Summary
Human breast tumors contain a breast cancer stem cell (BCSC) population with properties reminiscent of normal stem cells. We found 37 microRNAs that were differentially expressed between human BCSCs and non-tumorigenic cancer cells. Three clusters, miR-200c-141, miR-200b-200a-429 and miR-183-96-182 were down-regulated in human BCSCs, normal human and murine mammary stem/progenitor cells and embryonal carcinoma cells. Expression of BMI1, a known regulator of stem cell self-renewal, was modulated by miR-200c. MiR-200c inhibited the clonogenicity of breast cancer cells and suppressed the growth of embryonal carcinoma cells in vitro. Most importantly, miR-200c strongly suppressed the ability of normal mammary stem cells to form mammary ducts and tumor formation driven by human BCSCs in vivo. The coordinated down-regulation of three microRNA clusters and the similar functional regulation of clonogenicity by miR-200c provide a molecular link that connects breast cancer stem cells with normal stem cells.
Introduction
Cancers arise in tissues and organs which contain proliferating cells that regenerate old and damaged cells. Typically, these tissues are maintained by stem cells that have the ability to self-renew, a process by which the stem cells maintain the ability to undergo extensive proliferation while preserving the undifferentiated state. The stem cells also produce progeny that undergo a series of cell divisions in which they become progressively more differentiated before reaching maturation.
Like the tissues from which they arise, solid tumors are composed of a heterogeneous population of cells and many properties of normal stem cells are shared by at least a subset of cancer cells (Lobo et al., 2007; Stingl and Caldas, 2007). To maintain tissue homeostasis, normal stem cells must be able to undergo a large number of mitoses and in many tissues they must be able to migrate to different regions of the organ. Both of these properties are reminiscent of two hallmark properties of cancer cells, immortality and invasion.
The fact that tumors contain heterogeneous populations of cells at various stages of maturation and that cancer cells share properties of normal stem cells, led to the speculation that tumors may contain a cancer stem cell population that drives the growth of the tumor (Bruce and Gaag, 1963; Wu, 1968). Genetic studies in leukemia patients demonstrated that a primitive leukemia cell can give rise to fully mature non-replicating progeny, showing that not all cancer cells had the ability to form tumors (Fialkow, 1976a, 1976b, 1990). With improvements in isolation of both normal and cancer stem cells, there is now a growing body of evidence that in at least some cases of both human and mouse leukemia, as well as human and mouse epithelial tumors such as breast, colon, head and neck, skin, and brain cancer, a cancer stem cell population can be enriched based on phenotype (Al-Hajj et al., 2003; Cho et al., 2008; Dalerba et al., 2007; Lapidot et al., 1994; Malanchi et al., 2008; O'Brien et al., 2007; Prince et al., 2007; Ricci-Vitiani et al., 2007; Singh et al., 2004) MicroRNAs (miRNAs) are small noncoding regulatory RNAs that regulate the translation of mRNAs by inhibiting ribosome function, de-capping the 5′ Cap structure, deadenylating the polyA tail, and degrading the target mRNA (Filipowicz et al., 2008). MiRNAs are able to regulate expression of hundreds of target mRNAs simultaneously, thus controlling a variety of cell functions including cell proliferation, stem cell maintenance and differentiation. One of the best studied miRNAs, let-7 in Caenorhabditis elegans, was initially identified by genetic analysis of mutants with defects in developmental timing (Reinhart et al., 2000). Subsequently, Dicer was identified as a key enzyme of miRNA processing and function; Dicer null mutations result in embryonic lethality and depletion of stem cells (Bernstein et al., 2003). In addition, tissue specific deletion of Dicer affects self-renewal of embryonic stem cells, development of B lymphocyte lineage cells, and tissue morphogenesis (Chen et al., 2008; Davis et al., 2008; Koralov et al., 2008). In the skin, miR-203 is a critical regulator of stem cell maintenance (Yi et al., 2008). Deletion of DGCR8, another key enzyme for miRNA processing, alters silencing of self-renewal genes in embryonic stem cells (Wang et al., 2007). These findings demonstrate that miRNAs are critical regulators of self-renewal and differentiation.
Many of the common chromosomal amplifications and deletions seen in cancer contain miRNA coding sequences, and some miRNAs function as oncogenes or tumor suppressor genes (Calin et al., 2004; Esquela-Kerscher and Slack, 2006). For example, dysregulation of the miR-17-92 cluster can induce B-cell lymphoma and down-regulation of let-7 is associated with tumor progression and poor prognosis of lung cancer patients (He et al., 2005; Takamizawa et al., 2004). Expression of let-7 also prevents tumor sphere formation of breast cancer cell lines and inhibits tumorigenicity in an in vivo xenograft tumor assay (Yu et al., 2007). Finally, miRNA expression profiles are correlated with tumor stage, progression and prognosis of cancer patients (Calin et al., 2005; Iorio et al., 2005).
The ability to prospectively identify an enriched population of stem cells enables the interrogation of these cells for clues to the molecular regulators of key stem cell functions. In this report, we undertook a systematic comparison of the miRNAs in breast stem/progenitor cell populations and in their differentiated progeny that led to the identification of new regulators shared between normal and cancer stem cells.
Results
MiRNA Profiling of Human Breast and Embryonal Carcinoma Cells
As miRNAs are critical regulators of self-renewal and differentiation in both normal embryonic and adult tissue stem cells, we compared the miRNA expression profile between human CD44+CD24-/lowlineage- breast cancer cells (BCSCs) and the remaining lineage- non-tumorigenic breast cancer cells (NTG cells). In many patients with breast cancer, only a subset population of CD44+CD24-/lowlineage- cancer cells is highly tumorigenic in immunodeficient mice, as compared to the remaining lineage- breast cancer cells (Al-Hajj et al., 2003). The CD44+CD24-/lowlineage- cells have stem-cell-like properties such as self-renewal and differentiation, and can regenerate the original tumor from as few as 200 cells, whereas tens of thousands of the remaining lineage- non-tumorigenic cancer cells cannot. Multiplex real-time PCR was used to measure the expression of 460 miRNAs in BCSCs and NTG cells isolated from three human breast tumors. We found that 37 miRNAs were up-regulated or down-regulated in BCSCs compared to NTG cells in all three samples analyzed (Figure 1A). The expression of these 37 differentially expressed miRNAs was then measured in a total of 11 sets of human BCSCs and NTG cells, and this analysis confirmed that these 37 miRNAs were indeed differentially expressed (Figure 1B). Three clusters of miRNAs, the miRNA-200c-141 cluster located on chromosome 12p13, the miR-200b-200a-429 cluster located on chromosome 1p36, and the miR-183-96-182 cluster located on chromosome 7q32, were consistently down-regulated in human BCSCs (Figure 1B and 1C). For example, expression of miR-200a, miR-200b, and miR-200c was 2 to 218 times lower in BCSCs compared to NTG cells.
Figure 1. Profile of Human Breast Cancer Stem Cell miRNA Expression.
