Abstract
Neuroendocrine chromaffin granules of adrenal medulla represent regulated secretory vesicles that secrete neuropeptides and catecholamines which mediate cell-cell communication for physiological functions. This study addressed the identification of the major proteins in these secretory vesicles that provide dynamic storage and secretion of bioactive molecules. Proteins of the soluble compartment of the vesicles were separated by 2-dimensional gels and subjected to NH2-terminal Edman sequencing for identification and determination of NH2-termini. Results showed that proteolytic fragments of chromogranin A (CgA) and chromogranin B (CgB) represent the major proteins of these secretory vesicles. These fragments resulted from cleavage of their respective precursor proteins at the COOH-terminal side of dibasic and monobasic sites, which is consistent with the known cleavage specificities of prohormone processing enzymes. MALDI-TOF MS analyses of protein spots of similar molecular weight that possessed a range of pI values were represented by molecular forms of CgA and CgB proteins. These findings indicate the high prevalence of endogenous CgA and CgB proteolytic fragments that function in chromaffin secretory vesicles for release of bioactive molecules for cell-cell communication.
Chromaffin granules of sympothoadrenal medullary chromaffin cells represent secretory vesicles that synthesize, store, and secrete biologically active molecules consisting of neuropeptides and catecholamines (1–10) for neuroendocrine cell-cell communication. These biomolecules mediate cell-cell communication among multiple physiological systems including those that participate in stress responses (11–13), and in hormonal function including cardiovascular regulation (14,15).
Numerous neuropeptides are secreted from chromaffin cells that include the enkephalin opioid peptides, NPY (neuropeptide Y), galanin, VIP (vasoactive intestinal peptide), somatostatin, and others (1–5). These neuropeptides are produced from their respective proneuropeptide precursors by proteolytic processing enzymes that cleave at dibasic and monobasic sites (16–18). The smaller, active neuropeptides are stored in these secretory vesicles for regulated secretion. These neuropeptides are cosecreted with the catecholamines epinephrine, norepinephrine, and dopamine. Nicotinic cholinergic receptors stimulate the release of these bioactive molecules from chromaffin cells for regulation of neuroendocrine functions.
These secretory vesicles utilize numerous proteins to produce bioactive molecules for cell-cell communication that include secretory vesicle maturation, proteolysis of protein precursors for neuropeptide production, trafficking and fusion of vesicles to the plasma membrane for exocytosis. The soluble components of the secretory vesicle are especially important since they are released to the extracellular environment upon regulated secretion, resulting in cell-cell communication by secreted bioactive molecules. The secretion of these soluble neurohumoral components allow chromaffin cells to communicate with various target cells to coordinate physiological functions. Knowledge of the major proteins in the soluble compartment of chromaffin granules can provide insight into primary protein functions utilized in this secretory vesicle organelle. For this reason, the goal of this study was to identify the major proteins in the soluble compartment of chromaffin granules.
This study analyzed soluble proteins of purified chromaffin granules by separation on 2-dimensional gels with NH2-terminal Edman peptide sequencing. Results demonstrated that nearly all the proteins were identified as proteolytic fragments of chromogranin A (CgA) and chromogranin B (CgB). Moreover, all fragments were derived from parent precursor proteins by proteolytic processing at the COOH-terminal sides of dibasic and monobasic cleavage sites, consisent with prohormone processing sites. Complementary analyses of multiple 2-D gel spots by MALDI-TOF mass spectrometry of tryptic digests of proteins indicated the nature of numerous proteolytic fragments of CgA and CgB in chromaffin granules. These findings indicate the importance of specific proteolytic processing at basic residues of CgA and CgB proteins to generate numerous CgA- and CgB-derived fragments for chromaffin granule functions in mediating cell-cell communication among neuroendocrine systems.
Experimental Procedures
Preparation of chromaffin granule proteins from bovine adrenal medulla
Chromaffin granules were purified from fresh bovine adrenal medulla by sucrose density centrifugation, as described previously (19). The soluble fraction of chromaffin granules was prepared by lysis (by freeze-thawing) in the presence of a cocktail of protease inhibitors consisting of 1 mM EDTA, 10 µM pepstatin A, 10 µm leupeptin, 10 µm chymostatin, 10 µm E64c, and 1 mM AEBSF (4-(2-aminoethyl)benzene-sulfonylfluoride hydrochloride) in isotonic buffer (50 mM sodium acetate, pH 6.0, 150 mM NaCl). Samples were subjected to ultracentrifugation (Beckman L7–65 ultracentrifuge) at 100,000 × g in a SW60 rotor for 30 minutes at 4°C, and the resultant supernatant was collected as the soluble granule fraction, as we have described previously (19, 20). Protein concentrations were determined using Bradford assays (BioRad).
Two-dimensional gel electrophoresis of proteins
Chromaffin granule proteins (300 µg protein sample in 310 µl rehydration buffer) were separated by isoelectric focusing (IEF) followed by SDS-PAGE. IEF was conducted at pH 3–6 on 17 cm IPG strips (BioRad, CA) according to the manufacturer's protocols. Rehydration buffer for IPG strips consisted of 8 M urea, 2% CHAPS, 0.2% Biolytes 3/10, 2 mM TCEP (Pierce), and a pinch of bromophenol blue. Following the rehydration of IPG strips in Protean IEF cell (BioRad, CA) under active condition (50 Volt per strip) for 16 hrs at 15° C, proteins were subjected to IEF in steps of 250 V for 30 min, 10,000 V for 3 hrs, 50,000 volt hours, and 500 V. After IEF, the strips were equilibrated in buffer I (6 M urea, 2% SDS, 0.375 M Tris pH 8.8, 20% glycerol, and 10 mM TCEP) for 2 × 5 minutes on a shaker, and then in buffer II (6 M urea, 2% SDS, 0.375 M Tris pH 8.8, 20% glycerol, 2.5% iodoacetamide, and trace of bromophenol blue) for 2 × 5 minutes. The strips were then briefly placed in electrophoresis running buffer and blotted onto filter paper.
For SDS-PAGE, the IPG strips were cut to fit the top of a SDS-PAGE gel (1 mm thick, 6% SDS stacking layer and 12% SDS separation layer) on a Hoefer electrophoresis system. The strips in the sample wells were overlayed with 0.5% agarose solution in running buffer (25 mM Tris, 192 mM glycine, pH 8.3, 0.05% SDS). After 10 min, the gels were electrophoresed at constant current of 20 mA (stacking layer) and 25 mA (resolving gel) per gel.
Protein Analyses by NH2-terminal Edman Sequencing
The 2-D gels were equilibrated in semi-dry transfer buffer (48 mM Tris, 39 mM Gly, pH 9.2, 10% MeOH) for 15 min, followed by transfer of proteins to PVDF membranes at 20 volt for 1 hr according to the manufacturer's protocol (Sequi-Blot, BioRad, CA). The membranes were stained with amido black (0.1% in 10% acetic acid) for 2 minutes and destained in 5% acetic acid for 1 hr. Selected protein spots were excised and washed with distilled water five times, and air-dried before subjecting them to NH2-terminal peptide sequencing by Edman degradation on an Applied Biosystems Procise 494 protein sequencer at the Harvard Microchemistry and Proteomics Analysis Facility. (It is noted that the number of proteins subjected to Edman sequencing was limited by the cost of service fees for sequencing.) Protein sequences of most recent updates were retrieved from NCBI non-redundant protein databases for sequence analyses. NH2-terminal sequences determined by Edman degradation were queried to find matching proteins by using BLAST Alignment tool from NCBI. The BLAST tool was also used to align bovine proSAAS precursor sequence with that of human sequence to predict the signal sequence of the bovine protein.
