Abstract
While speciation can be found in the presence of gene flow, it is not clear what impact this gene flow has on genome- and range-wide patterns of differentiation. Here we examine gene flow across the entire range of the common sunflower, H. annuus, its historically allopatric sister species H. argophyllus and a more distantly related, sympatric relative H. petiolaris. Analysis of genotypes at 26 microsatellite loci in 1015 individuals from across the range of the three species showed substantial introgression between geographically proximal populations of H. annuus and H. petiolaris, limited introgression between H. annuus and H. argophyllus, and essentially no gene flow between the allopatric pair, H. argophyllus and H. petiolaris. Analysis of sequence divergence levels among the three species in 1420 orthologs identified from EST databases identified a subset of loci showing extremely low divergence between H. annuus and H. petiolaris and extremely high divergence between the sister species H. annuus and H. argophyllus, consistent with introgression between H. annuus and H. petiolaris at these loci. Thus, at many loci, the allopatric sister species are more genetically divergent than the more distantly related sympatric species, which have exchanged genes across much of the genome while remaining morphologically and ecologically distinct.
Keywords: EST, hybridization, introgression, microsatellites, speciation, sunflower
The most widely accepted species concept, the Biological Species Concept (BSC), states that “Species are groups of interbreeding natural populations that are reproductively isolated from other such groups” (Mayr 1963). However, it is now recognized that hybridization and gene flow exists between many recognized species, and that even highly divergent lineages show evidence of gene flow at some loci (Arnold 1992; Rieseberg 1997; Bergthorsson et al. 2004). These observations conflict with Mayr's emphasis on complete reproductive isolation. Many proponents of the BSC relax the definition to allow for some level of hybridization (e.g., Barton and Hewitt 1985; Coyne and Orr 2004), which makes the definition more realistic but also less rigorous.
Despite the flaws of the BSC, other proposed species definitions have failed to gain widespread acceptance among evolutionary biologists, and the BSC remains popular due to its simplicity and conceptual strengths. One of the most important strengths of the BSC is its emphasis on the dual nature of gene flow: within-species gene flow holds a species together, whereas barriers to gene flow allow species to diverge. However, for many species and species complexes, there is insufficient gene flow to homogenize the species at all loci (Morjan and Rieseberg 2004); only a fraction of the genome, in particular the fraction subject to geographically widespread selective sweeps, will have substantial gene flow across the species range. Similarly, in some species only a fraction of the genome will be closed to gene flow from closely related species (Barton 1979; Martinsen et al. 2001).
Thus, when interspecific barriers to gene flow remain incomplete, some level of exchange will often occur. In this case, theory predicts (Barton 1979; Barton and Hewitt 1985), and empirical studies confirm, that gene flow is restricted at sites with reduced recombination (Rieseberg et al. 1999; Noor et al. 2001; Stump et al. 2005; Turner et al. 2005; Geraldes et al. 2006; Yatabe et al. 2007) and/or strong selection against introgression (Rieseberg et al. 1999; Noor et al. 2001; Scotti-Saintagne et al. 2004). Outside of these regions, neutral or positively selected loci can cross the species boundary via hybridization and introgression. The consequences of this gene flow are only beginning to be understood. Gene flow between coexisting species has been the subject of many studies, particularly those focusing on the role of hybrid zones in allowing exchange of genetic material across some fraction of the genome (Rieseberg et al. 1999; Martinsen et al. 2001; Lexer et al. 2005; Llopart et al. 2005; Yatabe et al. 2007). These studies typically investigated a few dozen markers across pairs of populations. However, little is known about the effects of interspecific gene flow across the entire ranges of hybridizing species (although see Martinsen et al. 2001; Geraldes et al. 2006).
Here, we seek to expand our understanding of the nature of species by studying gene flow between species, as well as among populations within species. Sunflowers have been developed as a model system for speciation research (Rieseberg 2006; Rieseberg and Willis 2007) and numerous genetic and genomic tools are available (see http://cgpdb.ucdavis.edu/). In particular, the three sunflower species Helianthus annuus, H. argophyllus, and H. petiolaris are ideal study organisms for understanding the nature of species boundaries.
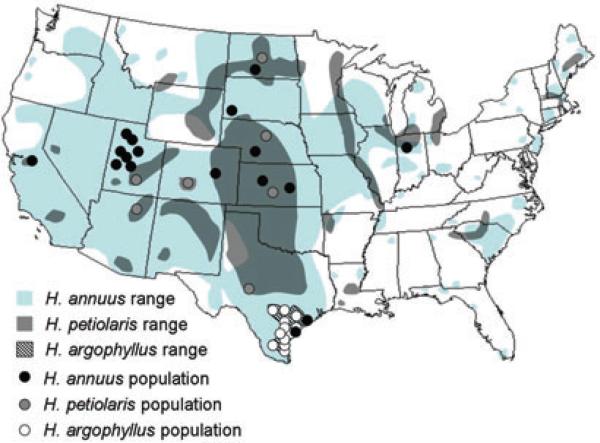
Helianthus annuus and H. petiolaris are widespread species (Heiser et al. 1969), whose ranges cover much of the continental United States (Fig. 1). The two species differ in many phenotypic traits, including reproductive timing, branching patterns, and height (Rosenthal et al. 2002, 2005), as well as having significant differences in expression patterns for many loci (Lai et al. 2006). Additionally, there are substantial differences in habitat preferences; for instance, H. annuus thrives in mesic clay soil whereas H. petiolaris requires drier, well-drained sandy soil (Heiser et al. 1969; Gross et al. 2004). Nonetheless, where the two soil types coexist, both species can often be found together, and form hybrid zones in which interbreeding between the species is very common (Rieseberg et al. 1998). Because of the widespread natural overlap in the species ranges (Fig. 1), and because of several ancient hybrid species whose origins have been estimated at between 60,000 and 210,000 years ago (Schwarzbach and Rieseberg 2002; Welch and Rieseberg 2002; Gross et al. 2003), or perhaps as long as 1.2 million years ago (J. Strasburg, pers. comm.), we know that this hybridization has been occurring for hundreds of thousands of years. Additionally, there is compelling evidence for high levels of gene flow between the two species at numerous loci (Rieseberg et al. 1999; Yatabe et al. 2007; Strasburg and Rieseberg 2008), despite large-scale karyotypic differences (Burke et al. 2004; Lai et al. 2005), strong conspecific pollen precedence (Rieseberg et al. 1995), and high levels of pollen and seed inviability in the hybrids (Rieseberg et al. 1999). Thus, despite this hybridization, the two species remain largely reproductively isolated and morphologically and ecologically distinct.
Figure 1.
Map of populations. Black circles: H. annuus populations; gray circles: H. petiolaris populations; white circles: H. argophyllus populations. Light gray shaded areas represent the natural range of H. annuus. Dark gray shaded areas represent the natural range of H. petiolaris. Areas shaded with diagonal lines represent the natural range of H. argophyllus.