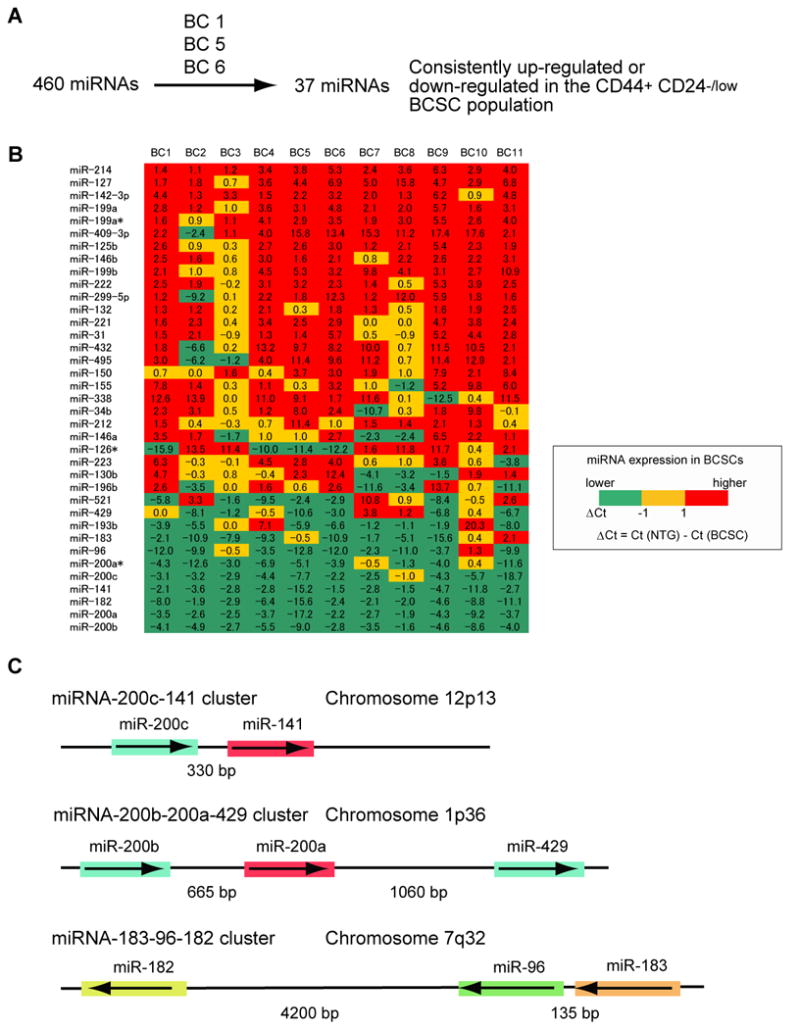
(A) Screening of 460 miRNA expression in human breast cancer stem cells. The details of the screen used to identify the 37 miRNAs differentially expressed by the CD44+CD24-/lowlineage- human breast cancer stem cells (BCSCs) and the remaining lineage- non-tumorigenic cancer cells (NTG cells) are shown schematically. (B) Expression profile of 37 miRNAs in human breast cancer stem cells. Flow cytometry was used to isolate BCSCs and NTG cells from 11 human breast cancer samples (BC1 to BC11). The amount of miRNA expression (Ct value) in 100 sorted cancer cells was analyzed by multiplex quantitative real-time PCR. Numbers represent the difference of Ct values (ΔCt) obtained from BCSCs and NTG cells. (C) A schematic representation of the three miRNA clusters down-regulated in human breast cancer stem cells. The miRNAs sharing the same seed sequence (from 2 to 7 base pairs) are marked by the same color.
It is thought that the CD44+CD24-/lowlineage- cells might be malignant counterparts of normal mammary stem or early progenitor cells (Al-Hajj et al., 2003; Mani et al., 2008). Similarly, embryonic carcinoma cells are malignant cells that arise from germ cells, which share many properties with pluripotent stem cells. Thus, the expression of these miRNAs was tested in Tera-2 embryonal carcinoma cells. Notably, Tera-2 cells either fail to express detectable levels of each of the miRNAs, or the level of expression is just at the level of detection (Figure S1, available on line).
The miRNA seed sequence serves to direct the miRNA to its mRNA targets (Lewis et al., 2005). Remarkably, the miR-200c-141 cluster and the miR-200b-200a-429 cluster are formed by two groups of miRNAs with essentially the same seed sequence (miR-200c/miR-200b/miR-429 miRNAs, and miR-200a/miR-141 miRNAs), which suggests that they might share some common target genes (Figure 1C). Given this similarity and the observed expression patterns, we suggest that down-regulation of all 3 of the clustered miRNAs in breast cancer CD44+CD24-/lowlineage- cells and Tera-2 embryonal carcinoma cells may be critical to maintain a stem cell function in cancer cells.
MiRNA Expression Connects Breast Cancer Stem Cell Differentiation with Normal Mammary Development
The functional similarities of cancer cells with normal tissue stem cells suggest that activation of normal stem cell self-renewal and/or differentiation pathways account for many of the properties associated with malignancies. We therefore tested early mammary stem and progenitor cells and more differentiated mammary epithelial progenitor cells for the expression of the miRNAs that are differentially expressed by BCSCs and NTG cells. We first performed this analysis in mouse where the mammary epithelium is better understood; CD24medCD49fhighCD29highSca-1- mouse mammary fat pad cells are enriched for mammary stem cells with an ability to regenerate a whole mammary gland in vivo. We collected the CD24medCD49fhighCD45-CD31-CD140a-Ter119- cells (MRUs, mammary repopulating units) that are enriched for mammary stem cells and the CD24highCD49flowCD45-CD31-CD140a-Ter119- cells (MaCFCs) that are enriched for more differentiated mammary epithelial progenitor cells (Figure 2A). We found that all three of the clustered miRNAs that were down-regulated in human breast cancer stem cells were also down-regulated in mouse MRU cells as compared to both MaCFCs as well as mature epithelial cells (Figure 2B, Figure S2).
Figure 2. Profile of Down-regulated miRNAs Shared Between Normal Mammary Stem Cells and Breast Cancer Stem Cells.

(A) Distribution of CD45-CD31-CD140a-Ter119- mouse mammary cells according to their expression of CD24 and CD49f MRU is a population enriched for mammary stem cells. MaCFCs are enriched for progenitors that do not regenerate a mammary duct in vivo. (B) Expression of miRNAs in MRUs as compared to MaCFCs. The expression of the miRNAs down-regulated in human breast cancer stem cells was analyzed in MRUs and MaCFCs isolated by flow cytometry from normal mouse mammary fat pads. The level of miRNA expression in 100 MRUs or MaCFCs was measured by quantitative real-time PCR. The analysis was repeated twice by using the two sets of samples derived from independently isolated populations of MRUs and MaCFCs. Numbers represent the difference of Ct values obtained from MRUs and MaCFCs. BC1, BC5 and BC6 represent miRNA expression of human breast cancer stem cells described in Figure 1.
An analysis of normal human breast epithelial cells isolated based both on CD44 and CD24, and on the markers recently described by Eaves and colleagues (Eirew et al., 2008) suggested that most of the 37 miRNAs are also differentially expressed by early human breast progenitors (Figure S3). This supports the notion that the differential expression of these 3 miRNA clusters between BCSCs and NTG cells is a part of the normal mammary cell developmental pathways.