Analyses of proteins from 2-D gels by MALDI-TOF mass spectrometry
Protein spots from the 2-D gels were subjected to analyses by MALDI-TOF mass spectrometry of trypsin digests of each spot to gain further information about the proteins. MALDI-TOF mass spectrometry of trypsin digests was performed as described previously (20). Briefly, tryptic digestion was performed by the automated robot digester ProGest (from Genomic Solutions) which provides a series of programmed steps to deliver and remove solutions via nitrogen pressure through perforated bottoms of 96 well plates. Gel pieces (~1.5 mm2) in 96-well plate were washed and destained with 2 cycles of 50 µl of NH4HCO3 (25 mM) and 50 µl of ACN. The proteins in gel pieces were then reduced with 40 µl DTT (10 mM) in NH4HCO3 (25 mM) for 10 min at 60 °C, followed by alkylation with 30 µl of iodoacetamide (100 mM) in NH4HCO3 (50 mM) for 45 min at room temperature. Gel pieces were then dehydrated with 2 cycles of 50 µl of NH4HCO3 (25 mM) and 50 µl of ACN prior to trypsin digestion. Trypsin digestion utilized 10 µl of sequence grade trypsin (250 µg, Promega) dissolved in 1 mM acetic acid mixed with 15 µl of NH4HCO3 (25 mM), and incubated at 37° C for 4 hrs, and stopped by the addition of 7 µl 10% formic acid. Peptides were collected by pressurizing with nitrogen into a collection plate (96-well).
For MALDI-TOF mass spectrometry, each protein digest sample (0.5 µl) was mixed with α-CHCA matrix (α-cyano-4-hydroxycinnamic acid, 0.5 µl) (Agilent Technologies, Inc.) and spotted onto MALDI target and air-dried. The mass spectra were acquired on a PE Biosystems Voyager DeSTR MALDI-TOF mass spectrometer with nitrogen laser, operating in delayed extraction and reflectron mode (20). The spectral analyses were made with internal calibration to masses of trypsin autolysis or external peptide peaks (bradykinin, ACTH fragment 18–39, and angiotensin I). The resulting peptide mass fingerprints were searched against the NCBI protein database using the Profound search engine within RADARS (Proteometrics, New York, NY) (21), and by MS-Fit (http://prospector.ucsf.edu) (22,23). In ProFound, “Expectation Value” (EV) scoring was used, which indicates the quality or significance of the match (Field, HI). EV scoring of less than 5 × 102 is equivalent to > 95% confidence and was considered significant. In MS-Fit, the “MOWSE” scoring method, a probability-based scoring system, was used to evaluate the protein match quality and significant identifications (22). A score greater than 70 is considered to represent significant matches. Data Explorer software (Applied Biosystems) was also used for generating peptide mass lists.
Results
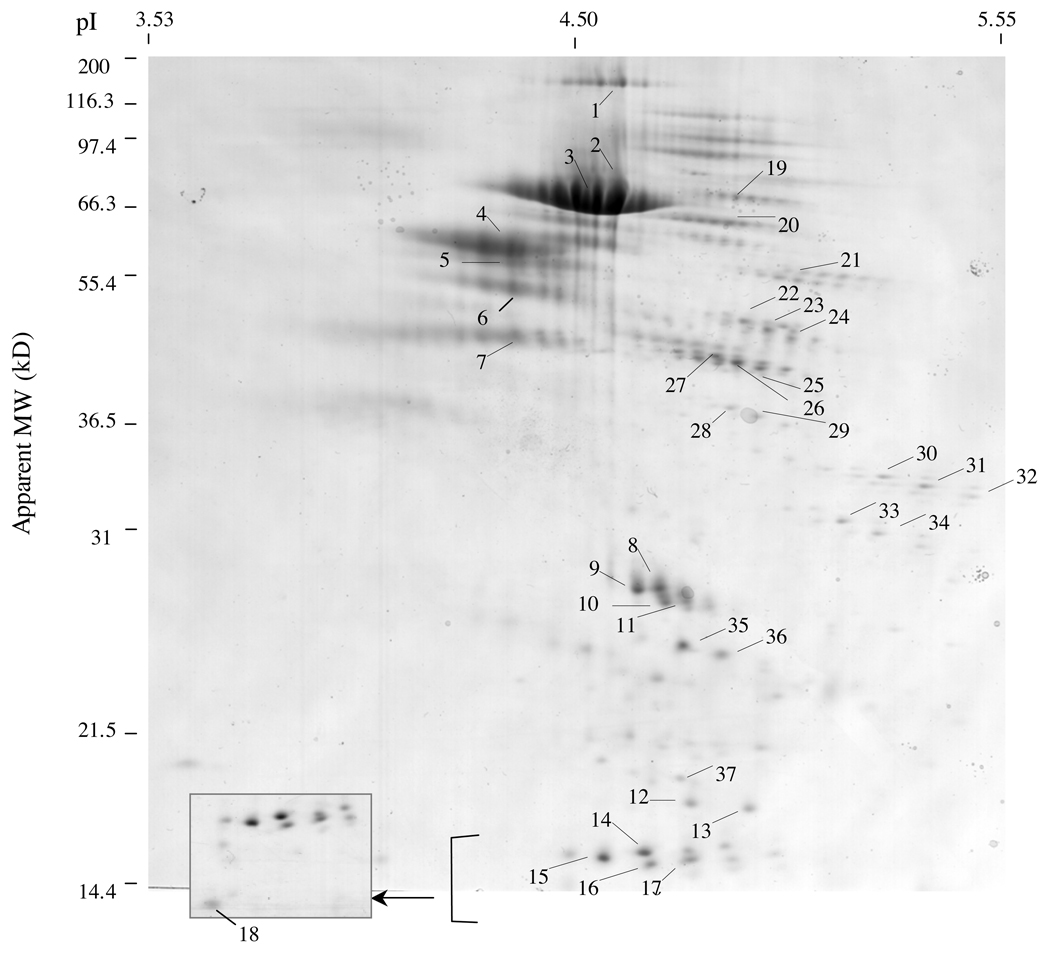
Chromaffin granule soluble proteins were subjected to 2-D electrophoresis by IEF (isoelectric focusing) and SDS-PAGE, followed by electrophoretic transfer to PVDF membranes stained with amido black to visualize proteins. Multiple proteins ranging from greater than 100 kDa to approximately 10 kDa were separated by the 2-D gel (figure 1). The major amido-black stained proteins were excised for Edman sequencing, indicated by numbers #1–37. Data for NH2-terminal sequences were grouped by protein identification information.
Figure 1. Two-dimensional gel separation of chromaffin granule proteins for N-terminal peptide sequencing.
Chromaffin granule proteins of the soluble fraction were subjected to separation by 2-D gels achieved by isoelectric focusing (first dimension) and SDS-PAGE (second dimension). Proteins were transferred to PVDF membranes and stained with amido black to visualize proteins. Selected protein spots were excised, subjected to NH2-terminal sequencing by Edman degradation, and were numbered #1–37. The inset (boxed) is provided to more clearly illustrate the indciateds group of proteins.