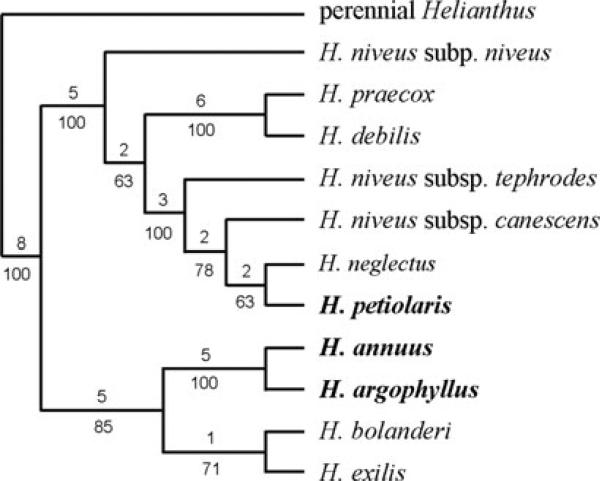
While H. annuus and H. petiolaris are common throughout much of North America, H. argophyllus is only found in southern Texas and a few introduced localities in Florida (Heiser et al. 1969). Due to this restricted geographic range, H. argophyllus is thought to have been largely or completely allopatric to both H. annuus and H. petiolaris for most of its history, and currently only overlaps with H. annuus (Fig. 1). This overlap is likely very recent, as H. annuus probably was absent from the H. argophyllus range until its introduction to this region within the past 100 years (Heiser 1951). In areas of contact, hybrid swarms have been reported (Heiser 1951), and this interbreeding is perhaps not unexpected, as H. argophyllus is the sister species of H. annuus (Schilling and Heiser 1981; Chandler et al. 1986; Schilling et al. 1998). Moreover, the two species have relatively high F1 fertility levels of 5−50% (Heiser 1951), as compared to less than 1% for crosses between H. annuus and H. petiolaris (Ungerer et al. 1998). The relationships between these and other annual sunflowers are illustrated in Figure 2.
Figure 2.
Phylogenetic relationships among the annual sunflowers, adapted from Rieseberg (1991).
We took advantage of the unique evolutionary and geographic history of these species to study the importance of genetic relatedness and geography in determining interspecific gene flow, sequence divergence and species boundaries. First, an FST-based approach was used to examine gene flow within and between the three species at 26 microsatellite markers, using 1015 individuals from 36 populations across the ranges of the three species. A complementary approach used available EST sequences developed by the Compositae Genome Project (compgenomics.ucdavis.edu). This second approach used only one genotype per species, but enabled us to look at sequence differences at 1420 loci simultaneously. We calculated divergence levels between the three species to identify loci with low divergence between H. annuus and H. petiolaris and extremely high divergence between the sister species H. annuus and H. argophyllus, a pattern consistent with introgression between H. annuus and H. petiolaris at these loci. This second approach enabled us to examine a larger fraction of the genome for signals of introgression.
Methods
MICROSATELLITE ANALYSIS
Plant material
Fresh leaf tissue was collected from 15 to 55 plants from each of 17 populations of H. annuus and 7 populations of H. petiolaris (see Table S1). Additionally, frozen tissue was used from each of four individuals from 12 populations of H. argophyllus (see Table S1). Populations were from across the range of each species (Fig. 1), including H. annuus populations from fields in California, Utah, Colorado, Kansas, Nebraska, North Dakota, South Dakota, Indiana, and Texas; H. petiolaris populations from Utah, Arizona, Colorado, Kansas, Nebraska, North Dakota, and Texas; and H. argophyllus populations from across the species natural range in Texas. These include the 12 populations of H. annuus from Kane and Rieseberg (2008) and the three H. annuus, three H. petiolaris, and 12 H. argophyllus populations from Yatabe et al. (2007), as well as two additional H. annuus populations and three additional H. petiolaris populations. All populations were quite large, with at least several hundred to thousands of plants in the immediate area. Plants were collected along one or more transects in each population, with individuals spaced at least 3 meters apart. DNA was extracted from 50 to 100 mg fresh or frozen leaf tissue using DNeasy 96-well plate kits (QIAGEN Inc., Valencia, CA).
Genotyping
Individuals were genotyped at the 26 most reliable microsatellite loci from previous studies (Kane and Rieseberg 2007; Yatabe et al. 2007; Kane and Rieseberg 2008). These microsatellites were developed from EST libraries (Heesacker et al. 2008) created by the Compositae Genome Project (http://cgpdb.ucdavis.edu/), which includes sequence data for over 94,115 H. annuus, 35,720 H. argophyllus, and 27,484 H. petiolaris ESTs (Barker et al. 2008). Microsatellites included di-, tri-, and tetra- nucleotide repeats. Primers flanking the microsatellite were designed using Primer3, with an additional tail added to the 5′ end of the forward primer for hybridization with fluorescent probes (Schuelke 2000) and synthesized by Integrated DNA Technologies Inc (www.idtdna.com).
During PCR, the 5′ end of the forward primer was labeled using fluorescent dye (Schuelke 2000). The polymerase chain reaction (PCR) reactions were performed in volumes of 10 mL containing 2 ng DNA, 2 units Taq DNA polymerase, using standard “touchdown” PCR protocols to reduce nonspecific primer binding and fragment amplification (Don et al. 1991).
Analysis of microsatellite fragment size was done on the Indiana University Institute for Molecular and Cellular Biology ABI 3730 (Applied Biosystems, Foster City, CA). Multiple, nonoverlapping PCR fragments were pooled and diluted 1:60 with ddH2O. One microliter of the diluted PCR product was added to 8.9 μL ddH2O and 0.1 μL of the GenScan-500 Liz Size Standard (Applied Biosystems). Samples were denatured at 95°C and snap cooled on ice, then centrifuged to remove bubbles before loading onto the ABI 3730. For each marker, genotypes were scored using GeneMapper 3.7 (Applied Biosystems). Markers were scored automatically, with all results inspected and manually corrected in cases of errors in the automatic scoring.
AMOVA
The genetic variance was partitioned into proportions due to genetic differences between species, among populations within species and within populations using analysis of molecular variance (AMOVA) in the program Arlequin 2.0 (Schneider et al. 2000), permuting individual genotypes, with 1000 permutations based on distance matrices.
Population structure and gene flow
Microsatellite Analyzer (MSA, Dieringer and Schlotterer 2002) was used to calculate heterozygosity and variance in microsatellite length, as well as pairwise genetic distances between populations (δμ)2; this measure was chosen because it is expected to be linearly related to time of divergence for microsatellites (Goldstein et al. 1995; Slatkin 1995). Global and pairwise FST were calculated using the Weir and Cockerham method (Weir and Cockerham 1984) using MSA. Neighbor joining trees illustrating population relatedness based on FST and (δμ)2 were created using PHYLIP (Felsenstein 2005).
Bayesian clustering was also used to assess population relatedness and gene flow, using the program Structure 2.2 (Pritchard et al. 2000). The number of populations (K) was estimated using the “admixture” ancestral model with correlated alleles, with K ranging from 2 to 27. Three independent runs of 50,000 MCMC generations and 20,000 generations of “burn-in” were used for each value of K. The true number of populations is expected to be the value of K that maximizes the estimated model log-likelihood, log(P(X|K)) (Falush et al. 2003).
To assess correlations between geographical and genetic distances between populations, Mantel tests (Mantel 1967) were performed in R 2.4.1 (R Development Core Team 2006) based on Pearson's product-moment correlation with 1000 permutations using the Vegan package (Oksanen et al. 2007). Mantel tests were performed separately for H. annuus and H. petiolaris. Because of the small number of individuals available for each H. argophyllus population, we could not perform similar analyses on this species. Traditional Mantel tests cannot be used to assess the relationship between geography and genetic distance between species, because the distance matrices must be symmetrical.