MiR-200c Targets BMI1
Potential molecular targets of miR-200bc/429 were predicted by TargetScan 4.2 (http://www.targetscan.org/) (Lewis et al., 2005). Among the potential targets, we focused on BMI1 because it possessed critically conserved nucleotides indicative of a legitimate target and is known to be essential in regulating self-renewal and differentiation of other stem cell types, including hematopoietic, brain and mammary stem cells (Molofsky et al., 2005; Park et al., 2003; Pietersen et al., 2008).
The ability of miR-200c to regulate the 3′UTR of BMI1 was evaluated via luciferase reporter assays. HEK293 cells, which did not express miR-200c and miR-429 and expressed barely detectable levels of miR-200b (data not shown), were used. The 3′UTR target sites of BMI1 were cloned into pGL3-Control vector, downstream of a luciferase minigene (Figure 3A). HEK293 cells were co-transfected with a pGL3 luciferase vector, pRL-TK Renilla luciferase vector and miR-200c precursor RNA. We observed that the co-transfection of the miR-200c precursor suppressed the luciferase activity of the vector with the wild-type BMI1 3′UTR by 35% (Figure 3B); moreover, mutation of the miRNA-200bc/429 seed region within the BMI1 3′UTR abrogated the repressive ability of the miRNA, demonstrating specificity of the target sequence for BMI1 (Figures 3A and 3B). The ability of miR-200c to regulate the endogeous BMI1 protein was also tested. To do this, HEK293T cells were transfected with a miR-200c precursor and cells were cultured for 7days. Western blotting showed that BMI1 protein expression was decreased in cells transfected by miR-200c (Figure 3C).
Figure 3. MiR-200c Targets BMI1.
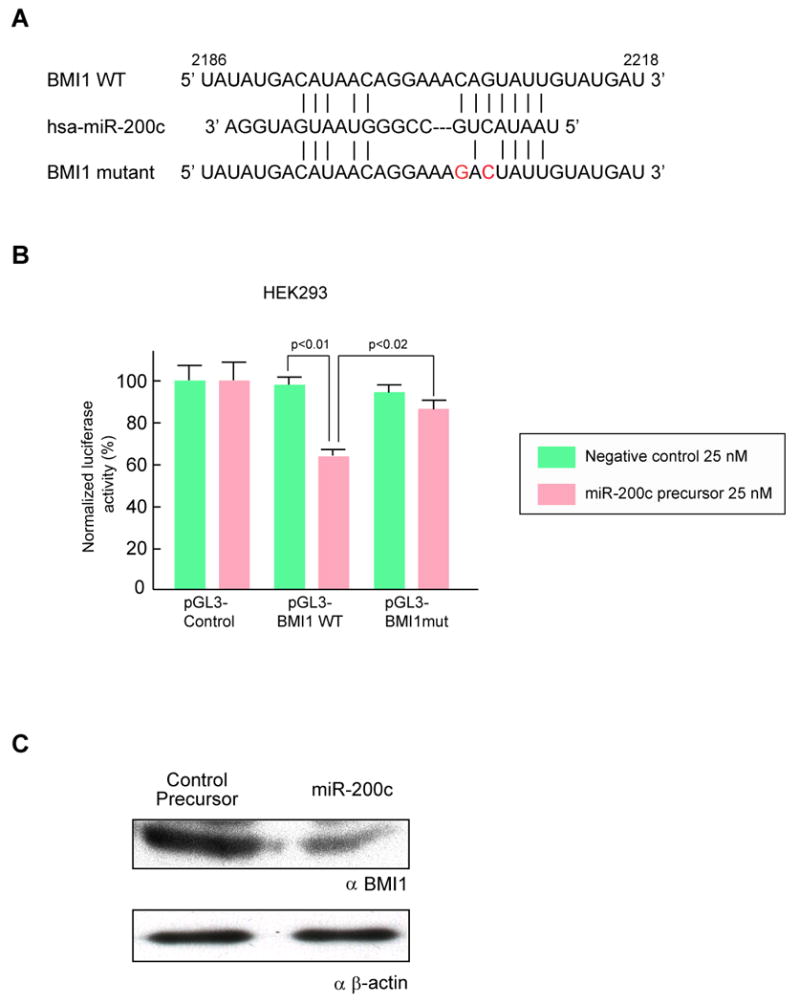
(A) Schematic representation of the miR-200bc/429 target sequence within the 3′ UTR of BMI1. Two nucleotides (complementary to nucleotide 6 and 8 of miR-200bc/429) were mutated in the 3′UTR of BMI1. The numbers indicate the position of the nucleotides in the reference wild-type sequences (NM_005180). (B) Activity of the luciferase gene linked to the 3′UTR of BMI1. The pGL3 firefly luciferase reporter plasmids with the wild-type or mutated 3′ UTR sequences of BMI1 were transiently transfected into HEK293 cells along with 25 nM miR-200c precursor or negative control, and a Renilla luciferase reporter for normalization. Luciferase activities were measured after 48 hours. The mean of the results from the cells transfected by pGL3 control vector was set as 100 %. The data are mean and S.D. of separate transfections (n=4). (C) Downregulation of endogenous BMI1 protein expression by miR-200c. HEK293T cells were transfected with 50 nM miR-200c precursor or negative control precursor. Lysates from 7 × 105 cells were loaded in each lane and BMI1 expression was analyzed by Western blotting. Expression of β-actin was used as a control. Replicate western blots from three independent experiments showed a similar down-regulation of BMI1.
MiR-200c Suppresses Cancer Cell Growth and Induces Differentiation
The observation that the same clusters of miRNAs were down-regulated in normal mammary stem cells, tumorigenic CD44+CD24-/lowlineage- breast cancer cells and embryonal carcinoma cells implies that these miRNAs are regulators of critical stem cell functions such as self-renewal and/or differentiation. In addition to suppressing the expression of BMI1, a gene critical for self-renewal in many types of stem cells, it has recently been shown that miR-200 family miRNAs prevent EMT (epithelial-to-mesenchymal transition) by suppressing expression of ZEB1 and ZEB2, two transcriptional repressors of E-cadherin (Christoffersen et al., 2007; Gregory et al., 2008; Park et al., 2008). EMT is a stem cell property that has been linked to both normal and cancer stem cells (Iwashita et al., 2003; Mani et al., 2008). To determine how expression of these miRNAs affects cells, we infected Tera-2 embryonal carcinoma cells with lentivirus that expresses miR-200c. The morphology of Tera-2 cells infected with miR-200c lentiviruses suggested that they had differentiated (Figure 4A). Indeed, staining with anti-neuron specific class III β tubulin (Tuj-1) antibody showed that miR-200c infected Tera-2 cells preferentially expressed the early post-mitotic neuron marker, Tuj1 antigen, suggesting that the miRNAs had induced neural differentiation (Figure 4B). Flow cytometry analysis confirmed that 36% of miR-200c expressing Tera-2 cells expressed Tuj-1 protein 10 days after infection, as compared to 0.9% of Tera-2 cells infected with the control lentivirus (Figure 4C). We found that Tera-2 cells infected with the miR-200c lentivirus, but not the control lentivirus, showed growth retardation (Figure 4D). Preliminary experiments suggest that miR-200c might also inhibit tumor formation in vivo (Figure S4).