Chromogranin A-derived proteolytic fragments
NH2-terminal peptide sequences indicated that the majority of proteins represented different molecular weight forms of chromogranin A (CgA) and chromogranin B (CgB). CgA-related proteins were identified for protein spots #1–18, and #36–37 (Table 1). Data for determined NH2-terminal peptide sequences, apparent molecular weight of protein spots on SDS-PAGE, apparent pI, and protein identification were compiled (Table 1). Some protein spots consisted of more than one peptide sequence (spots #4–6, and #36 and 37), as noted by primary and secondary sequences (1° and 2°) obtained from Edman sequencing of CgA (spots #4–6, 36)) and proSAAS (spot #37).
Table 1. Identification of CgA and ProSAAS Protein Forms by N-Terminal Peptide Sequencing.
Protein spots from 2-D gel separation of the soluble fraction of purified chromaffin granules were transferred to PVDF membranes, and membrane regions corresponding to designated protein spots were excised for N-terminal peptide sequencing by Edman degradation. The determined N-terminal amino acid sequences for each protein spot is indicated. If more than one protein sequence was identified, their deduced primary sequences are indicated. The properties of each protein spot are illustrated for apparent molecular weight (on 2-D gels), apparent isoelectric point (pI), identification of the N-terminal sequences as CgA (or ProSAAS for spot #37), and relative protein staining by amido black on PVDF membranes of the 2-D gel proteins.
| Spot #s | N-terminal Sequences by Edman Degradation |
Ratios | Apparent MW range (kD) |
Apparent pI | CgA/ Fragment ID | Relative Protein Staining |
|---|---|---|---|---|---|---|
| 1 | LPVNSPMNKG | 153 | 4.65 | CgA | Low | |
| 2,3 | LPVNSPMNKG | 66 | 4.55 – 4.62 | CgA | Very High | |
| 4 | 1°: HSSYEDELSE 2°: LPVNSPMNK |
4 to 1 | 59 | 4.39 | CgA | Medium |
| 5 | 1°: HSSYEDELSE 2°: LPVNSPMNKG |
1 to 1 | 56 | 4.39 | CgA | Low |
| 6 | 1°: DDFKEVEKSD 2°: LPVNSPMNKG |
3 to 1 | 50 | 4.39 | CgA | Medium to Low |
| 7 | LPVNSPMNKG | 45 | 4.39 | CgA | Medium to Low | |
| 8,9,10,11 | AAPGWPEDGA | 27 – 28 | 4.68 – 4.79 | CgA | Medium | |
| 12 | LPVNSPMNKG | 16 | 4.81 | CgA | Medium to Low | |
| 13 | LPVNSPMNKG | 15.5 | 4.95 | CgA | Medium to Low | |
| 14,15,16,17 | LEGEEEEEED | 14.5 – 15 | 4.60 – 4.81 | CgA | Medium | |
| 18 | GWRPNNNRED | Below 14.4 | 4.47 | CgA | Medium to Low | |
| 36 | 1°: ERGEVG_EER 2°: LPVNSPMNKG |
ND | 24 | 4.88 | CgB & CgA |
Low |
| 37 | 1°: LPVNSPMNKG 2°: AAPRGEAAGA |
1 to 1 | 18 | 4.79 | CgA & ProSAAS (PC1 inhibitor) |
Low Low |
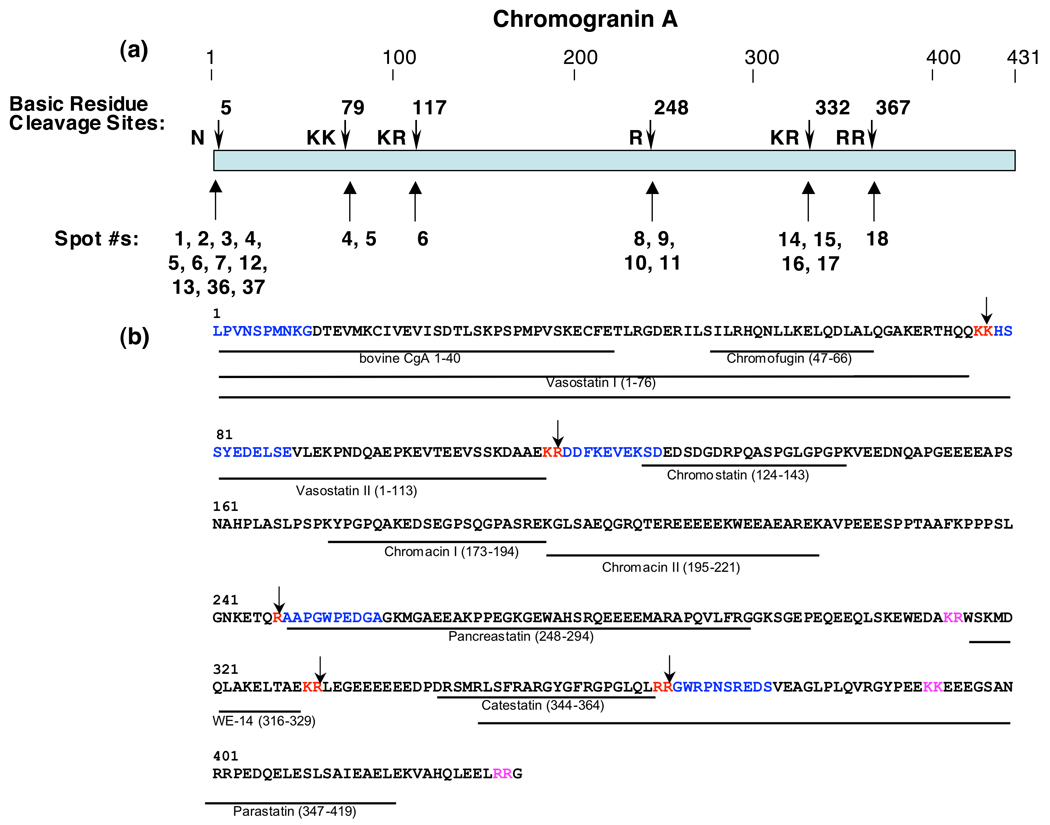
Importantly, analyses of the NH2-terminal sequences within the full-length primary sequence of the CgA protein illustrated its cleavage dibasic and monobasic cleavage sites that generated the NH2-termini of the isolated CgA fragments (figure 2). The CgA fragments were generated by cleavages at the COOH-terminal sides of dibasic KK, KR, and RR sites, as well as at the COOH-terminal side of a monobasic Arg site (figure 2a). The determined NH2-terminal peptide sequences (by Edman sequencing) at these cleavage sites within the full-length CgA protein are shown in blue in figure 2b. Thus, proteolytic processing at dibasic and monobasic cleavage sites generates multiple CgA fragments that represent abundant proteins in chromaffin secretory vesicles.
Figure 2. Cleavage sites of full-length CgA that generate proteolytic fragments in chromaffin granules.
a. Cleavage sites utilized to generate NH2-termini of CgA proteins in chromaffin granules. Cleavage sites of CgA representing NH2-termini of CgA fragments are shown by the arrows. The protein spots (numbered as in figure 1) whose NH2-termini correspond to the indicated cleavage sites are shown below the cleavage sites (shown by arrows).
b. Cleavage sites and biologically active peptides within full-length CgA. Cleavage sites utilized to generate CgA fragments in chromaffin granules are illustrated by the arrows at dibasic or monobasic residue sites shown in red font. NH2-terminal sequence determined by Edman degradation of CgA fragments are illustrated in blue color font, within the full-length CgA primary sequence; dibasic residues at cleavage sites are shown in red font. Biologically active peptides derived from CgA are shown as lines with peptide names under the bovine CgA primary sequence (NCBI Protein Database accession number P05059 (30)); cleavage sites predicted to generate these active fragments that were not identified in this study are shown in pink font.
