothesis is supported by Cdc7 kinase activity tests with synthetic peptide substrates and by comparison of effects of single and double alanine substitutions on Mer2 electrophoretic mobility (Wan et al. 2008). Thr28 may also be a Cdc7 target after Ser29 is phosphorylated. The CDK-S target site Ser271 may be regulated similarly, although this has not been tested (recall that Ser271 is not essential for DSBs; see above).
Previous attempts to mimic constitutive phosphorylation by CDK-S by mutating Ser30 to aspartate were unsuccessful, yielding greatly reduced DSB formation (Henderson et al. 2006). Remarkably, Wan et al. (2008) now show that DSB formation occurs at nearly normal levels when aspartate is substituted for all three phosphorylatable residues in this region (Thr28, Ser29, and Ser30). Even with this mutant, however, both Cdc7 and CDK-S are still required for DSB formation. Thus, phosphorylation of these residues can be bypassed if a suitably large patch of negatively charged residues is provided, but other essential targets of both kinases appear to exist.
Taken together, these results suggest the following scenario in which Cdc7 and CDK-S both act directly on Mer2 to promote DSB formation (Sasanuma et al. 2008; Wan et al. 2008). CDK-S targets two separate sites on Mer2 (one essential and one nonessential) and does so independently of Cdc7 activity (Fig. 2A). Cdc7 targets multiple residues that can be grouped into two separate categories: an essential target (Ser29) whose phosphorylation must be primed by prior action of CDK-S on Ser30, and another set of residues in the N-terminal region that are targeted independently of CDK-S. It remains unclear whether the CDK-S-independent targets are essential or only partially required for DSB formation, because there are three additional potentially phosphorylatable threonine residues in this region (all followed by acidic residues) that were not tested (Fig. 2A, gray arrows). Also, it is important to note that Cdc7 has not yet been shown to be able to directly phosphorylate any of these residues in the context of full-length Mer2 protein, so there remains the possibility that another kinase(s) may also be involved.
How does phosphorylation of Mer2 promote DSB formation?
Systematic two-hybrid and coimmunoprecipitation analyses demonstrated that Mer2 interacts with itself and with other DSB proteins—Xrs2, Mei4, and Rec114 (Arora et al. 2004; Henderson et al. 2006; Li et al. 2006; Maleki et al. 2007). CDK-S target site mutations in MER2 perturb these interactions (Henderson et al. 2006). Specifically, Mer2-S30A protein fails to interact with Rec114 and Xrs2 and shows decreased interaction with itself and with Mei4. Using chromatin immunoprecipitation, Sasanuma et al. (2008) now show that recruitment to a DSB site of Rec114, Mei4, and, most importantly, Spo11 itself is dependent on Cdc7, and that Spo11 recruitment is also impaired when putative Cdc7 phosphorylation sites on Mer2 are mutated. Taken together, these results suggest that CDK-S and Cdc7 exert their influence on Mer2 activity, at least in part, by modulating Mer2 interactions with other DSB proteins, and thereby control the loading of DSB proteins to chromatin (Fig. 2B; Sasanuma et al. 2008).
Back to the connection with DNA replication
Regulation of Mer2 by both CDK-S and Cdc7 is intriguing because it suggests how DSBs might be controlled relative to DNA replication (Sasanuma et al. 2008; Wan et al. 2008). One way to think about how this might work is as follows. Both meiotic replication and DSB formation depend on activity of both kinases, but interestingly, it also appears that the two processes are differently sensitive to quantitative changes in kinase levels. For Cdc7 in particular, it appears that much higher activity is required for DSB formation than is required for initiating DNA replication. This idea is based on the observation that DSB formation and Mer2 phosphorylation are blocked by cdc7ts alleles at restrictive temperature or cdc7-as3 in the presence of PP1, whereas replication proceeds in these conditions (Schild and Byers 1978; Wan et al. 2006). The situation is less clear for CDK-S, but similar differences may apply as well (Henderson et al. 2006).
Thus, when meiosis is induced and CDK-S and Cdc7–Dbf4 activities begin to rise, thresholds required for replication might be reached before thresholds required for DSB formation. Moreover, if kinase substrates compete with one another for available CDK-S and/or Cdc7, the distinction between replication and DSBs might be even sharper if replication substrates (e.g., origin-associated protein complexes) “win” this competition; e.g., if one or both kinases is sequestered by origins or replication forks.
Such a scenario could at least partially account for how replication normally precedes DSB formation: Mer2 (and other DSB-essential targets) may only be phosphorylated later, when kinase levels continue to climb and/or when competition with origins decreases as origins fire or are inactivated by replication fork passage. Such a scenario could also account for how DSB formation can occur if replication origins are never activated (e.g., in the absence of Cdc6 [Hochwagen et al. 2005]).
By itself, this model would not explain how DSB timing might be controlled locally relative to DNA replication (recall that delaying replication of a chromosomal region causes a parallel delay in DSB formation in that same region; see above). However, local coupling of the two processes could be achieved if Mer2 phosphorylation by CDK and/or Cdc7 is spatially linked to replication (Fig. 2B). Such preferential local targeting could be achieved if CDK-S and/or Cdc7 were physically associated with replication forks and tracked with them along chromosomes, or if kinase activity were preferentially targeted to some feature unique to replicated DNA (e.g., through recognition of newly assembled nucleosomes, sister chromatid cohesion complexes, or meiosis-specific axis components). Importantly, such preferential targeting would not be required once CDK-S and/or Cdc7 activity is high enough; e.g., late in S phase or if origins never fire.
This model does not require that Mer2 phosphorylation instantaneously triggers DSB formation. Instead, it need only be the case that phosphorylation is a rate-limiting step that sets in motion subsequent processes that culminate in DSB formation (e.g., recruitment of Mei4, Rec114, and other factors). Indeed, it is interesting to note that the delay between Mer2 phosphorylation after washout of Cdc7-as3 inhibitor (∼80 min) (Wan et al. 2008) is comparable with the time between replication fork passage and DSB formation measured in vivo (Borde et al. 2000).
It is important to stress that regulation of Mer2 is not the only means for controlling timing of DSB formation. For example, meiosis-specific transcription of DSB proteins (including Spo11 itself) probably contributes to establishing proper timing. Moreover, two other DSB proteins (Rec114 and Rec104) are phosphorylated, but it is not known what kinase(s) is responsible nor whether these modifications regulate DSB formation (Kee et al. 2004; Sasanuma et al. 2008).
Evolutionary conservation
The putative dual CDK-S/Cdc7 regulation site equivalent to Ser30 is conserved in most Mer2 orthologs in yeasts closely related to S. cerevisiae (Henderson et al. 2006; Wan et al. 2008). Wider conservation is difficult to address, however, because Mer2 orthologs have not been identified in more distantly related ascomycetes, including S. pombe, or in other lineages. Indeed, most of the DSB proteins are poorly conserved, such that orthologs have been difficult or impossible to find in evolutionarily distant organisms (Keeney 2007).
Nonetheless, at least some features of CDK- and Cdc7-dependent regulation of DSB formation are likely to be conserved. Most importantly, Hsk1 (the S. pombe ortholog of Cdc7) is essential for DSB formation in that organism (Ogino et al. 2006) and, similar to the situation in S. cerevisiae, Hsk1 activity is required for recruitment of Rec12 (the Spo11 ortholog) to DSB sites (Sasanuma et al. 2008). Moreover, meiotic recombination is reduced in mutants lacking a meiosis-specific cyclin, Rem1 (Malapeira et al. 2005). Rem1 is partially redundant with a vegetative cyclin (Cig2) for controlling meiotic replication (Borgne et al. 2002; Malapeira et al. 2005), so it is possible that these cyclins are also redundant for controlling DSB formation, in analogy to the partial redundancy of cyclins Clb5 and Clb6 in S. cerevisiae (Stuart and Wittenberg 1998; Smith et al. 2001). Direct targets of Hsk1 and/or CDK that are important for DSB formation remain to be determined.
Aside from Spo11, few DSB proteins are known in multicellular eukaryotes, and little information is available about possible roles of cell cycle regulatory factors in controlling DSB formation. It is interesting to note, however, that the Drosophila melanogaster protein MEI-P22 has nine matches to the CDK consensus phosphorylation site (Keeney 2007). MEI-P22 is essential for recombination initiation (Liu et al. 2002), so it is tempting to speculate that CDK may directly control MEI-P22 activity.
Concluding remarks
In order to initiate meiotic recombination, sexually reproducing organisms have evolved a dangerous mechanism that inflicts lesions on their own genomes, with the potentially lethal consequences of these lesions making it critical that cells tightly control their formation. Recent studies highlighted here are giving us our first glimpses of the detailed mechanisms by which this control operates.
Acknowledgments
We thank Nick Rhind, Nancy Hollingsworth, Kunihiro Ohta, and members of the Keeney laboratory for discussions and/or sharing of unpublished information. Work from our laboratory was supported by R01 GM58673 (to S.K.) and R01 HD40916 (to M. Jasin and S.K.). S.K. is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1642308
References
- Arora C., Kee K., Maleki S., Keeney S. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol. Cell. 2004;13:549–559. doi: 10.1016/s1097-2765(04)00063-2. [DOI] [PubMed] [Google Scholar]
- Benjamin K.R., Zhang C., Shokat K.M., Herskowitz I. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes & Dev. 2003;17:1524–1539. doi: 10.1101/gad.1101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V., Goldman A.S.H., Lichten M. Direct coupling between meiotic DNA replication and recombination initiation. Science. 2000;290:806–809. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- Borgne A., Murakami H., Ayte J., Nurse P. The G1/S cyclin Cig2p during meiosis in fission yeast. Mol. Biol. Cell. 2002;13:2080–2090. doi: 10.1091/mbc.01-10-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes M.D., Farah J.A., Smith G.R. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell. 2000;5:883–888. doi: 10.1016/s1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- Cho W.H., Lee Y.J., Kong S.I., Hurwitz J., Lee J.K. CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein. Proc. Natl. Acad. Sci. 2006;103:11521–11526. doi: 10.1073/pnas.0604990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo M.P., MacQueen A.J., Martinez-Perez E., McDonald K., Adamo A., La Volpe A., Villeneuve A.M. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Engebrecht J.A., Voelkel-Meiman K., Roeder G.S. Meiosis-specific RNA splicing in yeast. Cell. 1991;66:1257–1268. doi: 10.1016/0092-8674(91)90047-3. [DOI] [PubMed] [Google Scholar]
- Gerton J.L., Hawley R.S. Homologous chromosome interactions in meiosis: Diversity amidst conservation. Nat. Rev. Genet. 2005;6:477–487. doi: 10.1038/nrg1614. [DOI] [PubMed] [Google Scholar]
- Henderson K.A., Kee K., Maleki S., Santini P.A., Keeney S. Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell. 2006;125:1321–1332. doi: 10.1016/j.cell.2006.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochwagen A., Amon A. Checking your breaks: Surveillance mechanisms of meiotic recombination. Curr. Biol. 2006;16:R217–R228. doi: 10.1016/j.cub.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Hochwagen A., Tham W.H., Brar G.A., Amon A. The FK506 binding protein Fpr3 counteracts protein phosphatase 1 to maintain meiotic recombination checkpoint activity. Cell. 2005;122:861–873. doi: 10.1016/j.cell.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Jang J.K., Sherizen D.E., Bhagat R., Manheim E.A., McKim K.S. Relationship of DNA double-strand breaks to synapsis in Drosophila. J. Cell Sci. 2003;116:3069–3077. doi: 10.1242/jcs.00614. [DOI] [PubMed] [Google Scholar]
- Kee K., Protacio R.U., Arora C., Keeney S. Spatial organization and dynamics of the association of Rec102 and Rec104 with meiotic chromosomes. EMBO J. 2004;23:1815–1824. doi: 10.1038/sj.emboj.7600184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S. Spo11 and the formation of DNA double-strand breaks in meiosis. In: Lankenau D.H., editor. Recombination and meiosis. Springer-Verlag; Heidelberg, Germany: 2007. pp. 81–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Chiasma formation: Chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115:175–194. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- Lamb T.M., Mitchell A.P. Coupling of Saccharomyces cerevisiae early meiotic gene expression to DNA replication depends upon RPD3 and SIN3. Genetics. 2001;157:545–556. doi: 10.1093/genetics/157.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Hooker G.W., Roeder G.S. Saccharomyces cerevisiae Mer2, Mei4 and Rec114 form a complex required for meiotic double-strand break formation. Genetics. 2006;173:1969–1981. doi: 10.1534/genetics.106.058768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Jang J.K., Kato N., McKim K.S. mei-P22 encodes a chromosome-associated protein required for the initiation of meiotic recombination in Drosophila melanogaster. Genetics. 2002;162:245–258. doi: 10.1093/genetics/162.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah S.K., Turner J.M.A., Baudat F., Rogakou E.P., de Boer P., Blanco-Rodriguez J., Jasin M., Keeney S., Bonner W.M., Burgoyne P.S. Recombinational DNA double strand breaks in mice precede synapsis. Nat. Genet. 2001;27:271–276. doi: 10.1038/85830. [DOI] [PubMed] [Google Scholar]
- Malapeira J., Moldon A., Hidalgo E., Smith G.R., Nurse P., Ayte J. A meiosis-specific cyclin regulated by splicing is required for proper progression through meiosis. Mol. Cell. Biol. 2005;25:6330–6337. doi: 10.1128/MCB.25.15.6330-6337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleki S., Neale M.J., Arora C., Henderson K.A., Keeney S. Interactions between Mei4, Rec114, and other proteins required for meiotic DNA double-strand break formation in Saccharomyces cerevisiae. Chromosoma. 2007;116:471–486. doi: 10.1007/s00412-007-0111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A., Valsasina B., Brotherton D., Troiani S., Rainoldi S., Tenca P., Molinari A., Santocanale C. Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J. Biol. Chem. 2006;281:10281–10290. doi: 10.1074/jbc.M512921200. [DOI] [PubMed] [Google Scholar]
- Murakami H., Nurse P. Regulation of premeiotic S phase and recombination-related double-strand DNA breaks during meiosis in fission yeast. Nat. Genet. 2001;28:290–293. doi: 10.1038/90142. [DOI] [PubMed] [Google Scholar]
- Murakami H., Borde V., Shibata T., Lichten M., Ohta K. Correlation between premeiotic DNA replication and chromatin transition at yeast recombination initiation sites. Nucleic Acids Res. 2003;31:4085–4090. doi: 10.1093/nar/gkg441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino K., Masai H. Rad3-Cds1 mediates coupling of initiation of meiotic recombination with DNA replication. Mei4-dependent transcription as a potential target of meiotic checkpoint. J. Biol. Chem. 2006;281:1338–1344. doi: 10.1074/jbc.M505767200. [DOI] [PubMed] [Google Scholar]
- Ogino K., Hirota K., Matsumoto S., Takeda T., Ohta K., Arai K., Masai H. Hsk1 kinase is required for induction of meiotic dsDNA breaks without involving checkpoint kinases in fission yeast. Proc. Natl. Acad. Sci. 2006;103:8131–8136. doi: 10.1073/pnas.0602498103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore R., Cao L., Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- Peoples-Holst T.L., Burgess S.M. Multiple branches of the meiotic recombination pathway contribute independently to homolog pairing and stable juxtaposition during meiosis in budding yeast. Genes & Dev. 2005;19:863–874. doi: 10.1101/gad.1293605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Siomos M.F., Nasmyth K. Un menage a quatre: The molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- Prieler S., Penkner A., Borde V., Klein F. The control of Spo11’s interaction with meiotic recombination hotspots. Genes & Dev. 2005;19:255–269. doi: 10.1101/gad.321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasanuma H., Murakami H., Fukuda T., Shibata T., Nicolas A., Ohta K. Meiotic association between Spo11 regulated by Rec102, Rec104 and Rec114. Nucleic Acids Res. 2007;35:1119–1133. doi: 10.1093/nar/gkl1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasanuma H., Hirota K., Fukuda T., Kakusho N., Kugou K., Kawasaki Y., Shibata T., Masai H., Ohta K. Cdc7-dependent phosphorylation of Mer2 facilitates initiation of yeast meiotic recombination. Genes & Dev. 2008 doi: 10.1101/gad.1626608. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D., Byers B. Meiotic effects of DNA-defective cell division cycle mutations of Saccharomyces cerevisiae. Chromosoma. 1978;70:109–130. doi: 10.1007/BF00292220. [DOI] [PubMed] [Google Scholar]
- Sclafani R.A. Cdc7p–Dbf4p becomes famous in the cell cycle. J. Cell Sci. 2000;113:2111–2117. doi: 10.1242/jcs.113.12.2111. [DOI] [PubMed] [Google Scholar]
- Smith K.N., Penkner A., Ohta K., Klein F., Nicolas A. B-type cyclins CLB5 and CLB6 control the initiation of recombination and synaptonemal complex formation in yeast meiosis. Curr. Biol. 2001;11:88–97. doi: 10.1016/s0960-9822(01)00026-4. [DOI] [PubMed] [Google Scholar]
- Stuart D., Wittenberg C. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes & Dev. 1998;12:2698–2710. doi: 10.1101/gad.12.17.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonami Y., Murakami H., Shirahige K., Nakanishi M. A checkpoint control linking meiotic S phase and recombination initiation in fission yeast. Proc. Natl. Acad. Sci. 2005;102:5797–5801. doi: 10.1073/pnas.0407236102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax J.A., Woodbury E.L., Quang P.N., Paraz M., Blethrow J.D., Shah K., Shokat K.M., Morgan D.O. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- Valentin G., Schwob E., Della Seta F. Dual role of the Cdc7-regulatory protein Dbf4 during yeast meiosis. J. Biol. Chem. 2006;281:2828–2834. doi: 10.1074/jbc.M510626200. [DOI] [PubMed] [Google Scholar]
- Wan L., Zhang C., Shokat K.M., Hollingsworth N.M. Chemical inactivation of Cdc7 kinase in budding yeast results in a reversible arrest that allows efficient cell synchronization prior to meiotic recombination. Genetics. 2006;174:1767–1774. doi: 10.1534/genetics.106.064303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L., Niu H., Futcher B., Zhang C., Shokat K.M., Boulton S.J., Hollingsworth N.M. Cdc28–Clb5 (CDK-S) and Cdc7–Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes & Dev. 2008 doi: 10.1101/gad.1626408. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]