Figure 4. Growth Suppression of Embryonal Carcinoma Cells by miR-200c.
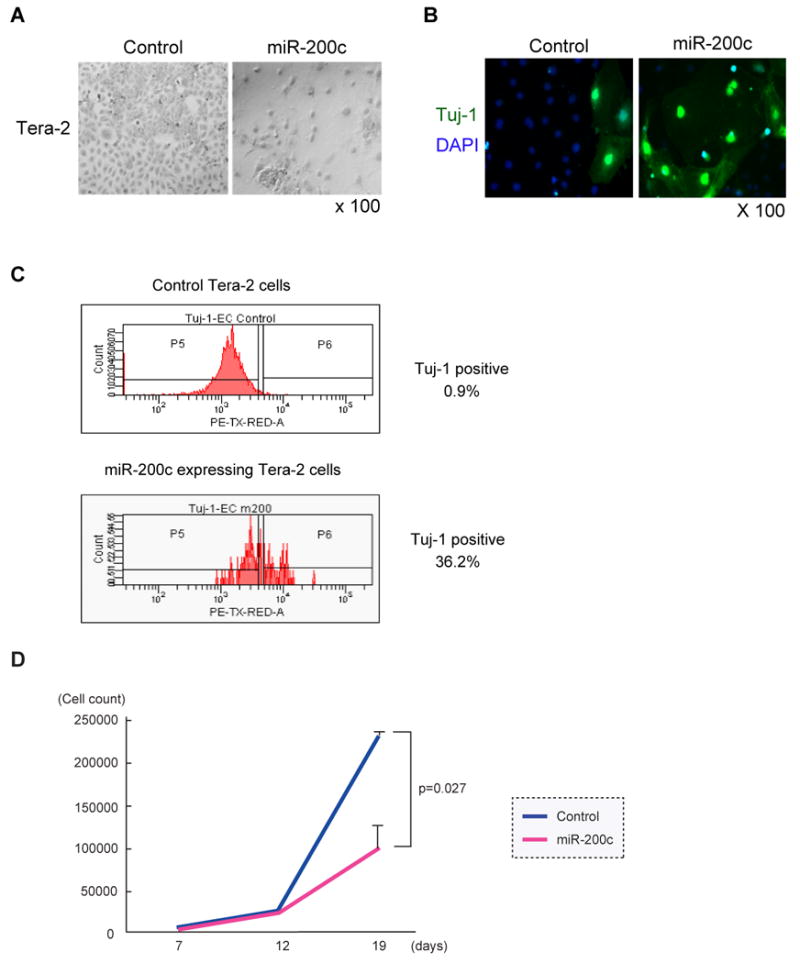
(A) Images of miRNA-expressing embryonal carcinoma cells. Tera-2 cells infected with the miRNA expressing lentivirus were collected by flow cytometry 4 days after infection. Tera-2 cells were cultured for 19 days and stained with Giemsa Wright staining solution. (B) MiR-200c enhanced differentiation of embryonal carcinoma cells. Tera-2 cells as described in (A) were stained with primary antibody against the early post-mitotic neuron marker, Tuj1 followed by Alexa-488 labeled secondary antibody. Cells were counterstained with DAPI. (C) Flow cytometry analysis of Tuj-1 expression. Tera-2 cells infected by miR-200c expressing lentivirus or control lentivirus were cultured for 6 days. Tera-2 cells were permeabilized and stained by anti-Tuj-1 antibody. Tuj-1 expression of GFP expressing Tera-2 cells was analyzed by flow cytometry. (D) MiR-200c inhibited the growth of embryonal carcinoma cells in vitro. 3000 miR-200c expressing or control Tera-2 cells were collected as described in (A) and cultured in a 96-well plate. Total cell numbers were counted on days 7, 12 and 19. The result is the average and S.D. from three independent wells.
MiR-200c Suppresses Clonogenicity of Breast Cancer Stem Cells
MMTV-Wnt-1 murine breast tumors are composed of both luminal and myoepithelial cells and an expanded mammary stem cell pool (Cho et al., 2008). We infected MMTV-Wnt-1 murine breast cancer cells with a miR-200c expressing lentivirus. Colony formation by the miR-200c infected cells was almost completely suppressed, reducing the number of colonies by 96% when compared to cells infected with the control lentivirus (Figure 5A). Flow cytometry can be used to isolate different populations of mammary cells that are enriched for stem cells, committed progenitor cells, or mature epithelial cells. When grown in tissue culture, the cell fraction that is enriched for normal mammary stem/progenitor cells (MRUs) or MMTV-Wnt-1 breast cancer stem cells form colonies that are biphenotypic, expressing both the myoepithelial cell cytokeratin CK14 and the epithelial cell cytokeratin CK8/18. (Neethan Lobo, unpublished data). Colonies that arise from the mature epithelial cell-enriched population express either CK8/18 or CK19 but not CK14. Culutured myoepithelial-enriched cells express CK14 but not CK8/18 or CK19 (Cho et al., 2008; Stingl et al., 2006). Breast cancer cells infected with the control virus formed large colonies and expressed CK14 and CK8/18, with an occasional cell that expressed CK19 (Figure 5B), whereas cells infected with the miR-200c expressing virus formed only small aggregates of cells that showed low levels of CK14 (Figure 5B). To prove functional relevance of BMI1 regulation by miR-200c, we constructed a BMI1 expressing lentivirus in which BMI1 cDNA does not contain the 3′UTR sequence that is targeted by miR-200c. Co-expression of this BMI1 transgene substantially rescued the defect in colony formation of breast cancer cells infected with the miR-200c lentivirus (Figure 5C and 5D). These results suggest that BMI1 is one of the key functional targets of miR-200c, at least with respect to the ability of miR-200c to suppress colony formation of breast cancer cells in vitro.
Figure 5. Effect of miR-200c on Clonogenicity of MMTV-Wnt-1 Murine Breast Cancer Cells.
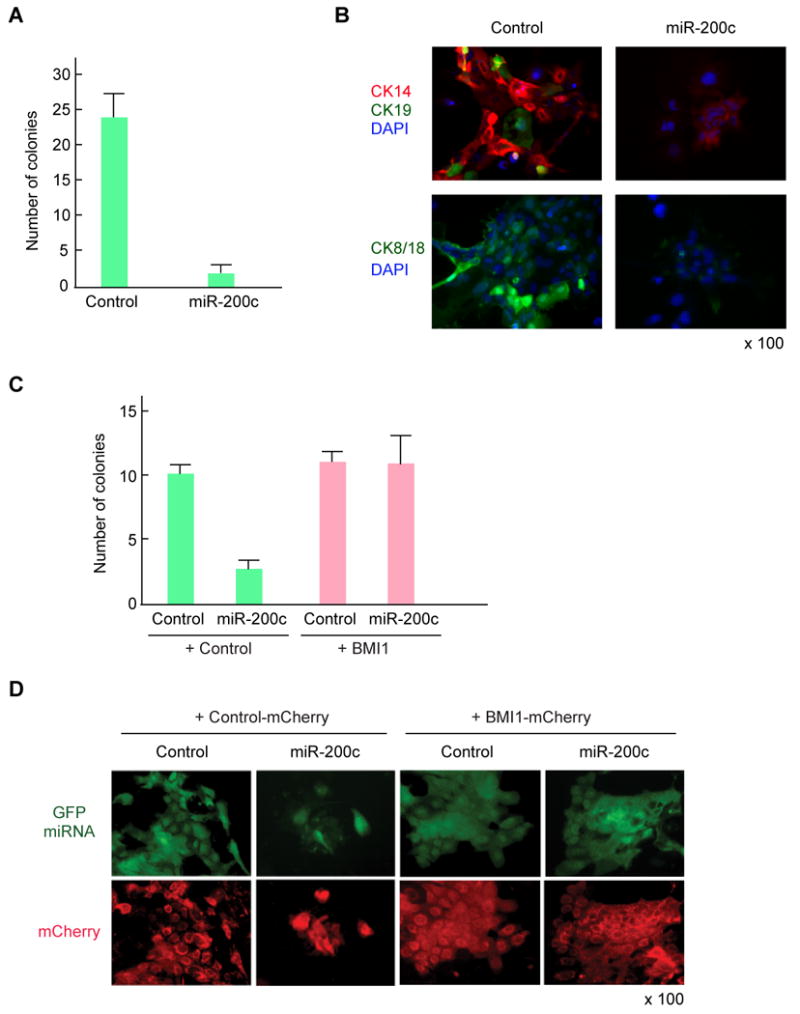
(A) The incidence of colony formation by MMTV-Wnt-1 breast cancer cells expressing miR-200c. MMTV-Wnt-1 breast cancer cells were dissociated and lineage positive cells were depleted using flow cytometry. 15,000 breast cancer cells were infected by miR200c-expressing lentivirus and cultured on an irradiated 3T3 feeder layer in a 24-well plate. After 7 days of incubation, the number of colonies with more than 10 GFP positive cells was counted. The result shows the average and S.D. from four independent wells. (B) Immunofluorescence images of colonies stained with antibodies against cytokeratin 14, 19, and 8/18. The GFP positive colonies were marked and stained with primary antibodies against cytokeratins followed by Alexa-488 and Alexa-594 labeled secondary antibodies. Cells were conterstained with DAPI. (C) BMI1 rescued the MMTV-Wnt-1 breast cancer cells expressing miR-200c. 10,000 breast cancer cells were co-infected by miR200c-expressing lentivirus (GFP) and BMI1-expressing lentivirus (mCherry), and cultured on irradiated 3T3 feeder layer in a 24-well plate. After 7 days of incubation, the number of colonies with more than 10 cells expressing both GFP and mCherry was counted. The result shows the average and S.D. from three independent wells. (D) Representative image of breast cancer colonies expressing both GFP and mCherry.
MiR-200c Suppresses Normal Mammary Outgrowth in Vivo
The observation that the same clusters of miRNAs were down-regulated both in normal mammary stem cells and in tumorigenic CD44+CD24-/lowlineage- breast cancer cells, and that miR-200c regulates the expression of the self-renewal gene BMI1 as well as EMT suggests that these miRNAs are regulators of normal and cancer stem cell functions such as self-renewal, proliferation, and/or differentiation. To clarify the role of miR-200c in normal mammary stem cells, we infected 50,000 lineage- murine mammary cells with the miR-200c expressing lentivirus and transplanted them into cleared mammary fat pads of syngeneic mice. Non-infected (mock) and control lentivirus infected mammary cells were transplanted as controls. Overall, 8 out of 18 transplants with non-infected mammary cells showed formation of a mammary tree, and 11 out of 20 transplants using cells infected with a control lentivirus formed a GFP-positive mammary tree, suggesting that lentivirus infection was highly efficient and did not perturb engraftment of mammary cells (Figure 6A). Histological and immunohistochemical analysis of mammary trees infected with control lentivirus showed normal structure and differentiation of both luminal and myoepithelial lineage mammary cells (Figure 6B). By contrast, only one GFP-positive mammary tree was formed out of 18 transplants using the mammary cells infected with miR-200c expressing lentivirus, while 6 transplants formed an aberrant, disorganized structure with a small cluster of mammary cells (Figure 6A and 6C). Similar to the miR-200c infected breast cancer cells, the engrafted miR-200c expressing mammary cells exclusively expressed CK14 but not CK8/18 (Figure 6C), suggesting induction of myoepithelial cell differentiation by miR-200c.
Figure 6. MiR-200c Suppresses Normal Mammary Outgrowth in Vivo.
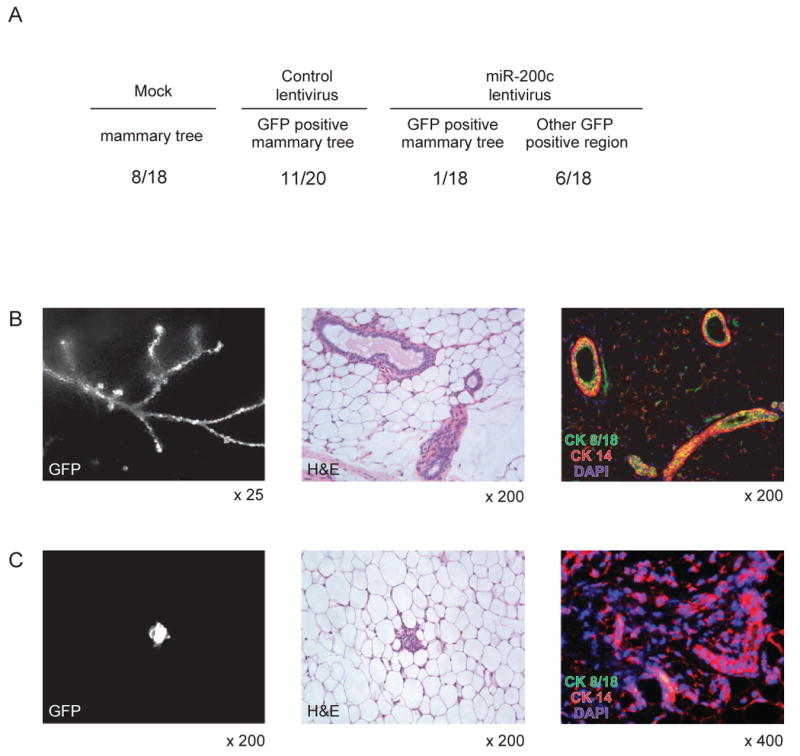
(A) Mammary outgrowths formed by control or miR-200c lentivirus infected mammary cells. Murine mammary cells isolated from FVB/NJ mouse were infected with a miR-200c expressing or control lentivirus. 5 × 104 infected mammary cells were injected into cleared fat pad of the same strain mouse. The frequency of the GFP-positive mammary trees is shown. Other GFP positive region means a small cluster of mammary cells formed by miR-200c infected mammary cells. The table is a summary of three independent experiments, each with essentially identical results. (B) Mammary tree outgrowth of control lentivirus infected mammary cells. GFP expression (left panel), Hematoxylin&Eosin staining (middle panel), immunostaining by cytokeratin 8/18 and cytokeratin 14 antibodies (right panel) are shown. (C) miR-200c expression perturbed mamamry cell differentiation. GFP expression (left panel), Hematoxylin&Eosin staining (middle panel), immunostaining by cytokeratin 8/18 and cytokeratin 14 antibodies (right panel) are shown.