Chromogranin B proteolytic fragments
Edman sequencing of multiple proteins revealed the identities of protein spots #19–36 (18 spots) as chromogranin B (CgB) fragments (Table 2). These CgB fragments ranged in apparent molecular weight (on SDS-PAGE) of approximately 69 to 24 kDa. Several spots (#27 and #36) showed primary and secondary peptide sequences obtained from Edman sequencing for CgB (spot #27), as well as for CgB and CgA (spot #36).
Table 2. Identification of CgB Protein Forms by N-Terminal Peptide Sequencing.
Protein spots from 2-D gel separation of the soluble fraction of purified chromaffin granules were transferred to PVDF membranes, and membrane regions corresponding to designated protein spots were excised for N-terminal peptide sequencing by Edman degradation. The determined N-terminal amino acid sequences for each protein spot is indicated. If more than one protein sequence was identified, their deduced primary sequences are indicated. The properties of each protein spot are illustrated for apparent molecular weight (on 2-D gels), apparent isoelectric point (pI), identification of the N-terminal sequences, and relative protein staining by amido black on PVDF membranes of the 2-D gel proteins.
| Spot #s | N-terminal Sequences by Edman Degradation |
Ratios | Apparent MW range |
Apparent pI | Prohormones/ Fragment ID |
Relative Protein Staining |
|---|---|---|---|---|---|---|
| 19 | ERGEVV_ _ EE | 69 | 4.92 | [CgB] | Low | |
| 20 | ERGEVGGEEE | 63 | 4.76 | CgB | Low | |
| 21 | ERGEVGREER | 53 | 5.06 | CgB | Low | |
| 22,23,24 | SSQEGNPPLE | 45 – 47 | 4.94 – 5.05 | CgB | Low | |
| 25,26 | SSQEGNPPLE | 41.5 – 41.8 | 4.91 – 4.97 | CgB | Low to Medium | |
| 27 | 1°: SSQEGNPPLE 2°: M(Q)PVDIR_ H_E |
5 to 1 | 42 | 4.86 | CgB | Low to Medium |
| 28,29 | ALEEGAEYGE | 36 −37 | 4.90 – 4.96 | CgB | Low | |
| 30,31,32 | SSQEGNPPLE | 32 −33 | 5.26 – 5.46 | CgB | Low | |
| 33,34 | SSQEGNPPLE | 30.5 – 31 | 5.16 – 5.24 | CgB | Low | |
| 35 | ERGEVG[S*]EER | 24.5 | 4.79 | CgB | Low to Medium | |
| 36 | 1°: ERGEVG_EER 2°: LPVNSPMNKG |
ND | 24 | 4.88 | CgB & CgA |
Low |
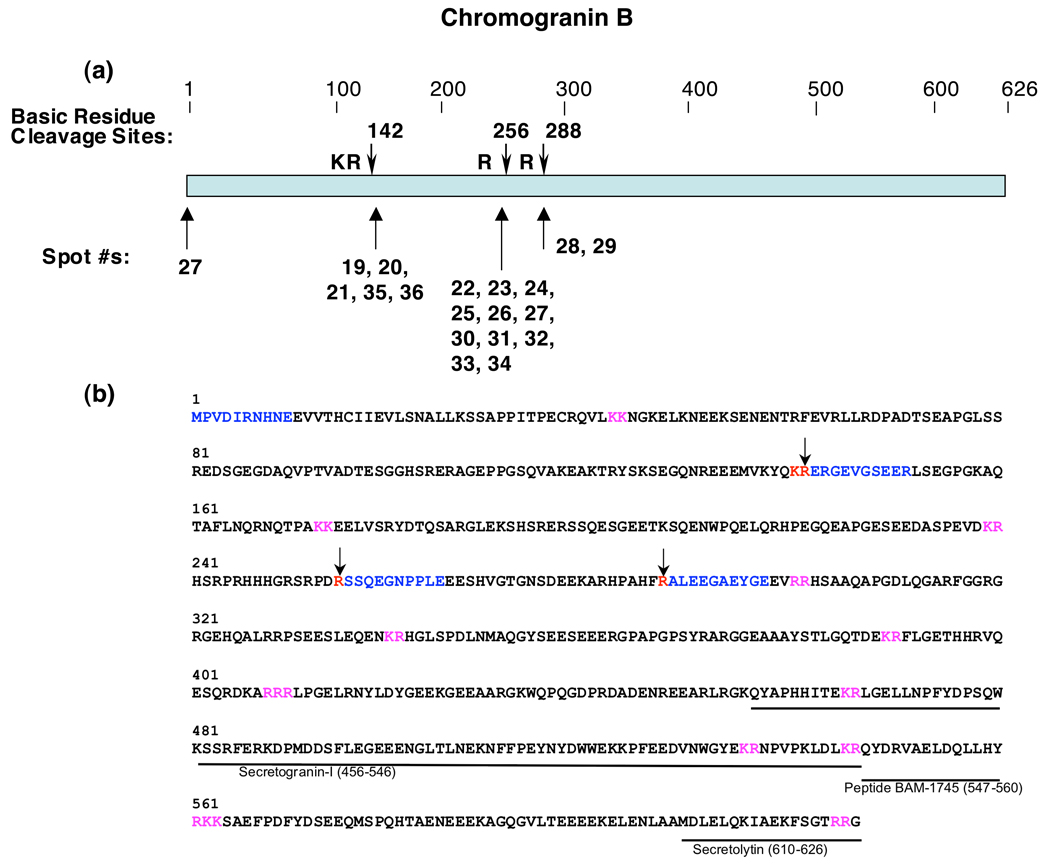
Mapping of the NH2-terminal sequences of CgB fragments showed that they resulted from proteolytic cleavages of CgB at dibasic and monobasic residue sites (figure 3). These fragments resulted from cleavage of CgB at the COOH-terminal sides of dibasic KR and monobasic Arg sites (figure 3a). Analyses of Edman sequencing results also indicated that one of the fragments (spot #27) contains the NH2-terminus of intact CgB (figure 3b). Interestingly, a larger number of CgB fragments (10 fragments) resulted from cleavage at a monobasic Arg site, compared to those resulting from cleavage at the dibasic KR site (5 fragments). These findings demonstrate that CgB undergoes specific proteolytic processing at dibasic and monobasic residue sites to generate multiple fragments present in chromaffin secretory vesicles.
Figure 3. Cleavage sites of full-length CgB that generate proteolytic fragments in chromaffin granules.
a. Cleavage sites utilized to generate NH2-termini of CgB proteins in chromaffin granules. Cleavage sites of CgB representing NH2-termini of CgB fragments are shown by the arrows. The protein spots (numbered as in figure 1) whose NH2-termini correspond to the indicated cleavage sites are shown below the cleavage sites (shown by arrows).
b. Cleavage sites and biologically active peptides within full-length CgB. Cleavage sites utilized to generate CgB fragments in chromaffin granules are illustrated by the arrows at dibasic or monobasic residue sites shown in red font. NH2-terminal sequences, determined by Edman degradation, of CgB fragments are illustrated in blue color font within the full-length CgB primary sequence; dibasic residues at cleavage sites are shown in red font. Biologically active peptides derived from CgB are shown as lines with peptide names under the bovine CgB primary sequence (NCBI Protein Database accession number P23389 (30)); cleavage sites predicted to generate these active fragments that were not identified in this study are shown in pink font.
