Abstract
Purpose
To evaluate an automatic 3D registration algorithm for serial high-resolution images of trabecular bone (TB) in studies designed to evaluate the response of the trabecular architecture to intervention or disease progression.
Materials and Methods
An efficient algorithm for registering high-resolution 3D images of trabecular bone is presented. The procedure identifies the six parameters of rigid displacement between two scans performed at different time points. By assuming a relatively small through-plane rotation, considerable time is saved by combining the results of a collection of regional 2D registrations throughout the TB region of interest (ROI). The algorithm was applied to 26 pairs of MR images acquired six months apart. Reproducibility of local TB structural parameters (plate, rod, and junction density) computed in manually selected regions were compared between baseline and registered follow-up images.
Results
All 26 registrations were completed successfully in less than 30 seconds per image pair. The resampled follow-up images agreed with baseline to around one pixel throughout the volume at 137×137×410 µm3 image resolution. Structural parameters in each region correlated well from baseline to follow-up with intraclass correlation coefficients ranging between 85–97% for TB plate density. Inter-regional variations in the parameters were large as compared with intra-region reproducibility.
Conclusion
The proposed algorithm was successful in automatically registering baseline and follow-up TB images in a translational study, and may be useful in regional analyses in longitudinal MR studies of TB architecture.
Keywords: Trabecular bone, registration, micro-MRI
Introduction
Trabecular bone (TB) remodels considerably faster than cortical bone. Further, the majority of osteoporotic fractures occur at locations where TB is dominant, i.e. the vertebrae, distal and proximal extremities, or ribs. There is now substantial evidence that the strength of bone is a function of the bone material composition, volume fraction and distribution, collectively summarized under the term “bone quality” 1. Further, a number of studies have shown a structural contribution to fracture risk independent of bone volume fraction or density 2–5. However, the most widely applied criterion for the assessment of osteoporotic risk remains apparent density, quantified by either two-dimensional (2D) projection imaging via dual-energy X-ray absorptiometry (DXA, see, for example, 6) or 3D imaging by quantitative computed tomography (qCT) 7.
Until recently, the only means for quantifying TB structure has been based on transiliac bone biopsies, which are analyzed by stereology, or more recently, on the basis of ex vivo 3D micro-computed tomography 8. Besides being invasive (and therefore rarely indicated clinically) these methods are ill suited for the assessment of treatment efficacy since the same site can be biopsied only once. At best, paired biopsies can be taken at the left and right ilium 8. However, there is no one-to-one correspondence between the measurement sites therefore leading to substantial sampling errors.
The emergence of high-resolution imaging modalities, key among which are micro-magnetic resonance imaging (µMRI) 9,10 and, more recently, high-resolution peripheral quantitative CT (HR-pQCT) 11, now permit in vivo structure analysis. The technology and applications of µMRI underlying the present work have been reviewed recently 12,13 and the method has demonstrated its potential in several laboratories for fracture discrimination 10,14–18. There is further evidence that µMRI can quantify the topological as well as conventional structural changes occurring in response to intervention in patients with various forms of osteoporosis 19,20.
Since the organization of the TB network is highly heterogeneous it is essential that precisely the same volume is examined in follow-up studies, which requires that the images be registered accurately. Both prospective and retrospective registration approaches have been pursued, each having a unique set of advantages. In prospective registration a localizer image is acquired prior to the high-resolution scan in the baseline and follow-up imaging session so as to ensure that the location and orientation of the high-resolution imaging slab matches that obtained at baseline 21,22. In the more commonly practiced retrospective registration approach no particular precautions are taken to ensure that the high-resolution scan acquisition volume and orientation in the follow-up scan precisely match that of the baseline scan, although rough landmarks are used to prescribe approximately the same volume. Typically, this is achieved on the basis of longitudinal localizer images (coronal or sagittal) from which the inferior boundary of the axial high-resolution imaging slab is selected as a fixed distance from the distal cortical endplate 18. The resulting high-resolution images are then matched to each other after they were acquired 23,24 . The proposed technique falls into the latter category of methods.
A complication in the registration task is that the periosteal soft tissue is not rigidly connected to the TB region of interest (ROI) (see Fig. 1a). Thus, a successful technique should isolate and register only the ROI. Segmentation errors caused by aliasing or other artifacts can make it difficult to perform automatic registration based on the shape of the ROI and manual intervention may be required 12,25. The proposed algorithm uses the local trabecular pattern near 100 randomly selected points in the ROI to register the 3D data sets. The method is thus fundamentally different from other 3D techniques (for example 26) which typically search the six directions of rigid body motion. A key assumption in our method is that the through-plane tilt is relatively small (10° or less), which is reasonable considering that the limb (lower leg or forearm) is easily positioned approximately parallel to the magnet bore.
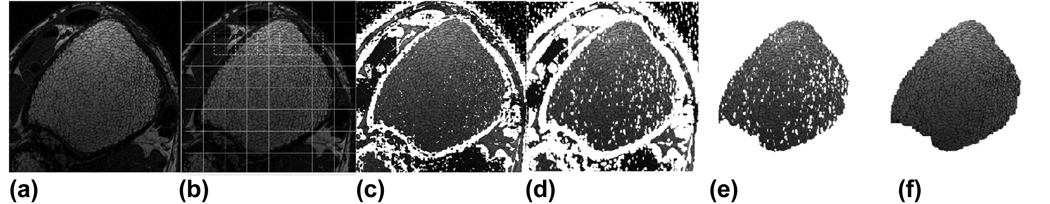
Fig 1.
Automatic trabecular bone ROI segmentation procedure illustrated for the distal tibia. (a) Manually selected rectangular region centered on the ROI; (b) Overlapping grid pattern for applying local threshold; (c) Initial local threshold mask, roughly identifying cortical shell; (d) Dilation of mask in (c) to avoid leakage; (e) Selection of main connected component; (f) Final ROI after filling small holes.
The proposed technique has the potential to facilitate the study of the implications of bone remodeling on topology and scale of the TB network at a finer spatial scale. Rather than using bulk parameters over the entire trabecular volume to track changes over time, it enables a region-by-region or even pixel-by-pixel analysis. This capability is of particular importance since trabecular bone loss or accrual is well known to be localized.
The registration procedure was applied to a collection of 3D MR scans of the distal tibia acquired as part of a six month serial pilot study. The advantages of the method were demonstrated by examining structural parameter reproducibility between time points, as well as spatial variation of parameters across the ROI.