MiR-200c Suppresses Tumorigenicity of Human Breast Cancer Stem Cells
We previously found that in many patients' tumors, the CD44+CD24-/lowlineage- cells (BCSCs) of human breast cancer are highly enriched for cells with the ability to form a transplantable xenograft tumor. To evaluate the effect of miR-200c on human BCSCs, we infected human BCSCs with the miR-200c expressing lentivirus. Then, 5,000 to 10,000 infected BCSCs were injected in the mammary fat pad of the NOD/SCID mice. Human breast cancer stem cells infected with control lentivirus formed 6 tumors out of 13 injections, whereas miR-200c expressing breast cancer stem cells formed only1 tumor out of 13 injections (Figure 7A and 7B). Tumors arising from cells infected with the control lentivirus expressed GFP, whereas the only tumor arising from the miR-200c infected cells did not (Figure 7C). This suggests that the tumor arose from cells that were not transduced with miR-200c. These results suggest that like their normal mammary cell counterparts, the miR-200c infected human breast cancer stem cells from this patient lost the ability to self-renew and proliferate extensively in vivo.
Figure 7. MiR-200c Suppresses Tumorigenicity of Human Breast Cancer Stem Cells.
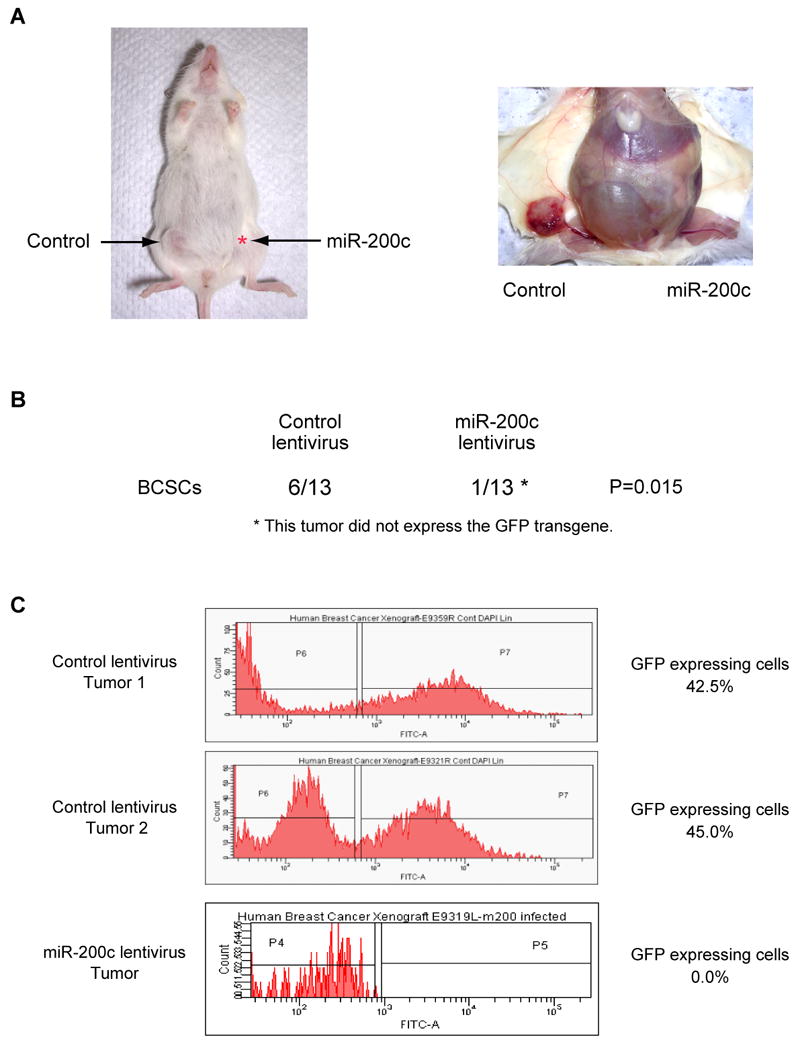
(A) A representative tumor in a mouse injected with human breast cancer stem cells is shown. CD44+CD24-/lowlineage- breast cancer stem cells were isolated from an early passage human breast xenograft tumor and infected by miR-200c expressing lentivirus or control lentivirus. 5 × 103 or 1 × 104 infected cells were injected into the breast of immunodeficient NOD/SCID mice. Tumor growth was monitored for five months after injection. (B) Tumor incidence of miRNA-expressing breast cancer stem cells (BCSCs). Six out of 13 control lentivirus infected BCSCs developed tumors after five months. One out of 13 miR-200c lentivirus infected BCSCs developed a tumor that did not expressed the GFP transgene. (C) GFP expression of tumors derived from control or miR-200c expressing lentivirus infected cells. Tumors were dissociated and GFP expression was analyzed by flow cytometry. The lentivirus contains a GFP minigene to mark virus infected cells.
Discussion
Two recent studies have shown that undifferentiated tumors and embryonic stem cells share expression of a subset of genes. However, neither study provided evidence for a functional link between the gene expression signature and adult stem cell biology, either normal or malignant (Ben-Porath et al., 2008; Wong et al., 2008). The results reported here show that miR-200c-141, miR-200b-200a-429 and miR-183-96-182 are down-regulated in normal mammary stem cells, in human breast cancer stem cells and in embryonal carcinoma cells and that miR-200c modulates expression of BMI1. In addition, our results provide a molecular explanation, at least in part, for the increased tumorigenicity displayed by the subpopulation of CD44+CD24-/lowlineage- breast cancer cells in many patients' tumors (Al-Hajj et al., 2003; Mani et al., 2008). The 5 down-regulated miRNAs shared similar seed sequences, and yet mapped to two clusters on different chromosomes. Although the miRNAs that share seed sequences do not always have completely overlapping targets, one could speculate that there might be functional redundancy of these families of miRNAs to maintain stem cell homeostasis and prevent tumors, by ensuring that a single mutation does not perturb the regulation of their targets.
The regulation of BMI1 by miR-200c is intriguing. Indeed, self-renewal and proliferation of both hematopoietic stem cells, normal mammary stem cells, and neural stem cells is defective in Bmi1-/- mice (Molofsky et al., 2003; Park et al., 2003; Pietersen et al., 2008). BMI1 is a member of the Polycomb-group proteins and is known to epigenetically repress the transcription of Hox genes and p16Ink4a p19Arf locus. Bmi1 represses apoptotic, senescence and differentiation pathways in stem cells (Park et al., 2003). Our results suggest that these same pathways might also be modulated by miR-200c at least in part through BMI1. Although the stress of transplantation could accentuate these effects on survival in miR-200c-expressing stem cells, the observation that Bmi1 mutant mice clearly show a loss of adult blood stem cells independent of the stresses of transplantation suggests that miR-200c expression will affect stem cell functions in the absence of this stress (Park et al., 2003). NOD/SCID mice have functional NK-cells that can modulate engraftment (Quintana et al., 2008) and it is formally possible that miR-200c also regulates NK-cell sensitivity. However, NK-cell activity cannot explain miR-200c inhibition of engraftment of normal mouse breast stem cells in syngeneic mice, the inhibition of colony formation of breast cancer stem cells by miR-200c in vitro (where NK cells are not present), or the ability of Bmi1 to partially rescue colony formation by the breast cancer cells expressing miR-200c in vitro. Mutations to downstream targets of Bmi1 such as TP53 can partially relieve stem cell dependence on Bmi1 for self- renewal (Akala et al., 2008; Park et al., 2003). It is therefore possible that in a subset of breast cancer patients, miR-200c alone might become unable to inhibit self-renewal because of particular mutations in the cancer cells.