ProSAAS protein fragment
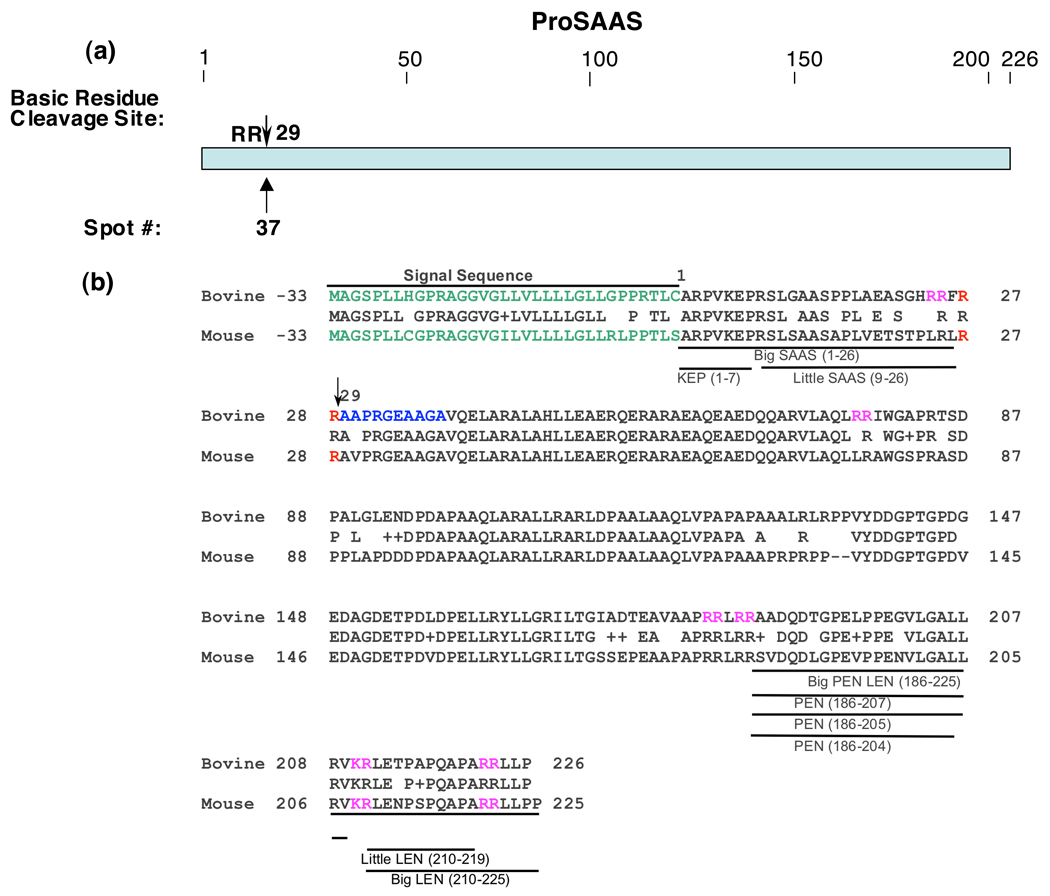
One protein was identified as a proteolytic fragment of proSAAS, which undergoes proteolytic processing to generate neuropeptides, as well as an endogenous inhibitor of prohormone convertase 1 (PC1/3) (spot #37, Table 1). Its NH2-terminal sequence indicated that this fragment resulted from cleavage at the COOH-terminal side of a RR dibasic site of proSAAS (figure 4). Because the majority of proSAAS-derived peptides have been defined in mouse species, and not in bovine, alignment of bovine proSAAS sequence with mouse proSAAS is provided to illustrate PEN, LEN, and related peptide products. Although a proteolytic fragment of proSAAS was identified, the results of this study show that fragments of CgA and CgB compose the major protein components in the soluble fraction of chromaffin secretory vesicles.
Figure 4. Cleavage site of proSAAS proteolytic fragment.
a. NH2-terminal cleavage site of proSAAS fragment. The Arg-Arg cleavage site utilized to generate the proSAAS fragment isolated by 2-D gels of chromaffin granules is illustrated by the arrow.
b. ProSAAS-derived peptides and cleavage sites. The arrow at residue #29 indicates the cleavage site of proSAAS determined by NH2-terminal Edman sequencing from its protein spot obtained from the 2-D gel (figure 1). The bovine sequence for proSAAS is aligned with the mouse proSAAS sequence, with active mouse peptides indicated. Since proSAAS-derived peptides have been characterized in mouse tissues (rather than bovine tissues), the proSAAS peptide fragments are shown for mouse proSAAS (NCBI Protein Database accession number Q9QXV0 (39–41)) and aligned to the bovine proSAAS sequence (accession number NP001077149 ).
Further analyses of 2-D gel proteins by MALDI-TOF mass spectrometry
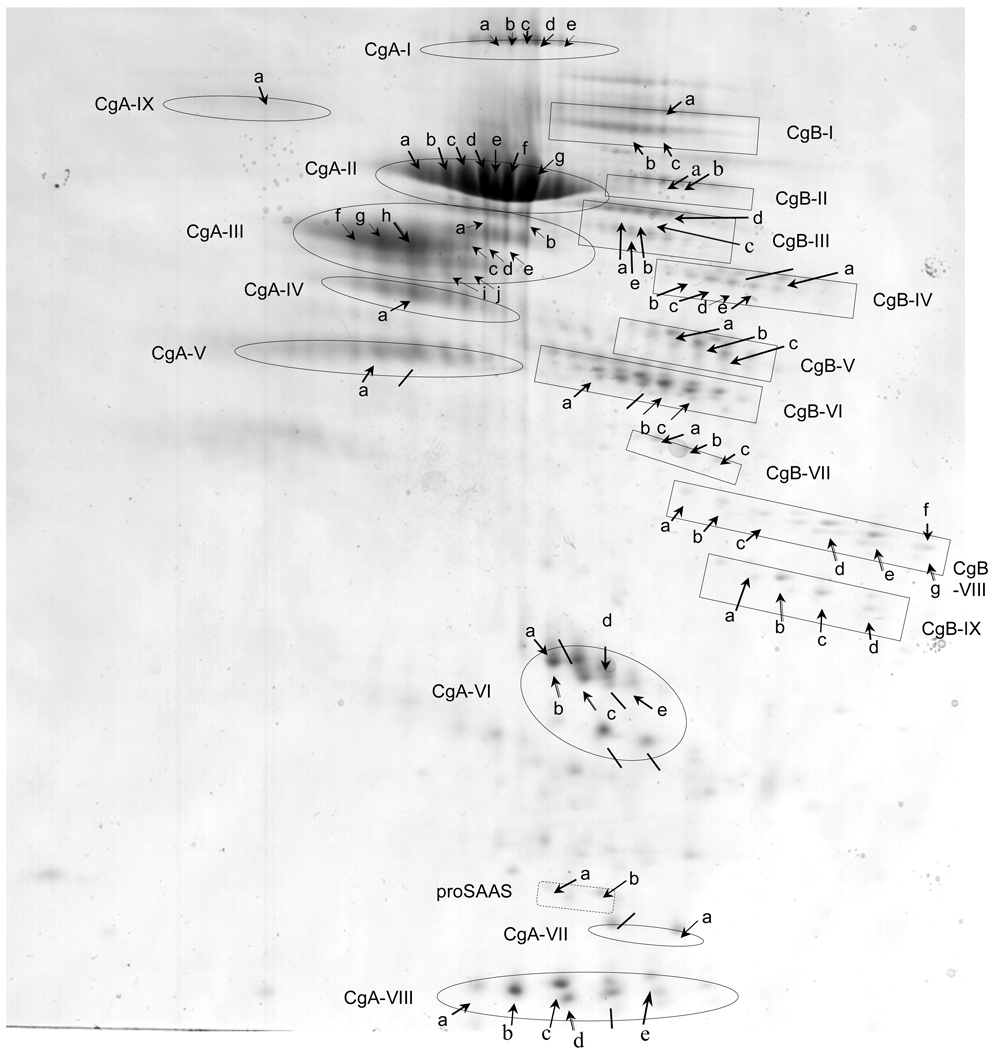
Analyses of the majority of the spots were also conducted by MALDI-TOF mass spectrometry. MALDI-TOF was used as further data supporting the identity of proteins of from the soluble component of chromaffin granules, combined with analyses by N-terminal Edman sequencing. Protein spots subjected to MALDI-TOF analyses, after tryptic digestion, are illustrated by groups of proteins representing forms of CgA or CgB (as well as proSAAS), and individual proteins designated by letters within each group (figure 5). Masses of tryptic peptides were found to represent sequences from CgA and CgB (supplemental Table A and supplemental Table B, respectively). These data indicated that groups of proteins with similar apparent molecular weight, yet with different pIs, represented multiple forms of CgA and CgB (illustrated in figure 5). Proteins that corresponded to CgA are indicated as groups of CgA-I to CgA-IX. Similarly, protein spots corresponding to CgB are indicated as groups CgB-1 to CgB-IX. Additionally, two protein forms of proSAAS (a and b) were identified (supplemental Table C). The combined NH2-terminal sequencing results and MALDI-TOF mass spectrometry data demonstrate that proteolytic fragments of CgA and CgB represent the abundant protein components of chromaffin granules.