Long-term migration rates
To assess long-term migration rates between the three species, we applied a coalescent approach using the likelihood and Bayesian implementations available in Migrate-n to determine the migration rates and θ (Beerli and Felsenstein 1999, 2001; Beerli 2004). For the purposes of the analysis, we group all populations of H. annuus into a single meta-population, as well as all populations of H. petiolaris and H. argophyllus for a total of three meta-populations. With this methodology we were more interested in the long-term gene flow between species, rather than the migration rate between each sampled population. Due to the high variability of the microsatellite loci, we used the Brownian microsatellite model. We ran two preliminary Migrate-n maximum likelihood analyses based on slightly different models, one model with a Gamma distributed mutation rate across loci, and one without. The inclusion of the Gamma distributed mutation rates did not result in a significantly better model, and therefore we used the model without a Gamma parameter.
We ran two identical maximum likelihood analyses, with different starting seeds, to assess whether the chains had sampled the parameter space properly. Additional parameters for the final maximum likelihood runs included 20 short chains with an increment of 20 and a sampling of 500; five long chains with an increment of 20 and a sampling of 1000; with a burn-in of 100; and the default heating scheme. The Bayesian analyses were run with 5 long-chains, with an increment of 20, and a sampling of 10,0000, with a burn-in of 20,000. Additional Bayesian parameters included an update frequency of 0.4, a Metropolis-Hastings sampling algorithm for both θ and M, uniform priors for θ of 0 to 4, which seemed reasonable according to the mutation rate of these loci and the effective population size determined by Strasburg and Rieseberg (2008), and a uniform prior distribution of M from 0 to 8.
Along with the maximum likelihood analyses we tested several models of migration using the likelihood ratio test available in Migrate-n. We tested all possible combinations of symmetric migration between the species, as well as testing all possible combinations of no migration between each species. We then tested these models against the null-model's maximum likelihood score using both the likelihood ratio test, and the Akaike information criterion (AIC) (Beerli and Felsenstein 1999). Both of these tests are implemented directly in Migrate-n.
Estimates of recent migration
Although estimating migration rates using Migrate-n and a coalescent methodology have many benefits, it also has the drawback of being susceptible to ancestral population genetic forces such as lineage sorting. To determine whether the migration rates estimated from Migrate-n are the result of postdivergence migration we conducted additional analyses that estimate recent migration rates. The program BayesAss uses a Bayesian approach to estimate migrants within multiple populations (Rannala and Yang 2003). The current implementation of BayesAss is limited to 20 populations, we therefore grouped the 12 populations of H. argophyllus into six populations based on geographic proximity. We also excluded four populations of H. annuus, UTR, UTT, UTD, UTC to bring the total under the limit of 20 populations. The added benefit of this method over a standard genetic distance method is the ability to estimate asymmetric migration rates. Because we are determining gene flow between two different species we may not necessarily assume migration is symmetric, or equivalent in either direction.
The BayesAss estimates of migration can also be used to determine if an association exists between migration rate and geographic distance. We calculated the linear regression, R2 coefficient, and then ran 10000 permutations on each regression to determine a P-value for the association. These analyses were done on datasets for gene flow from H. annuus to H. petiolaris, from H. petiolaris to H. annuus, from H. annuus to H. argophyllus, from H. argophyllus to H. annuus, from H. petiolaris to H. argophyllus, and from H. argophyllus to H. petiolaris. We hypothesize that migration rate will decline with increased geographic distance.
BIOINFORMATIC ANALYSES
Whole transcriptome analyses
A bioinformatic pipeline was used to identify all single-copy orthologs found in H. annuus, H. argophyllus, and H. petiolaris. Helianthus EST sequence databases generated by the Compositae Genome Project (http://cgpdb.ucdavis.edu/) were clustered into contigs for each species using TGICL (Quackenbush et al. 2000). These contigs were used in reciprocal all versus all megablast searches in all three possible pairwise comparisons between the three species (Altschul et al. 1990, 1997) to identify significant similarities in sequences found in the three databases. Orthologs were defined conservatively as reciprocal best hits between all pairwise combinations of all three databases, with a minimum cut-off of 90% sequence similarity over 300 base pairs. The best protein hit to available plant proteins in Genbank (as of 7 September 2006) was identified using BLASTX (Altschul et al. 1997), with a minimum e-value of 1e−10. Orthologs without significant protein hits were discarded. Predicted protein sequences for the remaining orthologs were made using the GeneWise hidden Markov models (Birney et al. 2004) implemented in the program Wise2.2, and aligned using MUSCLE3.6 (Edgar 2004). The corresponding coding DNA sequence alignment was found using the program RevTrans1.4 (Wernersson and Pedersen 2003), and formatted for PAML (Yang 1997, 2007) using the program SEALS (Walker and Koonin 1997). The codeml program from PAML3.15 (http://abacus.gene.ucl.ac.uk/software/paml.html) was used to calculate divergence at synonymous sites (Ks) for each ortholog set. Sequences were clustered based on a simple hierarchical clustering algorithm, so that the sequences with the smallest Ks values were paired together. Where Ks values were identical between several pairs of sequences, the relationship was classified as ambiguous. Additionally the best protein hit from Arabidopsis was identified for each ortholog using BLASTX (Altschul et al. 1997) so that genes could be annotated for possible functions categorized according to biological function and the cellular localization of protein products based on Gene Ontology, or GO, functional groups (Rhee et al. 2003). Differences in the distribution of genes into the various GO functional categories were evaluated statistically using chi-square tests, with categories representing 0−5% of the genes combined into an “other” category.
The CGP database contains ESTs for H. petiolaris (27,484 sequences), H. argophyllus (35,720 sequences), and H. annuus (94,115 sequences). For genes that follow the species phylogeny, genetic distances should be greatest between H. petiolaris, and the other two species and lowest between the two sister species H. annuus and H. argophyllus. However, lineage sorting should cause some fraction of the genes to have one of the two alternate topologies (Doyle 1992; Avise 1994, 2000), with the most distant species being either H. annuus or H. argophyllus. The two alternate topologies should be equally likely under lineage sorting. In contrast, recent introgression between H. annuus and H. petiolaris would have a more predictable effect on the phylogeny, reducing genetic distances between H. annuus and H. petiolaris and increasing genetic distances between H. annuus and H. argophyllus (Abbott 1992; Arnold 1992).
We tested for possible effects of rate variation on the observed topologies in 923 ortholog sets from H. annuus, H. petiolaris, H. argophyllus, and H. ciliaris. We used maximum likelihood approach in PAUP (Swofford 2003) to reconstruct a phylogeny for each dataset. We used a general time-reversible model that included a gamma parameter that accounted for rate heterogeneity and invariable sites. Each parameter was estimated separately for each dataset. A likelihood score was obtained with and without a molecular clock enforced. A likelihood ratio test was preformed to determine whether the tree with no molecular clock was significantly more likely.