The difference between a self-renewing normal stem cell and a self-renewing cancer cell is that unlike normal stem cells in which the total number of stem cells in a tissue is highly regulated and expansion beyond the normal level is restricted by genetic programs (Morrison et al., 2002), there is a continuous expansion of self-renewing cells in cancer resulting in the development of a tumor. From these observations, one would predict that cancer stem cells likely share some elements of the self-renewal machinery present in normal stem cells. Indeed the miRNA profile described in this study shows remarkable similarities between breast cancer stem cells and normal mammary stem cells, and some of these miRNAs have been linked to cancer (Esquela-Kerscher and Slack, 2006). Prediction programs such as Targetscan4.2 (Lewis et al., 2005) suggest that there are likely many genes other than BMI1 that are regulated by these differentially expressed miRNAs and are known to be functionally important for stem cells. The down-regulation of let-7 miRNAs in human breast cancer stem cells was previously reported (Yu et al., 2007). Only occasionally did we see differences in let-7 expression between breast cancer stem cells and non-tumorigenic cancer cells in the 11 breast cancer patients that we screened. The discrepancies in let-7 expression between these two studies might be related to differences in tumor histology or the genetic background of the patient populations analyzed. Alternatively, loss of let-7 expression could have occurred when the cell line used by Yu et al. (2007) was derived (Daniel et al., 2009).
EMT is a widespread, developmental program that regulates cell migration in many tissues and organs, and is associated with normal and malignant mammary stem cell function (Mani et al., 2008). Recent studies have shown that expression of components of the EMT pathway including SNAI2 is highest in the CD44+CD24-/lowlineage- breast cancer cells (Liu et al., 2007; Mani et al., 2008). Here we show that miR-200 family miRNAs were strongly suppressed in CD44+CD24-/lowlineage- human breast cancer cells. The miR-200 family of miRNAs taragets multiple sites in the 3′UTRs of ZEB1 that serve as EMT inducers. Suppression of ZEB1 and ZEB2 up-regulates expression of E-cadherin and inhibits EMT (Christoffersen et al., 2007; Gregory et al., 2008; Park et al., 2008) Collectively these findings begin to paint a picture of the miR-200 family miRNAs as important regulators of multiple stem cell functions, by controlling both EMT and self-renewal and/or proliferation in normal mammary stem cells and breast cancer stem cells.
We note that recently, the existence and relevance of the prospective isolation of cancer stem cells has been challenged (Kelly et al., 2007). Our results, however, clearly show that the ability to prospectively isolate cancer cells that preferentially engraft immunodeficient mice can in fact uncover valuable information about cancer biology. Indeed, a gene signature established from BCSCs was strongly associated with patient prognosis (Liu et al., 2007). Whereas a previous miRNA expression screen of all the cells in a tumor failed to uncover the 3 clusters of miRNAs described here, prospective isolation of the proposed cancer stem cells resulted in the demonstration of differential expression of miRNAs and revealed that miR-200c, one of the down-regulated miRNAs in the tumorigenic subset of human breast cancer cells, strongly suppressed the ability of normal stem cells to form mammary ducts and the breast cancer stem cells to form tumors, respectively. Our results support for the notion that pathways important for normal stem cells are also used by at least a subset of the cancer cells.
In summary, the findings in this paper provide a strong molecular link between normal breast stem/progenitor cells, the CD44+CD24-/lowlineage- breast cancer cells and embryonal carcinoma cells. The down-regulation of miR-200 family miRNAs suggests that normal and breast cancer stem cells share common molecular mechanisms that regulate stem cell functions such as self-renewal, proliferation and EMT.
Experimental Procedures
Additional methods can be found in the Supplemental Data.
Cell Culture
Human embryonal kidney (HEK) 293 and 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 250 ng/mL amphotericin B (Invitrogen) and incubated at 5% CO2 at 37°C.
The human embryonal carcinoma cell line Tera-2 (HTB-106) was purchased from ATCC, and grown in modified McCoy's medium (Invitrogen) with 100 units /ml of penicillin G and 100 μg / ml of streptomycin, supplemented with 15% fetal bovine serum and incubated at 5% CO2 at 37°C.
Multiplex Real-time PCR Assay
11 sets of CD44+CD24lowlineage- breast cancer stem cells and the remaining lineage- non-tumorigenic human breast cancer cells were isolated using a BD FACSAria sorter as previously described (Al-Hajj et al., 2003). For RNA preparation, 100 CD44+CD24lowlineage- human breast cancer stem cells and the other non-tumorigenic lineage- cancer cells were double-sorted into Trizol (Invitrogen) and RNA was extracted following the manufacturer's protocol. Glycogen (Invitrogen) was used as a carrier for precipitation. RT, pre-PCR and the multiplex real-time PCR for miRNA profiling was performed by multiplex real-time PCR method as described previously (Tang et al., 2006). Briefly multiplex reverse transcription reactions were performed with 466 sets of second strand synthesis primers. Then multiplex pre-PCR reactions were performed with 466 sets of forward primers and universal reverse primers. The multiplex pre-PCR product was diluted, aliquoted into 384 well reaction plates and the abundance of each miRNA was measured individually by using the 7900HT Fast Real-Time PCR System (Applied Biosystems). Results were normalized by the amount of small nuclear RNA expression, C/D box 96A and C/D box84. The difference of miRNA expression between two populations were calculated such as ΔCt = normalized Ct (non-tumorigenic cancer cells) – normalized Ct (breast cancer stem cells). The Ct value was set 40 for ΔCt calculation when expression of miRNA was undetectable.
Breast Cancer Cell Colony Formation Assay
Mouse MMTV-Wnt1 tumors were digested using 200 U/ml Liberase Blendzyme 2 (Roche) and dissociated as described (Cho et al., 2008). Cells were stained with anti-CD31, CD45, and CD140a antibodies and lineage positive cells were depleted by flow cytometry. 15,000 cells were infected with 20 MOI of miR-200c expressing lentiviruses by spin infection for 2 hours followed by incubation at 37°C for 2 hours in DMEM/F12 supplemented with 5% BSA, 2% heat inactivated FBS, 1:50 B27, 20 ng/mL EGF, 20 ng/mL βFGF, 10 μg/mL insulin, and 10 μg/ mL heparin. For co-infection experiments, 10,000 cells were infected with 10 MOI of miR-200c expressing lentivirus and 20 MOI of BMI1 expressing lentivirus. The infected cells were washed twice with the same medium and then the medium was replaced by Epicult medium (Stemcell technologies) with 5% FBS. The infected cells were seeded on the 30,000 irradiated 3T3 feeder cells in the 24-well plate. The medium was replaced again by Epicult medium without serum 24 hours after seeding and cells were incubated for 7 days at 5% CO2 at 37°C.
Statistical Analysis
When two groups were compared, the Student's t test was used. Fisher's exact test was used to analyze the significance of in vivo experiment results.
Mammary Cell Transplantation Assay
Murine breast tissue derived from FVB/NJ mice were digested and dissociated. Cells were stained with anti-CD31, CD45, CD140a, and Ter119 antibodies, and lineage- murine mammary cells were collected by flow cytometry or MACS magnetic separation columns (Miltenyi Biotec). Isolated cells were mixed with 5MOI of lentivirus and incubated for 16 hours at 5% CO2 at 37°C. 50,000 lentivirus infected cells were injected into cleared mammary fat pad of weaning age FVB/NJ female mouse. All experiments were carried out under the approval of the Administrative Panel on Laboratory Animal Care of Stanford University.
After 6.5 to 8 weeks, GFP expression of the transplanted mammary tissue was checked under the fluorescent microscope (Leica DMI 6000 B). For immunostaining, paraffin embedded murine mammary tissue were deparaffinized, and incubated with primary antibody (1:100 dilution for rabbit anti-cytokeratin 14 (Covance), and rat anti-cytokeratin 8/18 antibodies (Developmental Studies Hybridoma Bank, DSHB)), followed by staining with 1:200 diluted Alexa Fluor 488-conjugated anti-rat IgG antibody and Alexa Fluor 594-conjugated anti-rabbit IgG antibody (Invitrogen). GFP expression was analyzed by staining with 1:30 diluted Alexa Fluor 594-conjugated anti-GFP antibody (Invitrogen). The stained tissue was observed using a fluorescent microscope (Leica DM 4000 B).
Human Breast Cancer Xenograft Assay
The CD44+CD24lowlineage- human breast cancer stem cells (corresponding to 1.5% of cancer cells) were isolated by flow cytometry. Breast cancer stem cells were infected by 20 MOI of miR-200c expressing lentivirus or control lentivirus by spin infection for 2 hours followed by incubation at 37 °C for 2 hours. Infected cells were washed by PBS and were mixed with Matrigel (BD Biosciences). 5,000 or 10,000 infected cells were injected into mammary fat pad of female NOD/SCID mouse. All experiments were carried out under the approval of the Administrative Panel on Laboratory Animal Care of Stanford University.
Supplementary Material
Acknowledgments
We thank Andy Fire for helpful discussions and advice. We thank Jessica S. Lam for technical assistance. This work was supported by the California Breast Cancer Research Program of the University of California #12FB-0053 to Y.S., by the Fundacion Alfonso Martin Escudero and the Fulbright to M.Z.U., and by the grants from the NIH (NIH CA100225, CA104987, CA126524, CA139490), the Breast Cancer Research Foundation, the Morton Family Foundation and the Ludwig Foundation to M.F.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akala OO, Park IK, Qian D, Pihalja M, Becker MW, Clarke MF. Long-term haematopoietic reconstitution by Trp53-/-p16Ink4a-/-p19Arf-/- multipotent progenitors. Nature. 2008;453:228–232. doi: 10.1038/nature06869. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Bruce WR, Gaag H. A quantitative assay for the number of murine lymphoma cells capable of proliferation in vivo. Nature. 1963;199:79–80. doi: 10.1038/199079a0. [DOI] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RW, Wang X, Diehn M, Shedden K, Chen GY, Sherlock G, Gurney A, Lewicki J, Clarke MF. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells. 2008;26:364–371. doi: 10.1634/stemcells.2007-0440. [DOI] [PubMed] [Google Scholar]
- Christoffersen NR, Silahtaroglu A, Orom UA, Kauppinen S, Lund AH. miR-200b mediates post-transcriptional repression of ZFHX1B. Rna. 2007;13:1172–1178. doi: 10.1261/rna.586807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel VC, Marchionni L, Hierman JS, Rhodes JT, Devereux WL, Rudin CM, Yung R, Parmigiani G, Dorsch M, Peacock CD, et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 2009;69:3364–3373. doi: 10.1158/0008-5472.CAN-08-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, Ullian EM. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirew P, Stingl J, Raouf A, Turashvili G, Aparicio S, Emerman JT, Eaves CJ. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat Med. 2008;14:1384–1389. doi: 10.1038/nm.1791. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Fialkow PJ. Clonal origin of human tumors. Biochim Biophys Acta. 1976a;458:283–321. doi: 10.1016/0304-419x(76)90003-2. [DOI] [PubMed] [Google Scholar]
- Fialkow PJ. Human tumors studied with genetic markers. Birth Defects Orig Artic Ser. 1976b;12:123–132. [PubMed] [Google Scholar]
- Fialkow PJ. Stem cell origin of human myeloid blood cell neoplasms. Verh Dtsch Ges Pathol. 1990;74:43–47. [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Iwashita T, Kruger GM, Pardal R, Kiel MJ, Morrison SJ. Hirschsprung disease is linked to defects in neural crest stem cell function. Science. 2003;301:972–976. doi: 10.1126/science.1085649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly PN, Dakic A, Adams JM, Nutt SL, Strasser A. Tumor growth need not be driven by rare cancer stem cells. Science. 2007;317:337. doi: 10.1126/science.1142596. [DOI] [PubMed] [Google Scholar]
- Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri M, Dick J. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;17:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K, Clarke MF. The Prognostic Role of a Gene Signature from Tumorigenic Breast-Cancer Cells. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- Malanchi I, Peinado H, Kassen D, Hussenet T, Metzger D, Chambon P, Huber M, Hohl D, Cano A, Birchmeier W, et al. Cutaneous cancer stem cell maintenance is dependent on beta-catenin signalling. Nature. 2008;452:650–653. doi: 10.1038/nature06835. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Qian D, Jerebek L, Thiel BA, Park IK, Ford PS, Kiel MJ, Schork NJ, Weissman IL, Clarke MF. A Genetic Determinant That Specifically Regulates the Frequency of Hematopoietic Stem Cells. J Immunol. 2002;168:635–642. doi: 10.4049/jimmunol.168.2.635. [DOI] [PubMed] [Google Scholar]
- O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen AM, Evers B, Prasad AA, Tanger E, Cornelissen-Steijger P, Jonkers J, van Lohuizen M. Bmi1 regulates stem cells and proliferation and differentiation of committed cells in mammary epithelium. Curr Biol. 2008;18:1094–1099. doi: 10.1016/j.cub.2008.06.070. [DOI] [PubMed] [Google Scholar]
- Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles LE. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Stingl J, Caldas C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat Rev Cancer. 2007;7:791–799. doi: 10.1038/nrc2212. [DOI] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Tang F, Hajkova P, Barton SC, O'Carroll D, Lee C, Lao K, Surani MA. 220-plex microRNA expression profile of a single cell. Nat Protoc. 2006;1:1154–1159. doi: 10.1038/nprot.2006.161. [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AM, Till JE, Siminovitch L, McCulloch EA. Cytological evidence for a relationship between normal hematopoietic colony-forming cells and cells of the lymphoid system. J Exp Med. 1968;127:455–467. doi: 10.1084/jem.127.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al. let-7 Regulates Self Renewal and Tumorigenicity of Breast Cancer Cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