Figure 5. Series of chromaffin granule protein spots from 2-D gels subjected to analyses by MALDI-TOF mass spectrometry.
Two-dimensional gels of the soluble fraction of purified chromaffin granules yielded numerous protein spots (indicated by arrows) which were subjected to analyses by MALDI-TOF mass spectrometry after excision and trypsin digestion. Proteins that were identified by both MALDI-TOF MS and N-terminal sequencing are listed: CgA-I-(d), CgA-II-(e, g), CgA-III-(h), CgA-IV-(a), CgA-VI-(b, c), CgA-VII-(a), CgA-VIII-(b, c, d), CgB-III-(c), CgB-VI-(b, c), CgB-VIII-(d, e, g), CgB-IX-(b, c), proSAAS-(b). Also, proteins that were identified by N-terminal sequencing only, are indicated by lines (from fig. 1). MALDI-TOF results (see supplemental Table A, supplemental Table B, and supplemental Table C) showed that most of these spots represented CgA and CgB, while two minor spots represented proSAAS (supplemental Table C). Proteins of similar apparent molecular weight but possessing a range of isoelectric points (pI) are grouped, and indicated as proteins groups CgA-I to CgA-IX, and CgB-I to CgB-IX, and proSAAS (a,b). These data demonstrate the preponderance of CgA- and CgB-derived proteolytic fragments in the soluble fraction of chromaffin granules.
Discussion
Analyses of the major proteins in chromaffin granules by 2-D gels and NH2-terminal Edman peptide sequencing revealed that proteolytic fragments of CgA and CgB represent abundant proteins in this dense core secretory vesicle organelle. Notably, the NH2-terminal sequences of the identified proteins indicated that they were generated by proteolytic processing of full-length CgA or CgB at the COOH-terminal sides of dibasic and monobasic residues. These results demonstrate that the major chromaffin granule proteins, CgA and CgB, undergo specific proteolytic processing at designated basic residue cleavage sites that generates numerous fragments ranging in apparent molecular weight of approximately 70 to 10 kDa. It is significant that among the proteins analyzed, no other type of protease cleavage site was observed. These findings suggest that specific proteolytic processing at dibasic and monobasic residues of CgA and CgB are required for chromaffin granule functions.
The distinct specificity for proteolytic cleavage at the COOH-terminal sides of dibasic and monobasic residue sites of CgA and CgB is consistent with the known cleavage specificity of prohormone convertases 1 and 2 (PC1/3 and PC2) that reside within the chromaffin granules (16–18). The determined cleavage sites for proteolytic fragments of CgA and CgB suggest PC1/3 and PC2 as key processing enzymes of CgA and CgB in these secretory vesicles. Cleavage at the COOH-terminal sides of these dibasic and monobasic residues is typically followed by removal of the COOH-terminal basic residues by carboxypeptidase E (CPE, also known as CPH) which is present in chromaffin granules (16, 24–28). Utilization of only dibasic and monobasic residue cleavage sites of the major proteins of chromaffin granules suggest specific and ordered proteolytic mechanisms for posttranslational modification of a major portion of proteins in this secretory vesicle organelle.
The cleavage sites identified for CgA correspond to processing sites required for producing biologically active peptides derived from CgA, including catestatin that regulates blood pressure (20, 29). The 2-D Edman sequencing results show cleavage at the COOH-terminus of vasostatin (30), at the NH2-terminus of pancreastatin (30), at the COOH-terminus of the WE-14 peptide (31), and at the COOH-terminus of catestatin (figure 2b) (29,30). Thus, it is likely that the CgA fragments detected on the 2-D gels may represent intermediates leading to production of CgA-derived biologically active peptides. It is also possible that the identified CgA fragments themselves participate in chromaffin granule mediation of biological functions.
CgB undergoes proteolytic processing to generate the active peptides BAM-1745 (32) and secretolytin (33,34) that are located within the COOH-domain of CgB (30). The observed cleavage sites in the midregions of CgB in this study suggest that those processing sites may be involved in generating BAM-1745 or secretolytin. Alternatively, the CgB fragments identified from the 2-D gels may themselves have functional properties in the chromaffin granules.
Further analyses of 2-D protein spots by MALDI-TOF mass spectrometry of trypsin digests provided data in support of the multiple protein forms of CgA and CgB. CgA was found to be represented by nine groups of protein spots, designated CgA-I to CgA-IX, that each were of similar apparent molecular weight, yet differed in pI values. Likewise, CgB was also found to be present in the 2-D gels as groups of proteins designated CgB-I to CgB-IX, with each group consisting of proteins of similar apparent molecular weights that show a range of varying relative charge properties illustrated by their differing isoelectric points.
In addition to proteolytic processing of CgA to generate biologically active peptides, including catestatin that participates in the regulation of blood pressure, CgA regulates the biogenesis of dense-core secretory granules (35–37). Downregulation of CgA in the neuroendocrine PC12 cell line by antisense RNAs resulted in loss of secretory granules and impairment of regulated secretion of peptide hormones. Alternatively, overexpression of CgA induced formation of dense core secretory granules. Based on results from this study, the presence of proteolytic fragments of CgA in chromaffin granules raises the interesting question of possible roles for CgA polypeptide forms in biogenesis of dense core secretory granules.
While this study with Edman sequencing and MALDI-TOF approaches indicates that proteolytic fragments of CgA and CgB represent the majority of chromaffin granule protein components, our previous study using different proteomic approaches (microcapillary LC-MS/MS of tryptic digests of gel slides obtained from one-dimensional SDS-PAGE gels) for protein identifications in these chromaffin granules demonstrated the presence of a multitude of proteins with distinct functions for prohormones, proteases, catecholamine neurotransmitter metabolism, protein folding, redox regulation, ATPases, calcium regulation, signaling components, exocytotic mechanisms, and related functions (38). The proteins with these functions were identified as 63 proteins in the soluble fraction of chromaffin granules, and 80 proteins in the membrane fraction. It is likely that further analyses with different proteomic strategies will yield identification of further novel protein components of chromaffin granules.
In summary, analyses of chromaffin granule soluble proteins by 2-D gel proteomics using Edman sequencing have demonstrated CgA- and CgB-proteins as major protein components. Significantly, specific proteolytic processing at dibasic and monobasic residues is utilized to generate these protein fragments of CgA and CgB. Clearly, proteolysis of these chromogranins does not occur in a random manner, but occurs primarily at basic residue motifs. These findings illustrate that proteolytic fragments of CgA and CgB represent the major proteins of the chromaffin secretory vesicles that produce and secrete biologically active neuropeptides and catecholamines.
Supplementary Material
Acknowledgements
The authors appreciate Edman sequencing performed by the Harvard Microchemistry Facility, directed by Dr. William Lane, as well as MALDI-TOF mass spectrometry performed by the Chemistry Core at the Buck Institute for Age Research, directed by Dr. Bradford Gibson. In addition, technical assistance by Mr. Thomas Toneff, Skaggs School of Pharmacy and Pharmaceutical Sciences, Univ. of Calif., San Diego, is appreciated.
This research was supported by grants from the National Institutes of Health.
Abbreviations
- AEBSF
4-(2-aminoethyl)benzene-sulfonylfluoride hydrochloride
- CgA
chromogranin A
- CgB
chromogranin B
- EDTA
ethylenediaminetetraacetic acid
- IEF
isoelectric focusing
- MALDI-TOF MS
matrix assisted laser desorption ionization time-of-flight mass spectrometry
- NPY
neuropeptide Y
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TCEP
tris-92-chloroethyl)-phosphate
- VIP
vasoactive intestinal polypeptide
Footnotes
Supporting Information Available
Supporting information is provided by supplemental Table A, supplemental Table B, and supplemental Table C which show data for MALDI-TOF MS analyses of CgA proteins, CgB proteins, and proSAAS proteins, respectively. This mateiral is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Yasothornsrikul S, Greenbaum D, Medzihradszky KF, Toneff T, Bundey R, Miller R, Schilling B, Petermann I, Dehnert J, Logvinova A, Goldsmith P, Neveu JM, Lane WS, Gibson B, Reinheckel T, Peters C, Bogyo M, Hook V. Cathepsin L in secretory vesicles functions as a prohormone-processing enzyme for production of the enkephalin peptide neurotransmitter. Proc. Natl. Acad. Sci. USA. 2003;100:9590–9595. doi: 10.1073/pnas.1531542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Funkelstein L, Toneff T, Hwang S-R, Reinheckel T, Peters C, Hook V. Cathepsin L participates in the production of neuropeptide Y in secretory vesicles, demonstrated by protease gene knockout and expression. J. Neurochem. 2008;106:384–391. doi: 10.1111/j.1471-4159.2008.05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hook V, Toneff T, Baylon S, Sei C. Differential activation of enkephalin, galanin, somatostatin, NPY, and VIP neuropeptide production by stimulators of protein kinases A and C in neuroendocrine chromaffin cells. Neuropeptides. 2008;42:503–511. doi: 10.1016/j.npep.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laslop A, Mahata SK. Neuropeptides and chromogranins: session overview. Ann N Y Acad Sci. 2002;971:294–299. doi: 10.1111/j.1749-6632.2002.tb04483.x. [DOI] [PubMed] [Google Scholar]
- 5.Winkler H. The adrenal chromaffin granule: a model for large dense core vesicles of endocrine and nervous tissue. J Anat. 1993;183:237–252. [PMC free article] [PubMed] [Google Scholar]
- 6.Ait-Ali D, Turquier V, Grumolato L, Yon L, Jourdain M, Alexandre D, Eiden LE, Vaudry H, Anouar Y. The proinflammatory cytokines tumor necrosis factor-alpha and interleukin-1 stimulate neuropeptide gene transcription and secretion in adrenochromaffin cells via activation of extracellularly regulated kinase 1/2 and p38 protein kinases, and activator protein-1 transcription factors. Mol Endocrinol. 2004;18:1721–1739. doi: 10.1210/me.2003-0129. [DOI] [PubMed] [Google Scholar]
- 7.Mahapatra NR, Mahata M, Hazra PP, McDonough PM, O'Connor DT, Mahat SK. A dynamic pool of calcium in catecholamine storage vesicles. Exploration in living cells by a novel vesicle-targeted chromogranin A-aequorin chimeric photoprotein. J Biol Chem. 2004;279:51107–51121. doi: 10.1074/jbc.M408742200. [DOI] [PubMed] [Google Scholar]
- 8.Videen JS, Mezger MS, Chang YM, O'Connor DT, Videen JS, Mezger MS, Chang YM, O'Connor DT. Calcium and catecholamine interactions with adrenal chromogranins. Comparison of driving forces in binding and aggregation. J Biol Chem. 1992;267:3066–3073. [PubMed] [Google Scholar]
- 9.Burgoyn RD. Mechanisms of catecholamine secretion from adrenal chromaffin cells. J Physiol Pharmacol. 1995;46:273–283. [PubMed] [Google Scholar]
- 10.Holz RW. Control of exocytosis from adrenal chromaffin cells. Cell Mol Neurobiol. 1988;8:259–268. doi: 10.1007/BF00711168. [DOI] [PubMed] [Google Scholar]
- 11.Carrasco GA, Van de Kar LD. Neuroendocrine pharmacology of stress. Eur J Pharmacol. 2003;463:235–272. doi: 10.1016/s0014-2999(03)01285-8. [DOI] [PubMed] [Google Scholar]
- 12.Wortsman J. Role of epinephrine in acute stress. Endocrinol Metab Clin North Am. 2002;31:79–106. doi: 10.1016/s0889-8529(01)00024-x. [DOI] [PubMed] [Google Scholar]
- 13.Zukowska-Grojec Z. A novel sympathetic stress hormone and more. Ann N Y Acad Sci. 1995;771:219–233. doi: 10.1111/j.1749-6632.1995.tb44683.x. [DOI] [PubMed] [Google Scholar]
- 14.Fung MM, Viveros OH, O'Connor DT. Diseases of the adrenal medulla. Acta Physiol. 2008;192:325–335. doi: 10.1111/j.1748-1716.2007.01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Connor DT, Takiyyuddin MA, Print MP, Dinh TQ, Barbosa JA, Rozansky DJ, Mahata SK, Wu H, Kennedy BP, Ziegler MG, Wright FA, Schlager G, Parmer RJ. Catecholamine storage vesicle protein expression in genetic hypertension. Blood Press. 1999;8:285–295. doi: 10.1080/080370599439508. [DOI] [PubMed] [Google Scholar]
- 16.Hook V, Funkelstein L, Lu D, Bark S, Wegrzyn J, Hwang S-R. Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu. Rev. Pharmacol. Tox. 2008;48:393–423. doi: 10.1146/annurev.pharmtox.48.113006.094812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seidah NG, Prat A. Precursor convertases in the secretory pathway, cytosol and extracellular milieu. Essays Biochem. 2002;38:79–94. doi: 10.1042/bse0380079. [DOI] [PubMed] [Google Scholar]
- 18.Zhou A, Webb G, Zhu X, Steine,r DF. Proteolytic processing in the secretory pathway. J. Biol. Chem. 1999;274:20745–20748. doi: 10.1074/jbc.274.30.20745. [DOI] [PubMed] [Google Scholar]
- 19.Yasothornsrikul S, Toneff T, Hwang S-R, Hook VYH. Arginine and lysine aminopeptidase activities in chromaffin granules of bovine adrenal medulla: relevance to prohormone processing. J. Neurochem. 1998;70:153–163. doi: 10.1046/j.1471-4159.1998.70010153.x. [DOI] [PubMed] [Google Scholar]
- 20.Lee JC, Taylor CV, Gaucher SP, Toneff T, Taupenot L, Yasothornsrikul S, Mahata SK, Sei C, Parmer RJ, Neveu JM, Lane WS, Gibson BW, O’Connor DT, Hook VYH. Primary sequence characterization of catestatin intermediates and peptides defines proteolytic cleavage sites utilized for converting chromogranin A into active catestatin secreted from neuroendocrine chromaffin cells. Biochemistry. 2003;42:6938–6946. doi: 10.1021/bi0300433. [DOI] [PubMed] [Google Scholar]
- 21.Field HI, Fenyo D, Beavis RC. RADARS, a bioinformatics solution that automates proteome mass spectral analysis, optimises protein identification, and archives data in a relational database. Proteomics. 2002;2:36–47. [PubMed] [Google Scholar]
- 22.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Clauser KR, Hall SC, Smith DM, Webb JW, Andrew LE, Tran HM, Epstein LB, Burlingame A. Rapid mass spectrometric peptide sequencing and mass matching for characterization of human melanoma proteins isolated by two-dimensional PAGE. Proc. Natl. Acad. Sci, USA. 1995;92:5072–5076. doi: 10.1073/pnas.92.11.5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hook VYH, Eiden LE, Brownstein MJ. A carboxypeptidase processing enzyme for enkephalin precursors. Nature. 1982;295:341–342. doi: 10.1038/295341a0. [DOI] [PubMed] [Google Scholar]
- 25.Hook VYH, Mezey E, Fricker LD, Pruss RM, Siegel RE, Brownstein MJ. Immunochemical characterization of carboxypeptidase B-like peptide-hormone-processing enzyme. Proc. Natl. Acad. Sci. USA. 1985;82:4745–4749. doi: 10.1073/pnas.82.14.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hook VYH, LaGamma EF. Production inhibition of carboxypeptidase H. J. Biol. Chem. 1987;262:12583–12588. [PubMed] [Google Scholar]
- 27.Hook VYH, Eiden LE, Pruss RM. Selective regulation of carboxypeptidase peptide hormone-processing enzyme during enkephalin biosynthesis in cultured bovine adrenomedullary chromaffin cells. J. Biol. Chem. 1985;260:5991–5997. [PubMed] [Google Scholar]
- 28.Fricker LD. Carboxypeptidase E. Annu. Rev. Physiol. 1988;50:309–321. doi: 10.1146/annurev.ph.50.030188.001521. [DOI] [PubMed] [Google Scholar]
- 29.Taylor CV, Taupenot L, Mahata SK, Mahata M, Wu H, Yasothornsrikul S, Toneff T, Caporale C, Jiang Q, Parmer RJ, Hook VYH, O’Connor DT. Formation of the catecholamine release-inhibitory peptide catestatin from chromogranin A, Determination of proteolytic cleavage sites in hormone storage granules. J. Biol. Chem. 2000;275:22905–22915. doi: 10.1074/jbc.M001232200. [DOI] [PubMed] [Google Scholar]
- 30.Taupenot L, Harper KL, O'Connor DT. The chromogranin-secretogranin family. N Engl J Med. 2003;348:1134–1149. doi: 10.1056/NEJMra021405. [DOI] [PubMed] [Google Scholar]
- 31.Curry WJ, Shaw C, Johnston CF, Thim L, Buchanan KD. Isolation and primary structure of a novel chromogranin A-derived peptide, WE-14, from a human midgut carcinoid tumour. FEBS Lett. 1992;301:319–321. doi: 10.1016/0014-5793(92)80266-j. [DOI] [PubMed] [Google Scholar]
- 32.Flanagan T, Taylor L, Poulter L, Viveros OH, Diliberto EJ. A novel 1745-dalton pyroglutamyl peptide derived from chromogranin B is in the bovine adrenomedullary chromaffin vesicle. Cell Mol. Neurobiol. 1990;10:507–523. doi: 10.1007/BF00712845. [DOI] [PubMed] [Google Scholar]
- 33.Strub JM, Garcia-Sabone P, Lonning K, Taupenot L, Hubert P, Van Dorsselaer A, Aunis D, Metz-Boutique MH. Processing of chromogranin B in bovine adrenal medulla, identification of secretolytin, the endogenous C-terminal fragment of residues 614–626 with antibacterial activity. Eur. J. Biochem. 1995;229:356–368. doi: 10.1111/j.1432-1033.1995.tb20476.x. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Vandenberghe I, Depreitere J, Devreese B, Clerens S, Nouwen EJ, Van Beeumen J, de Potter W. Identification and characterization of novel chromogranin B-derived peptides from porcine chromaffin granules by liquid chromatography/electrospray tandem MS. Eu. J. Biochem. 2001;268:235–242. doi: 10.1046/j.1432-1033.2001.01864.x. [DOI] [PubMed] [Google Scholar]
- 35.Kim T, Tao-Chen JH, Eiden LE, Loh YP. Chromogranin A, an “on/off” switch controlling dense core secreotry granule biogenesis. Cell. 2001;106:499–509. doi: 10.1016/s0092-8674(01)00459-7. [DOI] [PubMed] [Google Scholar]
- 36.Kim T, Tao-Cheng JH, Eiden LE, Loh YP. Large dense core secretory granule biogensis is under the control of chromogranin A in neuroendocrine cells. Ann. N.Y. Acad. Sci. 2002;971:323–331. doi: 10.1111/j.1749-6632.2002.tb04487.x. [DOI] [PubMed] [Google Scholar]
- 37.Kim T, Zhang CF, Sun Z, Wu H, Loh YP. Chromogranin A deficiency in transgenic mice leads to aberrant chromaffin granule biogenesis. J. Neurosci. 2005;25:6958–6961. doi: 10.1523/JNEUROSCI.1058-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wegrzyn J, Lee J, Neveu JM, Lane WS, Hook V. Proteomics of neuroendocrine secretory vesicles reveal distinct functional systems for biosynthesis and exocytosis of peptide hormones and neurotransmitters. J. Proteome Research. 2007;6:1652–1665. doi: 10.1021/pr060503p. [DOI] [PubMed] [Google Scholar]
- 39.Fricker L, McKinzie AA, Sun J, Curran E, Qian Y, Yan L, Patterson SD, Courchesne PL, Richards B, Levin N, Mzhavia N, Devi LA, Douglass J. J. Neurosci. 2000;20:639–648. doi: 10.1523/JNEUROSCI.20-02-00639.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mzhavia N, Berman Y, Che FY, Fricker LD, Devi LA. ProSAAS processing in mouse brain and pituitary. J Biol Chem. 2001;276:6207–6213. doi: 10.1074/jbc.M009067200. [DOI] [PubMed] [Google Scholar]
- 41.Mzhavia N, Qian Y, Feng Y, Che FY, Devi LA, Fricker LD. Processing of proSAAS in neuroendocrine cell lines. Biochem J. 2002;361:67–76. doi: 10.1042/0264-6021:3610067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.