Materials and Methods
Registration Algorithm
The objective of the registration procedure is to compute the 3D transformation parameters (3 rotations and 3 translations) describing the misalignment between high-resolution baseline and follow-up images in serial MR studies. These parameters are then used either to resample the follow-up images to match those obtained at baseline, or to identify a consistent 3D mask used for the analysis. We focus here on the application of registering axial images of distal tibia, but the algorithm could also be applied to other anatomical locations and orientations, provided that (a) the pattern of the trabecular bone is resolved, and (b) only small through-plane rotational displacement is expected. The registration algorithm comprises four steps.
The first step is to apply an automatic segmentation algorithm 27 to both the baseline and follow-up images in order to separate the trabecular bone ROI from the surrounding anatomy. As mentioned above, this is important because the distal tibia is not rigidly connected to the surrounding fat and muscle. Therefore the registration must be solely based on features of the trabecular bone network itself. The accuracy of this segmentation step will not significantly impact subsequent steps, since only a very rough segmentation is required for the algorithm to succeed. Nevertheless, an accurate ROI determination is also desirable for consistent time point comparisons of structural parameters (subsequent to the registration step). Therefore, we provide here the details of this automatic segmentation procedure, keeping in mind that a less thorough but simpler procedure could also be used if image registration was the only goal.
The segmentation algorithm requires a single, minor user intervention where the operator draws a rectangle surrounding the region of interest, with the center of the rectangle occurring approximately at the center of the endosteal cavity, here illustrated for the tibia (Fig. 1a). The purpose of this operation is to minimize the risk of inadvertently selecting nearby anatomic feature (such as the fibula or a region of muscle in the case of tibia imaging). Isolation of the trabecular ROI is equivalent to identifying the surrounding cortical shell, appearing dark in the MR image. To achieve this, a local threshold is applied to each slice within small 20×20 pixel neighborhoods (Fig. 1b). In each neighborhood the mean intensity is computed, and pixels with intensity less than 70% of this mean are initially classified as belonging to the cortex (appearing white in Fig. 1c). A local (rather than global) threshold is needed here to compensate for intensity variations due to a spatially variant receive coil sensitivity profile. Fifty percent overlapping regions are also used throughout the volume (shown in Fig. 1b) to avoid discontinuities at neighborhood boundaries. In the case where two overlapping regions disagree with respect to the classification of a single pixel, the pixel is initially classified as belonging to the cortex.
After initial segmentation via local threshold, the cortical region is dilated by two pixels in all directions in order to minimize the chance of leakage in areas of thin cortex (Fig. 1d). Next, the largest connected component in the selected region is isolated (Fig. 1e). At this stage, the ROI typically contains a number of holes corresponding to dense patches of trabecular bone misclassified as cortical bone in the threshold step. These erroneous classifications are remedied by filling in all holes of diameter less than 30 pixels (this parameter was chosen empirically). Most slices (>90%) should be properly segmented at this point, but a small number of slices may have problems due to signal leakage, that is the step of selecting the main connected component may fail to select only the ROI when there are areas of thin cortical bone. The dilation step is designed to minimize this risk, but some slices may still be affected. Thus the final step is to ‘synchronize’ the slices, ensuring spatial consistency between the ROIs in neighboring slices, which is accomplished by iteratively removing voxels from the ROI that are not adjacent to ROI voxels in the two neighboring slices.
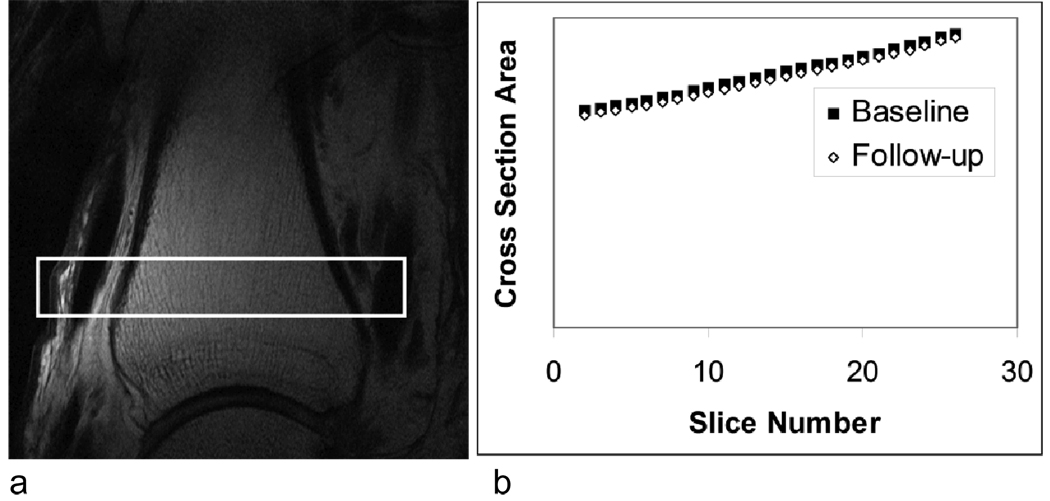
Following segmentation, the second step of registration is to approximately determine the longitudinal (slice) shift between baseline and follow-up images. As pointed out in the introduction, the through-plane rotational misalignment is assumed to be small, so there should be at least a rough correspondence between baseline and follow-up slices. While this step is not critical for the success of the overall algorithm, a good estimate of the slice shift can dramatically improve efficiency because it reduces the slice search radius in subsequent steps. To estimate this shift, we exploited the significant variation in cross-sectional area of the tibia along the slice direction within our analysis region (see Fig. 2). The cross-sectional area for each slice was estimated using pixel counts from the segmented images, and the slice shift was then approximated by comparing those areas between baseline and follow-up. In the example of Fig. 2b, the estimated slice shift is the horizontal shift between the two curves.
Fig 2.
a) Sagittal localizer of the distal tibia with high-resolution imaging slab indicated; b) results of step 2 of the registration procedure showing cross-sectional area versus slice number in baseline and follow-up scan.
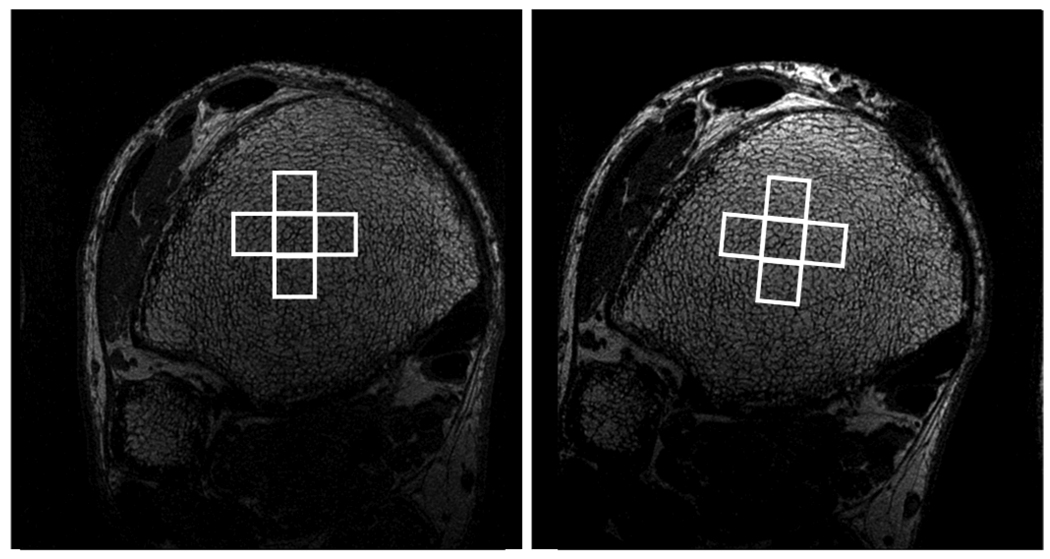
In the third step, initial estimates of in plane registration parameters (two translations and one rotation) are obtained. As with the previous step, the purpose of this operation is to improve the efficiency of the critical fourth step by significantly narrowing the necessary search range. A pixel p0 is chosen randomly toward the center of the ROI in the central slice of the baseline image. The 40×40 square region, R0 centered at p0 is matched to a 2D patch of the same size in the follow-up scan by searching the nearest ±3 slices, stepping through XY rotation angles ranging from −30 to +30 in increments of 3 degrees, and exhaustively searching for in-plane translations. The best match is determined by maximizing the cross correlation between rotated versions of R0 and 2D patches within the follow-up image. Rotations of R0 were implemented by applying a series of shear transformations. The success of the match is tested by checking for consistency with the same procedure applied to four adjacent 40×40 regions (Fig. 3). If these 5 test registrations cannot be fit to within 2 pixels using some 3D rigid transformation, the registration is labeled as having failed, and this step is repeated up to two more times using a different randomly selected central pixel.
Fig 3.
Baseline (left) and follow-up (right) images of the distal tibia. In steps 3 and 4 of the registration algorithm, two-dimensional rectangular regions in the baseline image are matched to patches in the follow-up image using the pattern of the trabecular bone network. Four adjacent patches are used to verify the success of the registration.
The full 3D rigid transformation (three translations and three rotations) is obtained in the fourth and final step. One hundred pixels (p1, p2, …, p100) are randomly chosen throughout all slices of the baseline ROI. For each pixel pi, the 40×40 region Ri (within a slice) centered at pi is matched to a 2D region centered at qi in the follow-up image using the same procedure as in the third step, except with a finer and significantly narrowed search range. As in step 3, each point is tested for success using adjacent test regions. If the number of successful regional registrations is less than 50 (i.e. 50%), then the registration is considered to have failed. After registration of these regions, the pixel pairs pi→qi for the successful registrations are used to fit the full 3D transformation. Specifically a 3D affine transformation (R,T) is chosen to minimize the least-squares distance:
| (1) |
where the sum is over only indices of the successful region registrations. Here, the rotation matrix and translation vector
| (2) |
are obtained by solving
| (3) |
where
| (4) |
and
| (5) |
Only the columns corresponding to successful regional registrations are included in P and Q. For computational reasons, we do not restrict R to be a rotation matrix, but consider arbitrary 3-dimensional linear transformations. In practice, the resulting transformation is expected to be very close to a rotation. While we could normalize R to be a true rotation (using singular value decomposition), such an adjustment may be undesirable if the imaging gradients are slightly miscalibrated causing the actual voxel dimensions to deviate from the prescribed resolution. In such a case, fitting a general linear transformation is more accurate.
Resampling Procedure
Once the 3D rotation and translation parameters have been obtained, there are at least two approaches for using these parameters for purposes of a consistent analysis. One approach is simply to resample the follow-up image to match the reconstruction grid of the baseline scan. In this case, care must be taken to avoid bias in the follow-up scan due to resampling effects. For example, linear interpolation in the context of a half-pixel shift is known to cause a decrease in resolution. But even when a sinc kernel (ideal for MRI) is used for resampling, subtle problems arise whenever there is non-zero through-plane rotation. This is a consequence of anisotropic resolution as it is commonly practiced in trabecular bone imaging, with lower resolution along the preferential direction of the trabeculae 28. Typically, the slice resolution is about a factor of three lower than that in the transverse plane, and therefore any small through-plane rotation in resampling will cause a slight decrease in transverse resolution. Although difficult to perceive in the image, this effect can bias the derived structural parameters.
An alternative is to resample the analysis mask rather than the image. That is, keep the follow-up image on its original reconstruction grid, but ensure that exactly the same 3D volume is analyzed by rotating and translating the baseline ROI mask obtained in the first step of the registration procedure, and intersecting it with the follow-up mask. For consistency, the follow-up mask must also be transformed and intersected with the baseline mask to yield a new baseline mask. In this study, both of the above mentioned resampling techniques were applied.
In Vivo Study
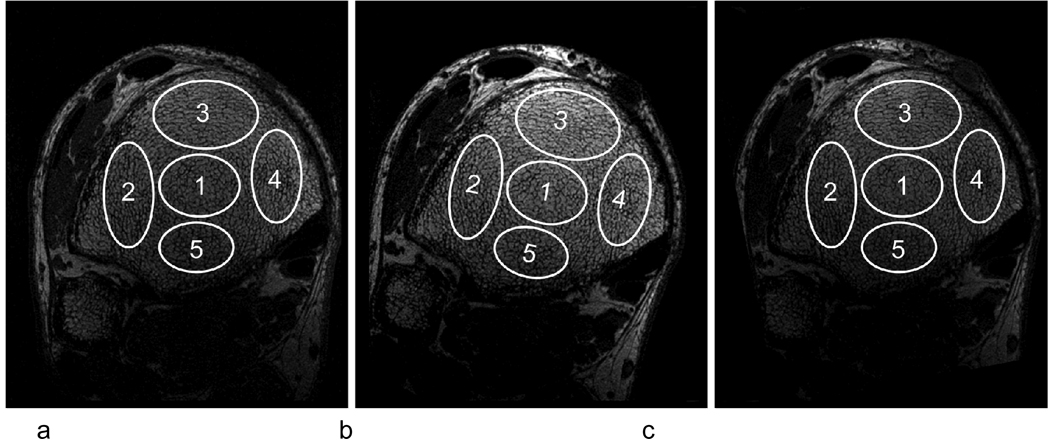
The registration algorithm was applied to data collected in a translational pilot study, in which 29 subjects were scanned at two time points, separated by six months. Three subjects were excluded due to unusable portions of the trabecular region. Although a significant portion of these images would be amenable to data analysis, these scans were excluded for the present study since we desired to evaluate five regions throughout the bone (see Fig. 4). The pilot study involved control and treatment groups, both of which were expected to experience some changes from baseline to follow-up. However, it is beyond the scope of this paper to investigate statistically significant changes between the two groups or between the two time points. Therefore, the results describing the effect of intervention on trabecular bone architecture will be reported elsewhere. It suffices to note that subjects were renal dialysis patients, half having been treated with low-frequency mechanical stimulation, a non-pharmacological intervention that may be anabolic to bone 29, the other half being subjected to placebo. Here we are solely interested in parameter reproducibility as well as correlations between various parameters. We point out that changes over time due to the intervention or due to the underlying renal osteodystrophy can only lower the measured reproducibility. We can therefore expect the reproducibility results to be at least as good as indicated here.
Fig 4.
Five regions manually selected in the baseline scan (a). Those regions were transformed to the non-resampled follow-up (b) and copied to the resampled follow-up (c).
The algorithm was programmed in C++ and run on a personal workstation (2.8 GHz Centrino Duo processor). As described above, two different strategies were used to apply the resulting 3D transformation parameters: (i) resampling of the follow-up image, and (ii) resampling of the ROI masks. In both cases, the images were processed using the Virtual Bone Biopsy 28, and the structural parameters derived.
All image data were acquired on a 1.5T Siemens Sonata scanner using a modified version 30 of the navigator-assisted 3D FLASE pulse sequence, 3D FLASE pulse sequence 31 with scan parameters TE/TR = 10/80ms, sampling frequency bandwidth = 17 kHz, field of view = 70×63×13 mm3 and image matrix size = 512×460×32, providing a voxel size of 137×137×l410 µm3 and scan time of 16 minutes.
To demonstrate the method’s potential for regional analysis (since in general the response to treatment may be anatomic location dependent), five ellipsoidal regions were manually selected in the baseline image, one toward the middle, and four toward the edges of the ROI (Fig. 4). These five regions were also copied to the resampled follow-up, and transformed (using the registration parameters) to the non-resampled follow-up. In addition to bulk parameters, selected topological parameters characterizing the trabecular network 32 were computed for each region. These include the topological surface, curve and junction densities. In digital topological analysis images of the trabecular structure are skeletonized thereby converting trabecular plates to surfaces and trabecular rods to curves 32 28 yielding the volume densities of these topological types, expressed in percent. The densities of junction-type voxels were also evaluated. These parameters are of particular interest since plates and rods and their mutual junctions are the basic building elements of the trabecular network and have been shown to be biomechanically relevant 33 34.
Results
The 3D registration parameters were computed for each baseline/follow-up pair in an average of 27 seconds per pair. After applying the estimate for in-plane translation and rotation from step 3, it was only necessary to search ±10 pixels for in-plane translation, ±3 slices, and ±3 degrees (in increments of 1 degree).
According to visual inspection, all 26 registrations were successful and accurate to around one pixel throughout the volume. This level of accuracy was confirmed in the 3D transformation fits, as the fitted transformations agreed with more than 90% of the baseline/follow-up point pairs within ±1 pixel in each direction.
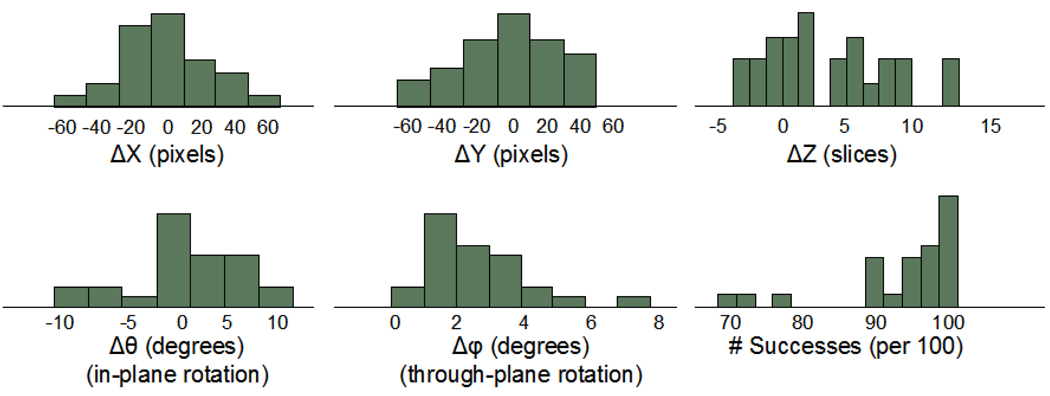
Fig. 5 shows the distribution of translation and rotation parameters in the registrations, as well as the success rate in the registration of the 100 randomly selected 2D regions for each baseline/follow-up pair. Despite efforts by the operator to choose consistent scan volumes, translational displacements between baseline and follow-up ranged between ±60 pixels (±8 mm) in the transverse plane and ±10 slices (±4 mm) in the longitudinal direction. In-plane rotation varied between ±10 degrees, whereas most through-plane rotational misalignments were less than 4 degrees. For all baseline/follow-up pairs, the algorithm used a minimum of 70% of the 2D regional registrations (recall that only successful registrations were used).
Fig 5.
Distributions of 3D registration parameters for 26 subjects scanned at two time points, and distribution of the number of successes per 100 regional registration trials.
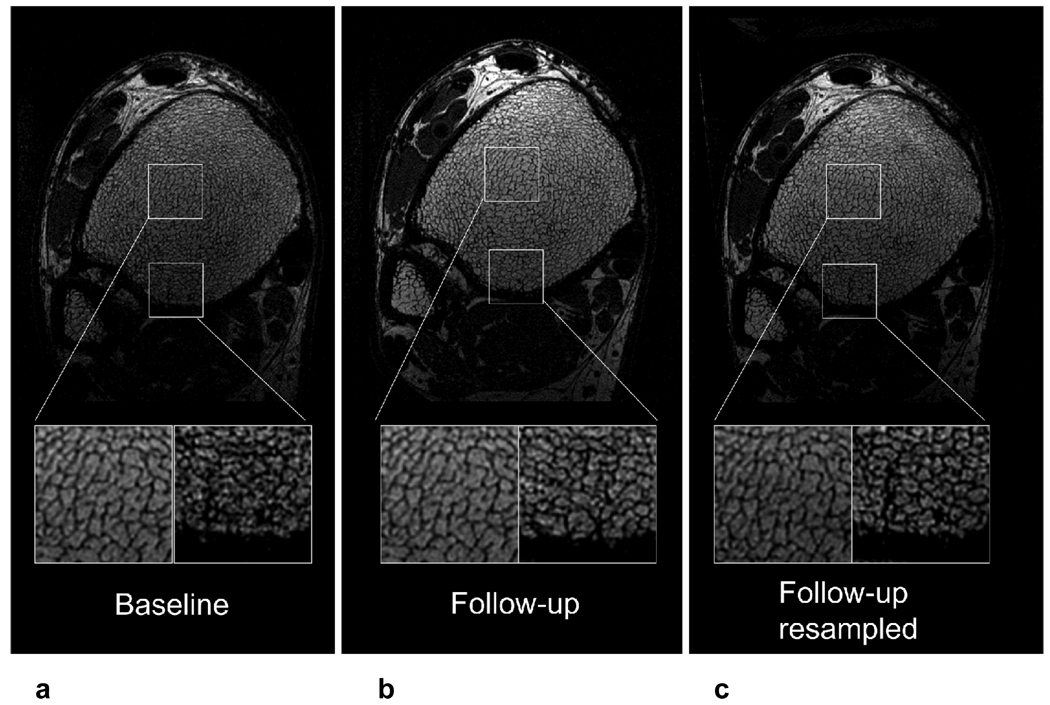
Fig. 6 shows the results of a single registration, illustrating the importance of correcting for through-plane tilt. The magnifications show good visual agreement in trabecular pattern between baseline and follow-up in one region of the bone, whereas the other region only agrees after 3D resampling. This is due to through-plane tilt (around 3 degrees in this case).
Fig 6.
Single slice at baseline (a), follow-up (b), and resampled follow-up (c). The left magnification shows a region that has matching trabecular pattern in all three images, whereas the right magnification shows a region where the trabecular network matches in the baseline and resampled follow-up, but not in the non-resampled follow-up. This behavior reflects the through-plane tilt (~3 degrees in this case).
The two methods mentioned above for follow-up analysis (i.e. resampling the image and resampling the mask) produced comparable results in terms of parameter correlations and distributions, although there were differences in individual parameter values. For simplicity, we present only the results for resampling of the follow-up mask, which we believe to be a more accurate technique, considering possible 3D resampling bias (which is of particular concern due to the anisotropic resolution in the case of through-plane tilt).
Table 1 shows the extent of baseline to follow-up reproducibility in terms of three structural parameters, as well as the variance in these parameters across the five regions. The average trabecular plate density (for all subjects) varied between 2.69% in Region 3 and 3.70% in Region 4, whereas there was comparatively little variation in this parameter from baseline to follow-up (less than 2% relative change for regions 1, 2, 3, and 5). Similarly, while there was significant regional variation in rod-density and junction density, the agreement between baseline and follow-up values for these parameters was on the order of 2–5% relative change for most regions.
Table 1.
Mean plate, rod and junction densities for each region across all 26 subjects, showing reproducibility from baseline to 6-month follow-up (follow-up in parentheses).
|
Parameter Means Baseline/(Follow-up) |
Region 1 | Region 2 | Region 3 | Region 4 | Region 5 | Mean |
|---|---|---|---|---|---|---|
| Plate Density | 2.96% (2.99%) |
3.31% (3.37%) |
2.69% (2.73%) |
3.70% (3.91%) |
3.70% (3.91%) |
3.27% (3.38%) |
| Rod Density | 0.387% (0.367%) |
0.395% (0.379%) |
0.391% (0.389%) |
0.402% (0.386%) |
0.402% (0.386%) |
0.395% (0.382%) |
| Junction Density | 0.335% (0.320%) |
0.362% (0.353%) |
0.294% (0.295%) |
0.397% (0.397%) |
0.397% (0.397%) |
0.357% (0.352%) |
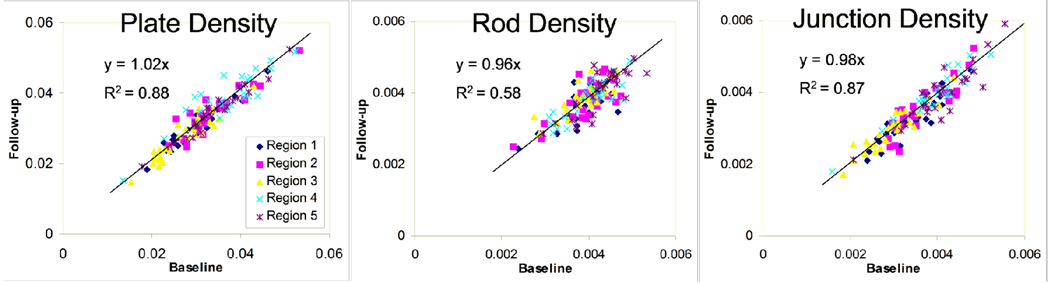
Reproducibility in mean parameters (averaged over all 26 subjects) was as expected better than baseline/follow-up reproducibility for individual subjects. Nevertheless, Fig. 7 shows the three topological parameters to be highly correlated between baseline and follow-up scans. Further, the slopes of the trend lines are close to unity, with moderate to high R-squared values, especially for plate and junction density. The regression lines were constrained to pass through the origin because all plotted parameters are densities, with absolute zero representing absence of the structure. Table 2 shows the intraclass correlation coefficient (ICC) 35 (a measure of reliability) for each parameter and analysis region. Table 3 shows the ICC for plate density between pairs of regions. The high values on the diagonal and comparatively low off-diagonal reflect the high degree of spatial variation in bone density, and demonstrate that the method can reproducibly distinguish between the varying levels of plate density.
Fig 7.
Follow-up vs. baseline correlation plots of plate, rod, and junction densities within each region.
Table 2.
Intraclass correlation coefficients for baseline vs. follow-up values of plate, rod, and junction density in each region.
| Intraclass Correlation | Region 1 | Region 2 | Region 3 | Region 4 | Region 5 | Mean |
|---|---|---|---|---|---|---|
| Plate Density | 96% | 90% | 93% | 85% | 85% | 90% |
| Rod Density | 65% | 70% | 79% | 80% | 80% | 75% |
| Junction Density | 88% | 88% | 93% | 94% | 94% | 91% |
Table 3.
Inter-regional intraclass correlation coefficients for baseline vs. follow-up values of plate density between pairs of spatial regions.
| Plate density Intraclass Correlation | Follow-up Region 1 | Follow-up Region 2 | Follow-up Region 3 | Follow-up Region 4 | Follow-up Region 5 |
|---|---|---|---|---|---|
| Baseline Region 1 | 96% | 53% | 41% | −16% | 46% |
| Baseline Region 2 | 49% | 90% | 42% | 24% | 70% |
| Baseline Region 3 | 39% | 39% | 93% | −9% | 20% |
| Baseline Region 4 | −4% | 44% | 9% | 85% | 54% |
| Baseline Region 5 | 45% | 82% | 24% | 40% | 97% |
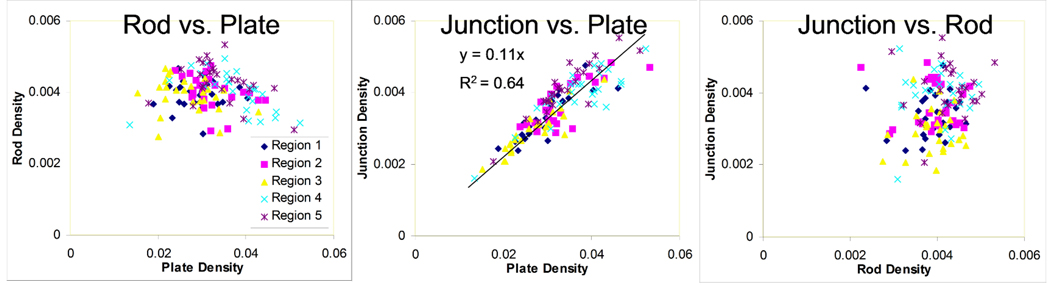
Correlations between pairs of VBB parameters (e.g. rod versus plate density) at baseline are shown in Fig. 8. While there is clear correlation between junctions and plates, there is no obvious relationship between rods and plates, or junctions and rods, indicating that these parameters carry independent information about the underlying bone structure.
Fig 8.
Pairwise comparison plots between plate, rod, and junction densities (baseline data only) for each region.
Discussion
The proposed 3D registration algorithm designed for time-course studies of trabecular bone architecture has several advantages over conventional 3D retrospective registration algorithms that search all six parameters of rigid motion for the best fit to the entire MR image. First, because only the local trabecular pattern within the bone of interest is used, the method is unaffected by changes in relative locations of soft-tissue structures surrounding the bone. The method is also largely immune to spatial variations in the received signal (which is a hallmark of most surface coils), since each registration involves only small patches in the baseline and follow-up images (see Appendix for details of the 2D regional correlation algorithm). Another advantage relates to computational efficiency. Since through-plane rotations are ignored in individual registrations of the 2D patches, only four (rather than six) degrees of displacement need to be searched at once (three translations and one in-plane rotation). Avoiding through-plane rotation in the 2D patches also obviates the need for resampling in three dimensions. As mentioned above, by combining the results of dozens of 2D registrations, we were able to indirectly obtain accurate measures of all six displacement parameters.
Due to small instrumental imperfections such as nonlinearities in the imaging gradients or small errors in the effective gradient amplitudes (not usually considered a problem), the true relationship between baseline and follow-up images may not exactly match a 3D rigid transformation. For example, a 5% inconsistency between readout and phase-encode resolution will not result in visible image distortion, but will cause problems when fitting a rigid transformation in the case of significant rotational displacement. An advantage of the present method is that we are able to detect and account for such inconsistencies. This is because the 3D transformation is fit as a general linear transformation rather than as a rigid rotation (there is no computational penalty for this since we have ample data, 100 pairs of points, to fit in 3D). In general we have found the resulting linear transformations to deviate slightly from rigid rotations, indicating slight errors in the imaging gradients.
Even in cases where prospective registration is used, retrospective techniques can be important for fine-tuning the registration. The prospective approach requires a pre-scan localizer, and there is a trade-off between registration accuracy and duration of the pre-scan. In 21, for example, pre-scan time is on the order of 2 minutes, with an accuracy of around 1 mm (corresponding to around 7 pixels in our case). Thus our retrospective correction (with accuracy to within 1 pixel) complements prospective registration techniques such as those described in 22 21. Conversely, even with a good retrospective registration algorithm, prospective registration is important to ensure maximal overlap between imaging slabs in the two scans.
Besides spatial variations in structure, the inter-regional differences in the measured structural parameters (see Table 1) may in small part be due to variations in signal-to-noise ratio (SNR). For example, region 3 is located in the area of highest SNR, close to the surface coil, whereas region 5 is in an area of lowest SNR. Interestingly, these two regions are toward opposite ends of the parameter spectrum in Table 1. Part of the difference could be due differences in SNR, although it is not possible to use this data to determine the extent of the effect. For serial studies examining small changes in parameter values over time, this type of effect could be a source of error if baseline and follow-up scans do not have the same spatial SNR distribution 24. Under these circumstances a bulk correction for SNR differences would not be sufficient. In these situations our registration technique will facilitate the use of spatially localized adjustments to compensate for parameter dependence on SNR, and thereby improving parameter reproducibility. Other potential sources of error may result from subject motion. Although the images were corrected for translational displacements during the scan via navigators, it has previously been shown that rotations of less than one degree can cause significant errors on the derived structural parameters 36 which can be corrected using retrospective motion correction techniques correcting for both translational and rotational motion 37.
Lastly, the method hinges on the veracity of the assumption that the rate of bone remodeling over the course of which structural measurements are conducted, does not alter the structure to an extent that spatial correlation between the two time points is significantly impaired. The remodeling rate in peri- and postmenopausal women, for example, is on the order of 0.13–0.24/year 38. There are virtually no data available in the literature whereby the same anatomic location was examined at two or more time points other than our own longitudinal studies in hypogonadal men and postmenopausal women 18,19, both suggesting the above conditions to be met. Nevertheless, it is possible that the actual performance of the algorithm is better than suggested by the data since the subjects on whom the study was conducted are not skeletally homeostatic (which could only be expected in young adults at peak bone mass). Thus, a portion of the error, defined as 1-rICC, where rICCis the intra-class correlation coefficient, could be ascribed to actual remodeling changes.
In conclusion, the proposed algorithm has been used to efficiently register baseline/follow-up pairs in a translational MR study of trabecular bone with good accuracy (on the order of 1 pixel), and a high rate of success. Because TB density and structure are heterogeneous, such registration is valuable for studying spatially localized changes in TB architecture over time.
Acknowledgments
Grant support:
National Institute of Health RO1 AR41443 and RO1 AR53156
Appendix A
Here we provide some implementation details for the basic registration step. The efficiency of the overall algorithm depends on the step of searching for a match of a small grayscale 2D patch, A, within a larger grayscale 2D array, B, by choosing the best XY-translation. Indeed, this critical procedure is repeated for every point pi and for each trial rotation angle and each trial slice offset. The translational offset (Δx,Δy) is chosen to maximize the cross-correlation between A and the corresponding 2D sub-array of B. If A and B were the same size (and wrapping was allowed), then this could be achieved efficiently by implementing a 2D convolution via FFT. However, in our situation, A is generally much smaller than B. One approach to remedy this would be to simply pad B with zeros to match the size of A. However, this is not adequate when there are variations in signal level across B since the procedure will tend to select offsets that would place A over the region of highest intensity of B. Thus we need a normalizing factor in the denominator. Fortunately, this normalizing factor can be achieved by implementing a single additional discrete convolution. For simplicity, we give the details for one dimension, as the method can easily be extended to two dimensions.
Let a0, a1, …, aM-1 and b0, b1, …, bN-1 be two arrays with M<N. We want to find a translation Δx (integer) that maximizes the normalized correlation:
| (6) |
Here, we consider b to be a periodic array. The sum in the numerator can easily be written as a discrete convolution:
| (7) |
where we simply extend ai to be zero for i≥M. The sum in the denominator can also be written as a discrete convolution:
| (8) |
where and
| (9) |
.
References
- 1.Heaney RP. Is there a role for bone quality in fragility fractures? Calcified Tissue International. 1993;53(S1):S3–S6. doi: 10.1007/BF01673394. [DOI] [PubMed] [Google Scholar]
- 2.Kleerekoper M, Villanueva AR, Stanciu J, Sudhaker Rao D, Parfitt AM. The Role of Three-Dimensional Trabecular Microstructure in the Pathogenesis of Vertebral Compression Fractures. Calcified Tissue International. 1985;37:594–597. doi: 10.1007/BF02554913. [DOI] [PubMed] [Google Scholar]
- 3.Aaron JE, Shore PA, Shore RC, Beneton M, Kanis JA. Trabecular architecture in women and men of similar bone mass with and without vertebral fracture: II. Three-dimensional histology. Bone. 2000;27(2):277–282. doi: 10.1016/s8756-3282(00)00328-8. [DOI] [PubMed] [Google Scholar]
- 4.Recker RR. Architecture and vertebral fracture. Calcified Tissue International. 1993;53(Suppl 1):S139–S142. doi: 10.1007/BF01673423. [DOI] [PubMed] [Google Scholar]
- 5.Legrand E, Chappard D, Pascaretti C, et al. Trabecular bone microarchitecture, bone mineral density and vertebral fractures in male osteoporosis. Journal of Bone and Mineral Research. 2000;15:13–19. doi: 10.1359/jbmr.2000.15.1.13. [DOI] [PubMed] [Google Scholar]
- 6.Wahner HW, Fogelman I. The evaluation of osteoporosis: dual energy X-ray absorptiometry in clinical practice. Cambridge: University Press; 1994. p. 296. [Google Scholar]
- 7.Genant HK, Cann CE, Ettinger B, Gordan GS. Quantitative computed tomography of vertebral spongiosa: a sensitive method for detecting early bone loss after oophorectomy. Annals of Internal Medicine. 1982;97:699–705. doi: 10.7326/0003-4819-97-5-699. [DOI] [PubMed] [Google Scholar]
- 8.Dempster DW, Cosman F, Kurland ES, et al. Effects of daily treatment with parathyroid hormone on bone microarchitecture and turnover in patients with osteoporosis: a paired biopsy study. J Bone Miner Res. 2001;16(10):1846–1853. doi: 10.1359/jbmr.2001.16.10.1846. [DOI] [PubMed] [Google Scholar]
- 9.Majumdar S, Genant HK, Grampp S, et al. Correlation of trabecular bone structure with age, bone, mineral density, and osteoporotic status: in vivo studies in the distal radius using high-resolution magnetic resonance imaging. Journal of Bone and Mineral Research. 1997;12:111–118. doi: 10.1359/jbmr.1997.12.1.111. [DOI] [PubMed] [Google Scholar]
- 10.Wehrli FW, Hwang SN, Ma J, Song HK, Ford JC, Haddad JG. Cancellous bone volume and structure in the forearm: noninvasive assessment with MR microimaging and image processing. Radiology. 1998;206:347–357. doi: 10.1148/radiology.206.2.9457185. [published erratum appears in Radiology 1998 Jun;207(3):833] [DOI] [PubMed] [Google Scholar]
- 11.Boutroy S, Bouxsein ML, Munoz F, Delmas PD. In Vivo Assessment of Trabecular Bone Microarchitecture by High-Resolution Peripheral Quantitative Computed Tomography″. J Clin Endocrinol Metab. 2005;90(12):6805–6815. doi: 10.1210/jc.2005-1258. [DOI] [PubMed] [Google Scholar]
- 12.Wehrli FW, Song HK, Saha PK, Wright AC. Quantitative MRI for the assessment of bone structure and function. NMR Biomed. 2006;19(7):731–764. doi: 10.1002/nbm.1066. [DOI] [PubMed] [Google Scholar]
- 13.Wehrli FW. Structural and functional assessment of trabecular and cortical bone by micro magnetic resonance imaging. J Magn Reson Imaging. 2007;(2):390–409. doi: 10.1002/jmri.20807. [DOI] [PubMed] [Google Scholar]
- 14.Boutry N, Cortet B, Dubois P, Marchandise X, Cotten A. Trabecular bone structure of the calcaneus: preliminary in vivo MR imaging assessment in men with osteoporosis. Radiology. 2003;227(3):708–717. doi: 10.1148/radiol.2273020420. [DOI] [PubMed] [Google Scholar]
- 15.Majumdar S, Link TM, Augat P, et al. Trabecular bone architecture in the distal radius using magnetic resonance imaging in subjects with fractures of the proximal femur. Magnetic Resonance Science Center and Osteoporosis and Arthritis Research Group. Osteoporos Int. 1999;10(3):231–239. doi: 10.1007/s001980050221. [DOI] [PubMed] [Google Scholar]
- 16.Wehrli FW, Gomberg BR, Saha PK, Song HK, Hwang SN, Snyder PJ. Digital topological analysis of in vivo magnetic resonance microimages of trabecular bone reveals structural implications of osteoporosis. J Bone Miner Res. 2001;16(8):1520–1531. doi: 10.1359/jbmr.2001.16.8.1520. [DOI] [PubMed] [Google Scholar]
- 17.Ladinsky GA, Vasilic B, Popescu AM, et al. Trabecular Structure Quantified With the MRI-Based Virtual Bone Biopsy in Postmenopausal Women Contributes to Vertebral Deformity Burden Independent of Areal Vertebral BMD. J Bone Miner Res. 2008;23(1):64–74. doi: 10.1359/JBMR.070815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benito M, Vasilic B, Wehrli FW, et al. Effect of testosterone replacement on bone architecture in hypogonadal men. J Bone Miner Res. 2005;20(10):1785–1791. doi: 10.1359/JBMR.050606. [DOI] [PubMed] [Google Scholar]
- 19.Wehrli FW, Ladinsky GA, Jones C, et al. In Vivo Magnetic Resonance Detects Rapid Remodeling Changes in the Topology of the Trabecular Bone Network Following Menopause and Protective Effect of Estradiol. J Bone Miner Res. 2008 doi: 10.1359/JBMR.080108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chesnut CH, 3rd, Majumdar S, Newitt DC, et al. Effects of salmon calcitonin on trabecular microarchitecture as determined by magnetic resonance imaging: results from the QUEST study. J Bone Miner Res. 2005;20(9):1548–1561. doi: 10.1359/JBMR.050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajapakse CS, Magland JF, Wehrli FW. Fast prospective registration of in vivo MR images of trabecular bone micro-structure in longitudinal studies. Magn Reson Med. 2008 doi: 10.1002/mrm.21593. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blumenfeld J, Carballido-Gamio J, Krug R, Blezek DJ, Hancu I, Majumdar S. Automatic prospective registration of high-resolution trabecular bone images of the tibia. Ann Biomed Eng. 2007;35(11):1924–1931. doi: 10.1007/s10439-007-9365-z. [DOI] [PubMed] [Google Scholar]
- 23.Newitt DC, van Rietbergen B, Majumdar S. Processing and analysis of in vivo high-resolution MR images of trabecular bone for longitudinal studies: reproducibility of structural measures and micro-finite element analysis derived mechanical properties. Osteoporos Int. 2002;13(4):278–287. doi: 10.1007/s001980200027. [DOI] [PubMed] [Google Scholar]
- 24.Vasilic B, Ladinsky GA, Saha PK, Wehrli FW. San Diego, CA: International Society for Optical Engineering; 2006. Micro-MRI-based image acquisition and processing system for assessing the response to therapeutic intervention; p. 61430W. [Google Scholar]
- 25.Vasilic B, Popescu AM, Bunker B, Wehrli FW. Kyoto, Japan: ISMRM; 2004. Semi-automatic method for 3D registration of trabecular bone images in serial studies; p. 2209. [Google Scholar]
- 26.Takao M, Sugano N, Nishii T, et al. Application of 3D-MR image registration to monitor diseases around the knee joint. J Magn Reson Imaging. 2005;22(5):656–660. doi: 10.1002/jmri.20435. [DOI] [PubMed] [Google Scholar]
- 27.Magland JF, Vasilic B, Wehrli FW. Berlin, Germany: ISMRM; 2007. Fully Automated Region of Interest Segmentation in the Virtual Bone Biopsy; p. 2629. [Google Scholar]
- 28.Wehrli FW, Saha PK, Gomberg BR, Song HK. Noninvasive assessment of bone architecture by magnetic resonance micro-imaging-based virtual bone biopsy. Proc IEEE. 2003;91:1520–1542. [Google Scholar]
- 29.Rubin C, Judex S, Qin YX. Low-level mechanical signals and their potential as a non-pharmacological intervention for osteoporosis. Age Ageing. 2006;35(Suppl 2):ii32–ii36. doi: 10.1093/ageing/afl082. [DOI] [PubMed] [Google Scholar]
- 30.Magland J, Jones C, Wald M, Wehrli FW. Berlin, Germany: ISMRM; 2007. Spin-Echo Micro-MRI of Trabecular Bone Using an Improved 3D FLASE Sequence; p. 382. [Google Scholar]
- 31.Ma J, Wehrli FW, Song HK. Fast 3D large-angle spin-echo imaging (3D FLASE) Magnetic Resonance in Medicine. 1996;35:903–910. doi: 10.1002/mrm.1910350619. [DOI] [PubMed] [Google Scholar]
- 32.Gomberg BG, Saha PK, Song HK, Hwang SN, Wehrli FW. Application of topological analysis to magnetic resonance images of human trabecular bone. IEEE Transactions on Medical Imaging. 2000;19:166–174. doi: 10.1109/42.845175. [DOI] [PubMed] [Google Scholar]
- 33.Stauber M, Rapillard L, van Lenthe GH, Zysset P, Muller R. Importance of individual rods and plates in the assessment of bone quality and their contribution to bone stiffness. J Bone Miner Res. 2006;21(4):586–595. doi: 10.1359/jbmr.060102. [DOI] [PubMed] [Google Scholar]
- 34.Liu XS, Sajda P, Saha PK, Wehrli FW, Guo XE. Quantification of the roles of trabecular microarchitecture and trabecular type in determining the elastic modulus of human trabecular bone. J Bone Miner Res. 2006;21(10):1608–1617. doi: 10.1359/jbmr.060716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shrout PE, Fleiss JL. Intraclass correlations: Uses in assessing rater reliability. Psychological Bulletin. 1979;86(2):420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- 36.Gomberg BR, Wehrli FW, Vasilic B, et al. Reproducibility and error sources of micro-MRI-based trabecular bone structural parameters of the distal radius and tibia. Bone. 2004;35(1):266–276. doi: 10.1016/j.bone.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 37.Lin W, Ladinsky GA, Wehrli F, Song HK. Image metric-based correction (autofocusing) of motion artifacts in high-resolution trabecular bone imaging. J Magn Reson Imaging. 2007;26:191–197. doi: 10.1002/jmri.20958. [DOI] [PubMed] [Google Scholar]
- 38.Recker R, Lappe J, Davies KM, Heaney R. Bone remodeling increases substantially in the years after menopause and remains increased in older osteoporosis patients. J Bone Miner Res. 2004;19(10):1628–1633. doi: 10.1359/JBMR.040710. [DOI] [PubMed] [Google Scholar]