Results
MICROSATELLITE ANALYSES
Genetic variation within and among populations and species
As expected for microsatellites, these markers exhibited high levels of diversity (Table S2). However, mean expected heterozygosity was significantly lower for H. argophyllus (0.48 ± 0.03; mean ± standard error) than H. annuus (0.68 ± 0.02) or H. petiolaris (0.71 ± 0.02) (P < 0.0001, two-tailed t-test), which were not significantly different from each other. Additionally, H. argophyllus had significant reduction in allele numbers and variance in allele size (P < 0.01, two-tailed t-test), with fewer alleles at most loci than in a typical population of either of the other species. Together, these data are consistent with a substantially lower effective population size for H. argophyllus, perhaps due to its restricted geographical range or population bottlenecks in its history. The two widespread species have probably had very large effective population sizes for most of their recent history (Strasburg and Rieseberg 2008), whereas the more restricted distribution of H. argophyllus, coupled with its more unusual habitat, should give H. argophyllus a much smaller effective population size. Thus, it is not surprising that we found much lower variation and heterozygosity for H. argophyllus at nearly all loci.
The AMOVA found that, as in previous studies of these species (Yatabe et al. 2007), most of the variation (81.1%) was within populations, with only 11.5% of the variation among populations and 7.4% of the variation distributed between species. Correspondingly, FST values between the species were relatively low, with averages of all pairwise FST of 0.19 ± 0.07 between H. annuus and H. petiolaris populations, 0.24 ± 0.03 between H. annuus and H. argophyllus populations, and 0.25 ± 0.06 between H. argophyllus and H. petiolaris populations. It should be pointed out that the FST values involving H. argophyllus are probably inflated by the lower variation present in that species, particularly if this lower variation is due to repeated bottlenecks (Whitlock and McCauley 1990).
Geographic clustering
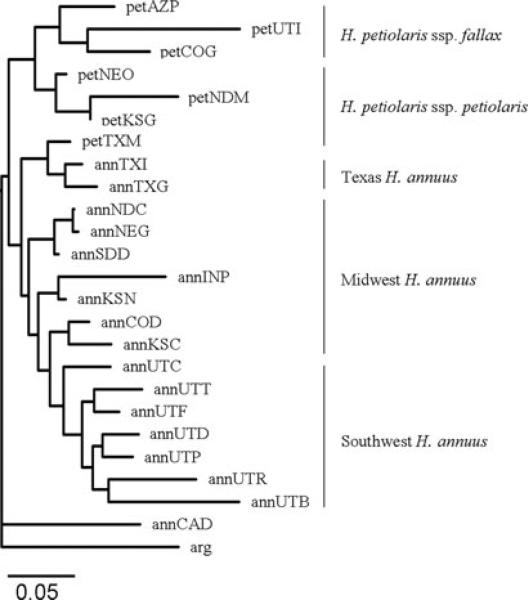
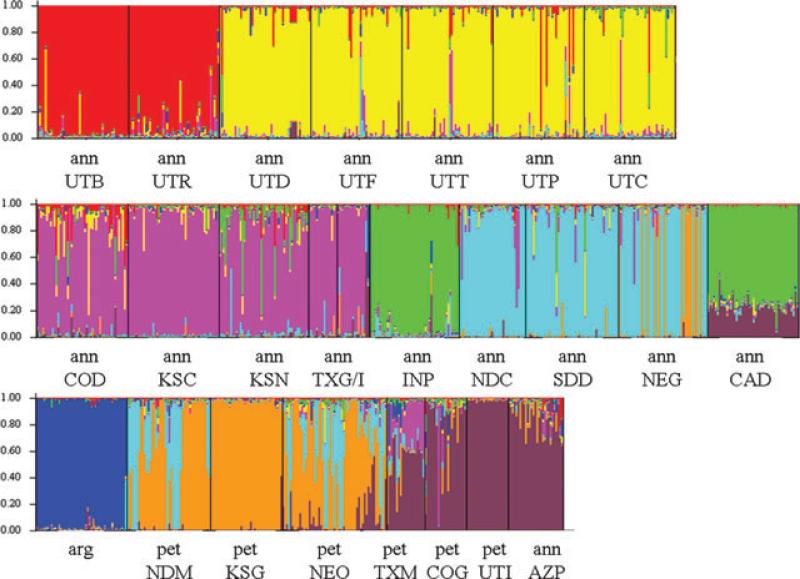
Both the Structure analysis and the FST values showed strong clustering by species and geographic region (Figs. 3 and 4). Populations of H. annuus from Utah tended to cluster together, as did populations from the Midwest. Populations of H. petiolaris also tended to cluster geographically into two groups, one from the Midwest and the other from the southwest, in accordance with standard subspecies classifications (Heiser et al. 1969). However, these analyses also show evidence of substantial levels of gene flow from nearby populations of other species. Interspecific gene flow was particularly striking in some populations. For example, petTXM showed such high levels of gene flow with nearby H. annuus populations that it was grouped with them in the neighbor-joining tree based on FST values (Fig. 3). Virtually identical results were obtained from trees based on (δμ)2. Other populations also had individuals that were classified as hybrids in the STRUCTURE analysis, particularly from the annNEG, petNDM, and petNEO populations. However, most populations showed only limited evidence for introgression. Furthermore, these populations had no noticeable intermediate or hybrid-like morphologies based on our observations, and showed typical traits of pure species.
Figure 3.
Unrooted neighbor joining tree showing the relationships between the 25 populations used, based on genetic distances, FST, using Phylip 3.63 program “neighbor.” Branch lengths represent FST distances (see scale).
Figure 4.
Structure bar plot representing assignments of genotypes to each population. Colors represent assigned genotype. This plot illustrates assignments assuming eight populations, demonstrating the pattern of clustering into regional groups within species.
Mantel tests revealed strong associations between geographical and genetic distances within H. annuus in particular (Mantel R2 = 0.21, P < 0.01). This association was slightly weaker in H. petiolaris, and only marginally significant (Mantel R2 = 0.15, P = 0.066), but there was less power because of the smaller number of populations examined.
Estimates of recent migration
The Migrate-n maximum likelihood analysis showed no evidence of asymmetric migration between H. annuus and H. argophullus, as the likelihood ratio test and AIC show symmetric migration between H. annuus and H. argophyllus is not significantly less likely than the null-model (lnL = 200.1199, P = 0.9646), but the other species pairs show significant asymmetries in the amount of gene flow between species (Table 1, P < 0.00010).
Table 1.
Maximum likelihood estimates and 95% confidence interval of θ and migration rates (M) from Migrate-N.
| 0.05 | MLE | 0.95 | |
|---|---|---|---|
| θann | 1.406184 | 1.420455 | 1.435013 |
| θarg | 0.959232 | 0.993273 | 1.029105 |
| θpet | 1.008772 | 1.028341 | 1.047956 |
| Marg_ann | 2.44585 | 2.579275 | 2.718596 |
| Mpet_ann | 9.78068 | 10.036586 | 10.297104 |
| Mann_arg | 2.449556 | 2.61424 | 2.785474 |
| Mpet_arg | 1.222902 | 1.337824 | 1.459331 |
| Mann_pet | 16.350713 | 16.803071 | 17.256924 |
| Marg_pet | 1.613208 | 1.75335 | 1.902092 |
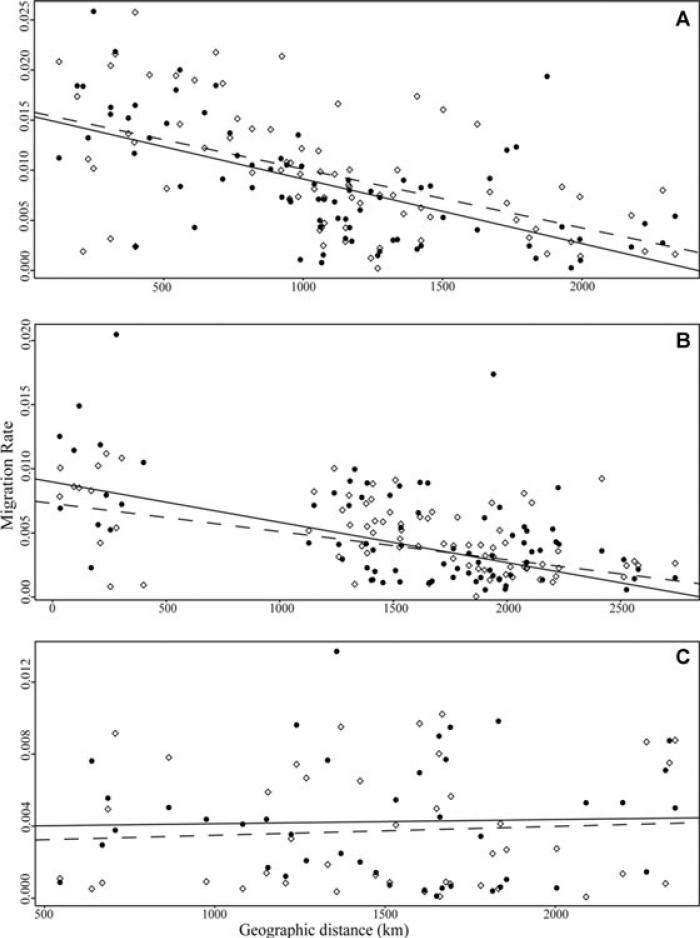
The BayesAss analyses support our estimates of migration rates from Migrate-n. The BayesAss analyses showed significant levels of gene flow for each asymmetric pairwise analysis summarized in Figures 5 and 6. Migration from H. annuus to H. petiolaris was significantly associated with geographic distance, R2 = 0.38 P < 0.0001; as was H. petiolaris to H. annuus, R2 = 0.28, P < 0.0001; and H. annuus to H. argophyllus, R2 = 0.29, P < 0.0001; and H. argophyllus to H. annuus, R2 = 0.28, P < 0.0001. However, the regression was not significant for either H. petiolaris to H. argophyllus, R2 = 0.001, P = 0.83; or from H. argophyllus to H. petiolaris, R2 = 0.005, P = 0.67.
Figure 5.
Scatter plots and regression lines showing the relationship between migration rate and geographic distance for: (A) asymmetric migration between populations of H. annuus and H. petiolaris, •H. petiolaris to H. annuus and solid line of best fit (y = −6.44E−6 x + 1.56E−2), ◇ H. annuus to H. petiolaris and dashed line of best fit (y = −5.87E−6 x + 1.59E−2); (B) asymmetric migration between populations of H. annuus and H. argophyllus, • H. annuus to H. argophyllus and solid line of best fit (y = −3.14E−6 x + 9.84E−3), ◇ H. argophyllus to H. annuus and dashed line of best fit (y = −2.20E−6 x + 8.17E−3); (C) asymmetric migration between populations of H. argophyllus and H. petiolaris, •H. petiolaris to H. argophyllus and solid line of best fit (y = 2.23E−7 x + 3.91E−3), ◇ H. argophyllus to H. petiolaris and dashed line of best fit (y = 4.95E−7 x + 2.99E−3).
Figure 6.
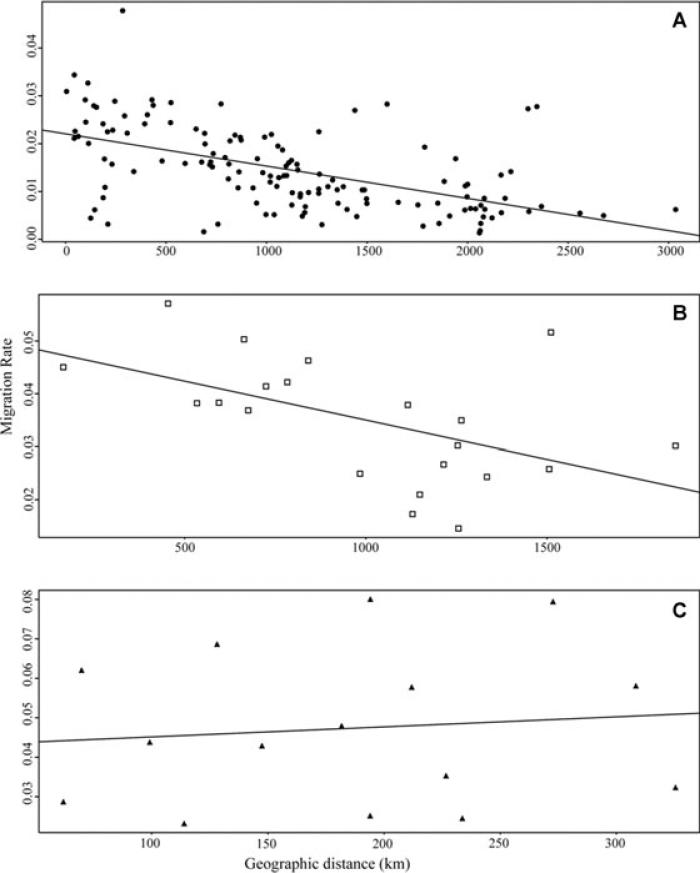
Relationship between migration rates and geographic distance within each species: (A) populations of H. annuus (y = −6.743E−6 x + 2.205E−2); (B) populations of H. petiolaris (y = −1.475E−5 x + 4.975E−2); (C) populations of H. argophyllus (y = 2.541E−5 x + 4.261E−2).
Analyses of single species also showed significant relationships between migration rate and geographic distance for H. annuus and H. petiolaris, Figure 6. However, the regression of migration rate and geographic distance for H. argophyllus populations was not significant, and this may have been due to the grouping of populations, which was necessary due to the low sampling within those populations. The intra-species analysis for H. annuus had a significant regression R2 = 0.3052, P = 0.001; H. petiolaris had a significant R2 = 0.2741, P = 0.0122; and H. argophyllus had an insignificant R2 = 0.01126, P = 0.6916.
BIOINFORMATIC ANALYSES
In the absence of introgression, orthologs from H. annuus are expected to be more similar to H. argophyllus, its sister species, which is thought to have diverged about 1.1 million years ago (90% HPD range 0.74−1.7 million years), than to H. petiolaris, which diverged about 600,000 years earlier and is a member of a more distant clade of annual sunflowers (Rieseberg 1991; Timme et al. 2007; Strasburg and Rieseberg 2008). Of the 1420 loci surveyed, 604 followed the expected topology. However, for many loci a very different pattern was found: H. annuus sequences were actually more similar to H. petiolaris than H. argophyllus for 374 orthologs, a pattern consistent with introgression. Another 278 loci clustered H. petiolaris and H. argophyllus. Finally, 164 orthologs were ambiguous, with differences in Ks that were too small to enable meaningful clustering (Ks below 0.005 between more than one species pair). Out of the 26 loci used in the FST comparisons, nine had ESTs from all three species, including four loci with genes from both domesticated and wild H. annuus. These nine genes showed a similar distribution of topologies, with four genes following the species tree, three showing greater similarity between H. annuus and H. petiolaris and none showing the third topology. The two remaining genes were ambiguous. Thus, while a plurality of the loci did show the expected species phylogeny, a substantial fraction did not, and many of the genes in this latter fraction showed a pattern consistent with recent introgression between H. annuus and H. petiolaris at many loci.
This pattern could also be explained by events other than introgression between H. annuus and H. petiolaris. Fixation (or sampling) of alternate ancestral alleles within the three species could cause this, if H. annuus and H. petiolaris happened to fix a more similar allele whereas H. argophyllus fixed a more divergent one. This is known as lineage sorting, and is expected to occur in young species such as these, based on coalescent theory (Pamilo and Nei 1988; Takahata 1989). This certainly explains some fraction of the loci that does not follow the species phylogeny. However, several factors suggest that this is unlikely to explain all of the loci for which H. annuus and H. petiolaris are closer than either species is to H. argophyllus. First, of the genes not following the species tree, there are substantially more genes that cluster H. annuus and H. petiolaris than genes clustering H. argophyllus and H. petiolaris, significantly more than would be expected by chance, if the two alternate topologies were equally likely (X2 = 14.4; df = 1; P < 0.001). This skewed distribution would be expected if H. annuus and H. petiolaris were still experiencing gene flow. Another potential explanation for the observed pattern could be that rate heterogeneity could cause H. annuus and H. petiolaris to cluster together spuriously. We tested for rate variation in 923 ortholog sets that had clear orthologs in H. annuus, H. petiolaris, H. argophyllus, and H. ciliaris, using maximum likelihood models that included a gamma parameter that accounted for rate heterogeneity. For 417 of the ortholog trees, the likelihood score was significantly higher when the molecular clock was not enforced, indicating significant rate heterogeneity. Slightly more of these trees produced the H. argophyllus/H.petiolaris pairing than H. annuus/H. petiolaris, whereas the 505 gene trees with no significant rate heterogeneity produced more trees with the H. annuus/H. petiolaris relationship. Although these differences are not significant, the trend is in the opposite direction from anything that would create spurious clustering of H. annuus and H. petiolaris. Thus, if anything, rate variation appears to be acting to obscure the signal of introgression between H. annuus and H. petiolaris. In sum, the observed patterns of clustering in the gene trees can be explained by ongoing gene flow between H. annuus and H. petiolaris, but not by other scenarios.
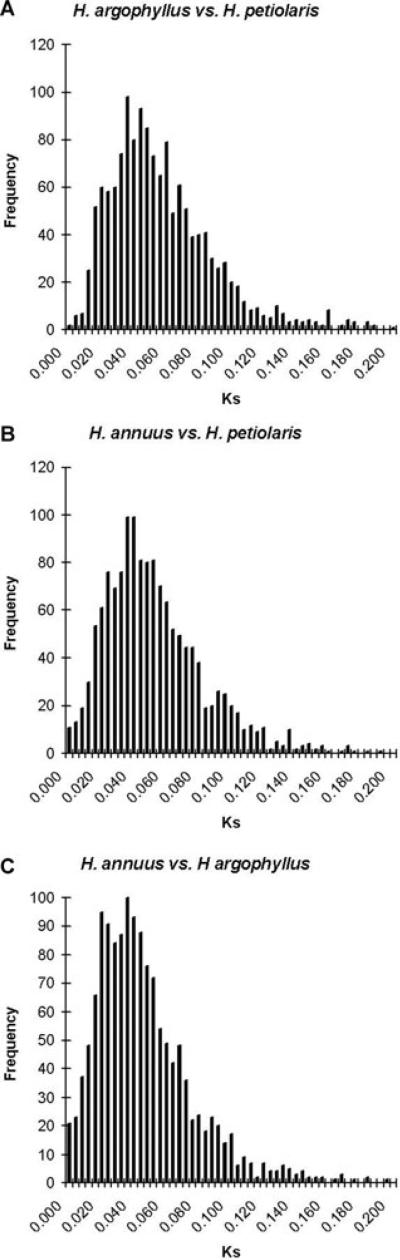
Ks data support this interpretation of the ortholog data. Significantly more genes (54 ortholog sets) had a less than 1% divergence between H. annuus and H. petiolaris, compared to only 26 genes with such low divergence between H. argophyllus and H. petiolaris (X2 = 15.3; df = 1; P < 0.001). The distribution of Ks values over all loci for each species pair also suggest high levels of introgression between H. annuus and H. petiolaris (Fig. 7). In the absence of introgression, comparisons between H. annuus and H. petiolaris should look similar to H. argophyllus and H. petiolaris, because H. annuus and H. argophyllus are sister species. However, Ks is significantly lower (t = 3.5, P < 0.001) between H. annuus and H. petiolaris (0.051 ± 0.01) than Ks between H. argophyllus and H. petiolaris (0.055 ± 0.01). This reduction in average Ks appears to be mainly due to an increase in the number of very low Ks values, some of which are likely introgressed between the two species. For instance, an ortholog similar to the Arabidopsis gene AT3G24360, involved in fatty acid metabolism, shows no synonymous or nonsynonymous changes at all within the coding region when H. annuus and H. petiolaris are compared. However, both species show about 4.8% divergence at synonymous sites compared to H. argophyllus.
Figure 7.
Distribution of Ks values for the three species comparisons. (A) H. annuus versus H. petiolaris. (B) H. argophyllus versus H. petiolaris. (C) H. annuus versus H. argophyllus.
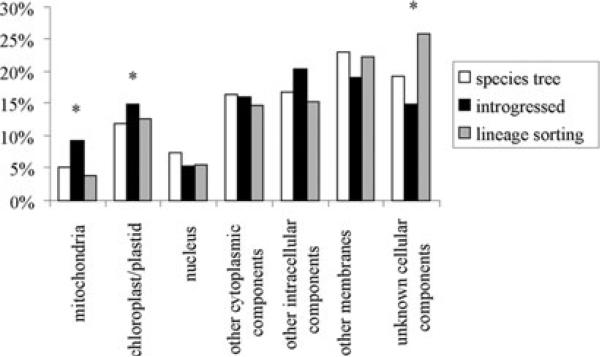
To evaluate whether the genes adhering to the species tree were of different functions than the other genes, we compared the GO functional categories of genes from the three possible topologies (Fig. 8). The cellular localization of gene products differed significantly between the groups when all categories were considered (X2 = 14.8.1; df = 6; P < 0.05). Significantly more genes (X2 = 6.0; df = 2; P < 0.05) targeted to the chloroplast, undifferentiated plastids and mitochondria had lowest Ks between H. annuus and H. petiolaris than in the other two topologies, indicating that introgression between these species may be particularly important for genes targeted to the organelles. For biological function, genes following the species tree appeared to be over-represented in the “response to abiotic or biotic stimulus” and “response to stress” categories. These categories were underrepresented in genes for which H. argophyllus and H. petiolaris alleles are most similar (X2 = 5.2; df = 1; P < 0.05). Of course, for any one gene, it is impossible to determine whether the relationship is caused by lineage sorting or introgression. However, the significant deviations from expectations under neutrality suggest that either introgression is elevated for these gene categories or lineage sorting is for some reason affecting these genes differently from other loci.
Figure 8.
Bar chart showing the GO Cellular Component categories for genes with topology consistent with the species tree (black), introgression between H. annuus and H. petiolaris (white), or lineage sorting resulting in clustering of H. argophyllus and H. petiolaris (gray).
Discussion
GENE FLOW BETWEEN ANNUAL HELIANTHUS SPECIES
Porous boundaries may be expected between recently diverged species (Abbott 1992; Arnold 1992; Rieseberg and Burke 2001). However, the retention of ancestral polymorphisms can be difficult to distinguish from introgression, particularly when using molecular markers (such as microsatellites or AFLPs) rather than DNA sequences. The study presented here used two approaches to get around this issue, and demonstrated the utility of each. In particular, our results show the strength of a population genetic approach across the species range: gene flow between species can be high enough to cause local reductions in interspecific genetic distances. Additionally, our second approach took advantage of available EST sequences to make comparisons of interspecific distances between allopatric and sympatric species, and found that a substantial fraction of the genome appears to have experienced recent gene flow. While the first method examines the geographic scale of gene flow, the second measures the impact of gene flow on a large fraction of the genome. The two approaches are complementary, and the fact that both show evidence of high levels of gene flow confirms that substantial gene flow has certainly occurred between these species.
Importantly, the sympatric species H. annuus and H. petiolaris showed evidence of high levels of gene flow at some loci, whereas other loci exhibited no evidence of gene flow. Genomic evidence for some history of hybridization in phenotypically normal individuals of these (Buerkle and Rieseberg 2001; Yatabe et al. 2007; Strasburg and Rieseberg 2008) and other species (e.g., Arnold et al. 1991; Whittemore and Schaal 1991) has previously been reported and should not be surprising, particularly in species which are known to hybridize widely. However, this paper differs from previous work by analyzing populations from across the species range, giving a broader perspective on the effects of gene flow. Moreover, our novel analysis examining interspecific genetic distances using EST sequences shows that gene flow affects a large proportion of expressed genes.
Previous work on these species has looked at introgression across the genome but only at a small spatial scale (e.g., Yatabe et al. 2007), giving a local rather than a species-wide perspective. Strasburg and Rieseberg (2008) looked at a more regional scale, with sequence data from 18 anonymous loci in a much smaller number of individuals: 25 individuals of H. annuus and 16 individuals of H. petiolaris from the southwestern portion of their range in the United States. Their estimates of gene flow are somewhat lower than ours (Table 1) perhaps due to the more restricted geographic range, or their higher proportion of H. annuus populations in portions of the range far from H. petiolaris populations. Nevertheless, the results presented here are largely consistent with these and other previous results, but give a much broader picture, confirming the effects of introgression across both species’ ranges and across much of the genome.
The extent of gene flow between the species is surprising, particularly given the morphological (Rosenthal et al. 2002, 2005), ecological (Gross et al. 2004), reproductive (Rieseberg et al. 1999), and karyotypic (Rieseberg et al. 1995; Burke et al. 2004) differences. Of particular interest, the significant association between geographical proximity and relatedness in the between-species comparison of sympatric H. annuus and H. petiolaris indicates substantial, long-term local gene flow between nearby populations of the two species, as confirmed by the Migrate analysis. It is presumably the more divergent loci that are responsible for the maintenance of strong reproductive barriers and substantial differences in ecology, morphology and life history between these species despite hundreds of thousands of years of widespread sympatry and gene flow. The recent gene flow levels estimated using BayesAss are higher than the historical gene flow levels estimated by the coalescent approaches of our Migrate analysis and the historical gene flow estimates from Strasburg and Rieseberg (2008). This may represent a substantial increase in the recent gene flow between all these populations, possibly due to human activities, particularly dispersal and disturbance.
In contrast to the relatively high gene flow between H. annuus and H. petiolaris, the more recently diverged species H. annuus and H. argophyllus showed a lower degree of contemporary gene flow, consistent with other evidence that H. argophyllus has been geographically separated from H. annuus for most of its evolution (Heiser 1951). Pairwise FST values between populations were generally higher in this comparison than for H. annuus versus H. petiolaris, although this may be inflated by H. argophyllus's lower level of sequence diversity and smaller effective population size. At the sequence level, most loci show fairly low divergence (Ks 0.044 ± 0.001). However, there are also loci with relatively high sequence divergence between the two. A number of factors may be responsible for this divergence. Presumably, introgression from H. petiolaris into H. annuus explains high divergence levels at many of these loci. While there appears to be significant geographic signature in the gene flow between H. annuus and H. argophyllus, there is no geographic signal in H. argophyllus alone. The low levels of gene flow between H. argophyllus and H. petiolaris could explain the lack of geographic signal from H. petiolaris to H. argophyllus.
Additionally, lineage sorting (fixation of divergent alleles inherited from the common ancestor to the three species) is likely to play a role. The level of lineage sorting apparently observed here would not be at all unexpected given the high effective population sizes and recent origins of these lineages (Strasburg and Rieseberg 2008). As previously discussed, however, the observation that far more genes cluster H. petiolaris with H. annuus than with H. argophyllus can be accounted for by gene flow between the former pair, but not by other scenarios. Thus, although the bioinformatic analysis used many genes but few individuals and the population genetic analyses used many individuals but fewer genes, both approaches show patterns that can only be explained by high levels of gene flow between H. annuus and H. petiolaris, with evidence of less substantial recent gene flow between either species and the more geographically distant species H. argophyllus.
It should be emphasized that all of the populations used appeared to be purely of one species or another, and contained no obvious hybrids based on morphology. None of the individuals were recent hybrids, which are quite easy to identify based on morphology. Thus, although the genetic evidence reveals mixed ancestry in some individuals, the phenotype appeared identical to that of pure species, suggesting that genes involved in species-specific traits are not able to cross the species boundaries. These results indicate that taxa can retain important morphological and ecological differences despite high levels of gene flow across much of the genome.
ECOLOGY AND SYMPATRY
The importance of ecological differences in maintaining species barriers in Helianthus is well appreciated. For example, in the absence of ecological differentiation, hybridization between sympatric Helianthus species can result in merging of the two species, or “genetic assimilation,” as occurred when H. annuus was introduced into the native range of two more narrowly distributed species, H. bolanderi and H. exilis (Heiser 1949; Stebbins and Daly 1961). In many places, interbreeding has resulted in the disappearance of pure H. bolanderi, which has similar habitat requirements to H. annuus, as it has been replaced by H. annuus and hybrid populations (Carney et al. 2000). This is a possible incipient case of extinction by hybridization (Rhymer and Simberloff 1996). However, H. exilis, the sister species of H. bolanderi, which grows in harsh serpentine soils too edaphically stressful for H. annuus (Madhok and Walker 1969), shows little evidence of hybridization or competitive displacement from H. annuus. Because highly porous barriers to interbreeding in these and other sunflower species permit relatively high levels of gene flow, ecological differences may be crucial in maintaining distinct taxa. This is certainly true for maintaining locally adapted ecotypes within Helianthus species (e.g., Sambatti and Rice 2007) but may also be important at the species level.
Selection also acts to hold individual species together as a cohesive unit (Rieseberg and Burke 2001), in addition to creating and maintaining interspecific boundaries (Filchak et al. 2000; Rundle et al. 2000; Via et al. 2000). Many species have levels of intraspecific gene flow that are too low to prevent divergence of distant populations due to drift (Ehrlich and Raven 1969). However, genes under selection can sweep across the range of a species fairly readily (Slatkin 1976). Selective sweeps may therefore contribute to the pattern we observed, in which genes involved in responses to stress or environmental stimuli were least affected by lineage sorting and closely tracked the species tree, as genes in these categories have been shown to be subject to population-wide and regional-wide selective sweeps (Kane and Rieseberg 2007, 2008). In contrast, genes expressed in or targeted to organelles are most susceptible to introgression, an interesting pattern in light of the fact that several previous studies found disproportionally high levels of introgression in chloroplast and mitochondrial genes relative to genes encoded in the nucleus (Rieseberg et al. 1990, 1991; Martinsen et al. 2001), although see Sambatti et al. 2008 for an alternative interpretation. Our findings suggest that nuclear-encoded genes targeted to these organelles also show high levels of introgression.
CONCLUSIONS
Our results indicate that a substantial fraction of the genome can move across species boundaries in Helianthus taxa. This makes sense given the extensive documented hybridization between H. annuus and H. petiolaris (Heiser 1947), and the long period of time over which it has occurred, both of which are expected to lead to gene flow at numerous loci, particularly if barriers to recombination occur at only a few loci (Rieseberg and Wendel 1993; Wu 2001). Their porous genomes show evidence of extensive gene flow at some loci, whereas other loci remain differentiated, presumably due to ecological selection or intrinsic postzygotic barriers. This combined pattern of introgression and sequence divergence supports models of speciation that explicitly incorporate both genetic barriers between species (Feder 1998; Wu 2001; Gavrilets and Vose 2005) and genetic sweeps holding species together (Rieseberg and Burke 2001). A crucial issue for future work will be to examine the nature of the barriers between species at loci showing high levels of diverence: are the low levels of gene flow at particular loci due to karyotypic differences in that genomic region, ecologically mediated selection, intrinsic selection against introgression, or other factors?
In addition to our biological findings, our results demonstrate the utility of two new approaches for the robust demonstration and quantification of interspecific gene flow. The isolation by distance approach using interspecific genetic distances provides a simple, but effective means for identifying interspecific gene flow that can be applied to many preexisting datasets for sympatric species. In contrast, the EST approach provides a means for estimating the fraction of loci across the genome that are affected by introgression, as well as for identifying candidate gene and functional groups that are least likely to be under divergent natural selection or negatively selected because of genetic background effects. Given recent advances in high-throughput sequencing, we anticipate that this latter approach will be increasingly widely employed to study the genomic consequences of introgression in the coming decade.
Supplementary Material
Table S1. Collection locations for populations used.
Table S2. Names, locations, and other information for microsatellite markers used.
ACKNOWLEDGMENTS
The authors would like to thank S. Elmendorf, E. Baack, H. Dempewolf, and two anonymous reviewers for helpful advice and comments. Also, the authors are very grateful to D. Ortiz-Barrientos and B. Gross for providing plant material and DNA from their collections. Additionally, we are indebted to R. Michelmore, A. Kosik, M. Matvienko, Z. Lai, JGI, S. Tang, and other members of the Compositae Genome Project, who generated the cDNA libraries in collaboration with Celera AgGen. This research was funded by the National Science Foundation (DBI0421630) and the National Institutes of Health (GM059065) grants to LHR, as well as National Science Foundation predoctoral and Integrative Graduate Education and Research Traineeship fellowships to NCK.
Footnotes
Supporting Information
The following supporting information is available for this article:
Supporting Information may be found in the online version of this article.
(This link will take you to the article abstract).
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article. Additional results and discussion can be found in a document at http://www.repository.naturalis.nl/record/289893.
LITERATURE CITED
- Abbott RJ. Plant invasions, interspecific hybridization and the evolution of new plant taxa. Trends Ecol. Evol. 1992;7:401–405. doi: 10.1016/0169-5347(92)90020-C. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ML. Natural hybridization as an evolutionary process. Ann. Rev. Ecol. Syst. 1992;23:237–261. [Google Scholar]
- Arnold ML, Buckner CM, Robinson JJ. Pollen-mediated introgression and hybrid speciation in Louisiana irises. Proc. Natl. Acad. Sci. U.S.A. 1991;88:1398–1402. doi: 10.1073/pnas.88.4.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC. Molecular markers, natural history and evolution. Chapman and Hall; New York: 1994. [Google Scholar]
- Avise JC. Phylogeography: The history and formation of species. Harvard Univ. Press; Cambridge, MA: 2000. [Google Scholar]
- Barker MS, Kane NC, Matvienko M, Kozik A, Michelmore RW, Knapp SJ, Rieseberg LH. Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Mol. Biol. Evol. 2008;25:2445–2455. doi: 10.1093/molbev/msn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH. Dynamics of hybrid zones. Heredity. 1979;43:341–359. [Google Scholar]
- Barton NH, Hewitt GM. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 1985;16:113–148. [Google Scholar]
- Beerli P. Migrate: Documentation and program, part of LAMARC. Version 2.4.4. 2004 Available at: http://evolution.gs.washington.edu/lamarc.html.
- Beerli P, Felsenstein J. Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics. 1999;152:763–773. doi: 10.1093/genetics/152.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Richardson AO, Young GJ, Goertzen LR, Palmer JD. Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17747–17752. doi: 10.1073/pnas.0408336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerkle CA, Rieseberg LH. Low intraspecific variation for genomic isolation between hybridizing sunflower species. Evolution. 2001;55:684–691. doi: 10.1554/0014-3820(2001)055[0684:livfgi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Burke JM, Lai Z, Salmaso M, Nakazato T, Tang S, Heesacker A, Knapp SJ, Rieseberg LH. Comparative mapping and rapid karyotypic evolution in the genus Helianthus. Genetics. 2004;167:449–457. doi: 10.1534/genetics.167.1.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney SE, Gardner KA, Rieseberg LH. Evolutionary changes over the fifty-year history of a hybrid population of sunflowers (Helianthus). Evolution. 2000;54:462–474. doi: 10.1111/j.0014-3820.2000.tb00049.x. [DOI] [PubMed] [Google Scholar]
- Chandler JM, Jan CC, Beard BH. Chromosomal differentiation among the annual Helianthus species. Syst. Bot. 1986;11:354–371. [Google Scholar]
- Coyne J, Orr HA. Speciation. Sinauer; Sunderland, MA: 2004. [Google Scholar]
- Dieringer D, Schlotterer C. MICROSATELLITE ANALYSER (MSA): A platform independent analysis tool for large microsatellite data sets. Mol. Ecol. Notes. 2002;3:167–169. [Google Scholar]
- Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. Touchdown PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ. Gene trees and species trees: Molecular systematics as one-character taxonomy. Syst. Bot. 1992;11:373–391. [Google Scholar]
- Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich PR, Raven PH. Differentiation of populations. Science. 1969;165:1228–1232. doi: 10.1126/science.165.3899.1228. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL. The apple maggot fly, Rhagoletis pomonella: Flies in the face of conventional wisdom about speciation?. In: Howard DJ, Berlocher SH, editors. Endless forms: Species and speciation. Oxford Univ. Press; New York: 1998. pp. 130–144. [Google Scholar]
- