As soon as the Hox genes were isolated in Drosophila and identified in mammals more than two decades ago, genetic and molecular data started to accumulate, suggesting a conserved function for these clustered genes in the specification of positional identity of rostro–caudal embryonic structures (Duboule and Dollé 1989; Graham et al. 1989). Temporal and spatial collinearity of expression of the Hox genes was shown to correlate with the anterior to posterior progression of development, with 3′ genes being expressed earlier and more anteriorly than successively more 5′ genes. Shortly after these discoveries, overlapping expression patterns of the 5′ subset of Hoxd genes in the developing mouse limb buds suggested that these genes were involved in patterning the limbs as well (Dollé et al. 1989). A strikingly high expression level of the last gene of the cluster, Hoxd13, in the genital bud and in the distal part of the limb buds opened the possibility that a “quantitative collinearity” might be playing a role in genital and limb development, obeying rules distinct from those of axial collinearity (Dollé and Duboule 1989; Dollé et al. 1991). The analysis of early and late phases of Hox gene expression in the chick limb bud further strengthened the suggestion that a “reversed,” 5′-nested Hox collinearity was at work at late stages in distal limb buds (Morgan and Tabin 1994; Nelson et al. 1996). The relationship between the two different types of collinearity was approached experimentally in the mouse, thanks to the design of a powerful genetic tool, the targeted meiotic recombination (Hérault et al. 1998), making it possible to engineer rearrangements at will within the Hoxd cluster. It appeared that 5′-located Hoxd genes that display spatial and temporal collinear expression in the developing trunk and early limb bud undergo a subsequent distinct regulation in the distal part of the limbs at later stages (Zakany et al. 2004; Tarchini and Duboule 2006). The later phase of Hoxd expression modulates morphogenesis of the digital plate, distally extending the limb buds. This distal limb expression was found to depend on remote enhancers on the centromeric (5′) side of the HoxD cluster (Spitz et al. 2003). Information from comparative genome analysis recently shed additional light on the relationship between Hox expression and function along the axis and in the growing limbs, in the context of evolutionary developmental biology. Vertebrate Hox genes are arranged in much more condensed and organized clusters than their homologs in all other clades of the evolutionary tree (Duboule 2007). As compact clusters are conceivably more easily controlled in a coordinated way from distant enhancers, this may have provided an evolutionary opportunity for the recruitment of efficient regulatory regions from outside the HoxD cluster. The novel regulatory input would be at the origin of the late expression phase of Hoxd genes in the distal limb margin, in a suitable context to shape the vertebrate limb terminal elements, the digits (Duboule 2007; Freitas et al. 2007).
In this issue of Genes & Development, Montavon et al. (2008) report on their quantification of the expression of the 5′ members of the HoxD cluster in normal and rearranged configurations, and on their design of a mathematical model of the regulatory circuit driving the late phase of Hoxd gene expression. They propose an additional, exquisite outcome of the quantitative regulation of the 5′ Hoxd genes: the ontogeny of a distinct morphology for digit 1, the thumb.
Hoxd gene expression: late limb collinearity opposite to axial collinearity
Transcriptional control of the linear arrangement of the Hox genes by regulatory elements outside the cluster obligatorily leads to polarized expression of the gene series (Tarchini and Duboule 2006). Early onset of Hox gene expression during embryogenesis is thought to depend on a remote regulatory region located on the 3′ telomeric side of the cluster, possibly coinciding or overlapping with the Early Limb Control Region proposed to control Hoxd gene expression in the early limb bud (Zakany et al. 2004; Tarchini and Duboule 2006). Sequential expression of 3′ to 5′ genes generates posteriorly nested overlapping sets of Hox expression domains along the antero–posterior (A–P) axis, and in the early limb buds that are therefore A–P polarized. The late phase of Hoxd expression in limb buds was long recognized to follow a collinearity opposite to axial collinearity, called “reverse” collinearity (Nelson et al. 1996) or quantitative collinearity (Dollé et al. 1991). Experimental rearrangement among 5′ Hoxd genes elegantly demonstrated that the gene order was responsible for this peculiar expression behavior of the 5′ Hoxd genes (Kmita et al. 2002). Any gene present at the first, 5′-most position of the cluster is expressed at the strongest level and this reverse collinearity is essential for correct digit morphogenesis (Kmita et al. 2002). A distal limb-specific regulatory region outside the Hoxd cluster on its 5′ side was identified, and this led to the characterization of the remote Global Control Region (GCR) and the Prox element (Spitz et al. 2003, 2005; Gonzalez et al. 2007), both essential for correct Hoxd expression in distal limbs (Fig. 1). Montavon et al. (2008) now unravel the mechanism underlying the tight transcriptional control of the four 5′-most Hoxd genes in the digital plate, which has a critical impact on digit morphogenesis.
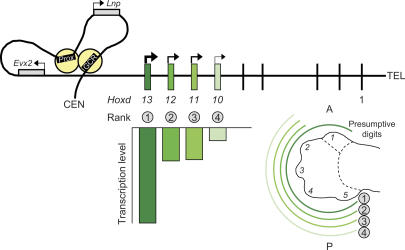
Figure 1.
Schematic representation (modified from Montavon et al. 2008) of the quantified expression of the four 5′-most members of the HoxD cluster, and of their expression domains in an E12.5 mouse distal limb bud. The more proximal expression domain of these Hoxd genes is not shown for the sake of simplicity, since it does not concern the digits. The thick black line above represents the chromosome with its telomeric (TEL) and centromeric (CEN) ends. The genomic region centromeric to the HoxD cluster is shown to form a loop, allowing the protein complexes (in yellow), bound to the enhancers GCR and Prox, to interact with the Evx2-Hoxd13 intergenic region. Hoxd13 to Hoxd10 and their transcriptional output are shown in green, with color intensity proportional to transcriptional activity. The rank of the Hoxd genes relative to the enhancers is indicated both above the expression bars on the left and flanking the curved lines showing the digital extent of the expression domains on the right. Presumptive digit 1 is singularized by its nonexpression of genes with rank 2–4. The transcription level of each Hoxd gene depends on both its rank and the specific affinity of its promoter for the enhancer complexes (see the text for more details). (A) Anterior; (P) posterior.
Unique Hox expression status and morphology of the thumb
Genes of the HoxA and HoxD clusters are important players during limb development and were proven to exert a growth-promoting role on the distal limb (Kmita et al. 2005; Tarchini et al. 2006). 5′ members of HoxA and HoxD are expressed early on with the same posterior restriction, but their late expression patterns differ. While Hoxa13 is expressed across the whole digital plate, the late phase of Hoxd expression maintains a posterior expression preference (Fig. 1). Strong activation of Hoxd13 elicited by the remote transcriptional enhancers leads to a farther extension of its transcription domain as compared with the other 5′ Hoxd genes, such that only the Hoxd13 domain encompasses presumptive digit 1, the future thumb (Fig. 1; Nelson et al. 1996; Kmita et al. 2002). Consequently, the dosage of Hoxa and Hoxd proteins is higher in digits 2–5 than in digit 1, resulting in a distinct thumb morphology. Posterior skewing of Hoxd expression domains and preferential activation of the 5′-most Hoxd gene are therefore responsible for the relative growth deficit in the anterior-most digit and the unique morphology of the thumb (Montavon et al. 2008). In agreement with this conclusion, ectopic expression of mouse posterior Hoxd genes in anterior-most distal cells leads to a digit 1 with an additional phalange, transforming the thumb into a digit with a more posterior identity. The fact that the relative posterior restriction of the late expression domain of bat versus mouse Hoxd13 in the forelimb bud is accompanied by a growth reduction of digits I and II compared with the other digits (Chen et al. 2005) may as well be in line with the demonstrated impact of the reverse collinearity of 5′ Hoxd genes on digital morphologies in the mouse.
The molecular mechanism behind the growth-stimulating activity of Hoxd genes in the digital plate remains to be elucidated. It may involve growth-promoting signaling pathways such as BMP, downstream from Shh (Drossopoulou et al. 2000), and sustained Fgf signaling (Sanz-Ezquerro and Tickle 2003) from the apical ectodermal ridge.
Strict quantitative control of 5′ Hoxd transcription
While the collinear 5′-to-3′ decrease in transcription levels of 5′ Hoxd genes had been observed upon in situ hybridization and challenged experimentally (Kmita et al. 2002), the new approach of measuring the steady-state RNA levels of the five 5′-most Hoxd genes (Montavon et al. 2008) validates and quantifies the decrease in Hoxd gene expression. In addition, it brings new features to light that had escaped detection so far. First, this approach reveals that promoter specificity definitely plays a role in the transcriptional output of a particular Hoxd configuration, in addition to gene rank, and that 5′ Hoxd genes are not equivalent in their efficiency of interaction with the global “digit” enhancers. This situation was not anticipated since deletion of Hoxd13 led to such an increase in Hoxd12 transcription that this gene acquired an expression domain and a function resembling those of Hoxd13 (Kmita et al. 2002). Importantly as well, it became clear, after decreasing or augmenting the number of Hoxd genes, that a quasilinear relationship exists between the gene number and the total mRNA transcribed from the 5′ part of the Hoxd cluster. This behavior, privileging some mechanisms rather than others, proves that the regulatory potential of the system is not saturated in the normal situation but rather adjusts to the gene number. An additional novel finding concerns the precise incidence of 5′ Hoxd deletions, insertions, and mutations on the transcription of Evx2 and Lunapark (Lnp). These genes, the function of which is unrelated to limb morphogenesis, are located between the GCR and 5′ Hoxd genes (Fig. 1). Quantification of the transcriptional responses of the seven genes considered, to alterations in and around the 5′ Hoxd region, suggests that the “digit” enhancers probably interact with the Hoxd13-Evx2 intergenic region, and subsequently scan the nearby Hoxd genes preferentially (Fig. 1).
A mathematical model able to account for the Hoxd expression features
Because digit morphology was known to be very sensitive to variations in the dosage and combination of 5′ Hox genes expressed during embryonic limb development (Zakany et al. 1997), understanding the complex regulatory control of these essential players in this process was expected to help define the conditions for correct autopod patterning. In the elaboration of a mathematical model capable of accounting for appropriate expression of each of the four 5′ Hoxd genes driving the late phase of distal limb outgrowth and patterning, Montavon et al. (2008) defined a first set of parameters utilizing elements from previous studies (Kmita et al. 2002) and from the novel outcome of their quantitative assays. These parameters correspond to the affinities of the enhancer complex for each of the individual promoters and to the fact that total transcription linearly depends on the number of promoters. A single class of models appeared to fit the experimental data, involving a first step of recognition of the topology of the complex by the cis-acting elements and associated protein complexes, followed by a second step of subsequent local scanning and interaction with each promoter. An additional parameter was introduced for each gene to include the probability that the complex did not stop at an upstream promoter already. Several rounds of parameter refinement and validation followed, using the large collection of Hoxd mutant alleles available in the laboratory. Montavon et al. (2008) finally came up with the “best-fit” model, generating predicted expression values for the individual genes matching very well the expression data in wild types and mutants. Interestingly, from the final parameter values of the “best-fit” model, it appears that the affinity of the Hoxd9 promoter is as low as that of Evx2, suggesting that only the four most 5′ Hoxd genes have evolved more robust affinities for the enhancer complex.
Hoxd expression in the genital bud fits the limb model
Montavon et al. (2008) set out to verify the adequacy of their model to account for the tightly controlled expression of 5′ Hoxd genes in the genital bud. “Quantitative” collinearity of 5′ Hoxd gene expression was observed in that precursor structure for the external genitalia at the same time as it was found in the distal limbs (Dollé and Duboule 1989; Dollé et al. 1991). Remarkably, 5′-nested Hoxd deletions and duplications induced comparable regulatory reallocations in the genital bud as in digits. With the exception of a few mutants with internal deletions, the mathematical model designed on the basis of parameters for the limbs was found to correctly predict transcriptional activity of the seven genes assayed during genital bud development, both in wild-type and mutant contexts. This suggests that both structures not only utilize the same patterning genes (Cobb and Duboule 2005), but do so by using the same regulatory strategy. This demonstrates the similarity in ontogenic principles and phylogenetic history of both structures, as proposed earlier (Dollé et al. 1991; Kondo et al. 1997; Cohn 2004; Suzuki et al. 2004).
Untangling multiple control strategies of the clustered Hox genes
Montavon et al. (2008) found that the transcriptional output in the genital buds of some internal HoxD deletion mutants could not be accommodated by the limb model. They mention the possibility that the deletions impaired an additional regulatory circuit by removing gene proximal sequences involved in genital bud but not in limb development. The use of a mathematical model of transcriptional behavior therefore may help to recognize the occurrence, in some instances, of an additional controlling circuit superimposed on the regulatory mechanism under study.
The quantitative expression assays of 5′ Hoxd genes and the successful modeling of their complex transcriptional output during limb bud development definitely establishes the role of the regulatory constraints of the Hox linear topology, and of promoter-specific affinities in the generation of a main distinction in digital morphologies. This strategy may help to further understand the determinants of the various digital features in mammals and other species. Given the likely impact of more numerous parameters in the process, such as the exact timing of expression of the Hox genes in all presumptive digital territories, and the influence of signaling factors involved in differential digit outgrowth, it is too early to say whether integrating the different genetic systems, each with their own time and space constraints, will not be too complex to be usefully modeled.
Acknowledgments
I thank Marie Kmita (IRCM, Montreal, Canada) and Frits Meijlink (Hubrecht Institute, Utrecht, The Netherlands) for critical reading of the manuscript. J.D. is supported by the Dutch Research Organization NWO ALW, the Dutch Bsik Program “Stem Cells in Development and Disease” and the EU Network of Excellence (framework 6) “Cells into Organs.”
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1641708
References
- Chen C.H., Cretekos C.J., Rasweiler J.J.T., Behringer R.R. Hoxd13 expression in the developing limbs of the short-tailed fruit bat, Carollia perspicillata. Evol. Dev. 2005;7:130–141. doi: 10.1111/j.1525-142X.2005.05015.x. [DOI] [PubMed] [Google Scholar]
- Cobb J., Duboule D. Comparative analysis of genes downstream of the Hoxd cluster in developing digits and external genitalia. Development. 2005;132:3055–3067. doi: 10.1242/dev.01885. [DOI] [PubMed] [Google Scholar]
- Cohn M.J. Developmental genetics of the external genitalia. Adv. Exp. Med. Biol. 2004;545:149–157. doi: 10.1007/978-1-4419-8995-6_9. [DOI] [PubMed] [Google Scholar]
- Dollé P., Duboule D. Two gene members of the murine HOX-5 complex show regional and cell-type specific expression in developing limbs and gonads. EMBO J. 1989;8:1507–1515. doi: 10.1002/j.1460-2075.1989.tb03535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollé P., Izpisua-Belmonte J.C., Falkenstein H., Renucci A., Duboule D. Coordinate expression of the murine Hox-5 complex homeobox containing genes during limb pattern formation. Nature. 1989;342:767–772. doi: 10.1038/342767a0. [DOI] [PubMed] [Google Scholar]
- Dollé P., Izpisùa-Belmonte J.C., Brown J.M., Tickle C., Duboule D. HOX-4 genes and the morphogenesis of mammalian genitalia. Genes & Dev. 1991;5:1767–1776. doi: 10.1101/gad.5.10.1767. [DOI] [PubMed] [Google Scholar]
- Drossopoulou G., Lewis K.E., Sanz-Ezquerro J.J., Nikbakht N., McMahon A.P., Hofmann C., Tickle C.2000. . A model for anteroposterior patterning of the vertebrate limb based on sequential long- and short-range Shh signalling and Bmp signalling Development 1271337–1348. [DOI] [PubMed] [Google Scholar]
- Duboule D. The rise and fall of Hox gene clusters. Development. 2007;134:2549–2560. doi: 10.1242/dev.001065. [DOI] [PubMed] [Google Scholar]
- Duboule D., Dollé P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989;8:1497–1505. doi: 10.1002/j.1460-2075.1989.tb03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas R., Zhang G., Cohn M.J. Biphasic Hoxd gene expression in shark paired fins reveals an ancient origin of the distal limb domain. PLoS ONE. 2007;2:e754. doi: 10.1371/journal.pone.0000754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F., Duboule D., Spitz F. Transgenic analysis of Hoxd gene regulation during digit development. Dev. Biol. 2007;306:847–859. doi: 10.1016/j.ydbio.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Graham A., Papalopulu N., Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989;57:367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- Hérault Y., Rassoulzadegan M., Cuzin F., Duboule D. Engineering chromosomes in mice through targeted meiotic recombination (TAMERE) Nat. Genet. 1998;20:381–384. doi: 10.1038/3861. [DOI] [PubMed] [Google Scholar]
- Kmita M., Fraudeau N., Herault Y., Duboule D. Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature. 2002;420:145–150. doi: 10.1038/nature01189. [DOI] [PubMed] [Google Scholar]
- Kmita M., Tarchini B., Zakany J., Logan M., Tabin C.J., Duboule D. Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature. 2005;435:1113–1116. doi: 10.1038/nature03648. [DOI] [PubMed] [Google Scholar]
- Kondo T., Zakany J., Innis J.W., Duboule D. Of fingers, toes and penises. Nature. 1997;390:29. doi: 10.1038/36234. [DOI] [PubMed] [Google Scholar]
- Montavon T., Le Garrec J.-F., Kerszberg M., Duboule D. Modeling Hox gene regulation in digits: Reverse collinearity and the molecular origin of thumbness. Genes & Dev. 2008 doi: 10.1101/gad.1631708. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan B., Tabin C. Hox genes and growth: Early and late roles in limb bud morphigenesis. Dev. Suppl. 1994;1994:181–186. [PubMed] [Google Scholar]
- Nelson C.E., Morgan B.A., Burke A.C., Laufer E., DiMambro E., Murtaugh L.C., Gonzales E., Tessarollo L., Parada L.F., Tabin C. Analysis of Hox gene expression in the chick limb bud. Development. 1996;122:1449–1466. doi: 10.1242/dev.122.5.1449. [DOI] [PubMed] [Google Scholar]
- Sanz-Ezquerro J.J., Tickle C. Fgf signaling controls the number of phalanges and tip formation in developing digits. Curr. Biol. 2003;13:1830–1836. doi: 10.1016/j.cub.2003.09.040. [DOI] [PubMed] [Google Scholar]
- Spitz F., Gonzalez F., Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- Spitz F., Herkenne C., Morris M.A., Duboule D. Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nat. Genet. 2005;37:889–893. doi: 10.1038/ng1597. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Shiota K., Zhang Y., Lei L., Yamada G. Development of the mouse external genitalia: Unique model of organogenesis. Adv. Exp. Med. Biol. 2004;545:159–172. doi: 10.1007/978-1-4419-8995-6_10. [DOI] [PubMed] [Google Scholar]
- Tarchini B., Duboule D. Control of Hoxd genes collinearity during early limb development. Dev. Cell. 2006;10:93–103. doi: 10.1016/j.devcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Tarchini B., Duboule D., Kmita M. Regulatory constraints in the evolution of the tetrapod limb anterior–posterior polarity. Nature. 2006;443:985–988. doi: 10.1038/nature05247. [DOI] [PubMed] [Google Scholar]
- Zakany J., Fromental-Ramain C., Warot X., Duboule D. Regulation of number and size of digits by posterior Hox genes: A dose-dependent mechanism with potential evolutionary implications. Proc. Natl. Acad. Sci. 1997;94:13695–13700. doi: 10.1073/pnas.94.25.13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakany J., Kmita M., Duboule D. A dual role for Hox genes in limb anteriorposterior asymmetry. Science. 2004;304:1669–1672. doi: 10.1126/science.1096049. [DOI] [PubMed] [Google Scholar]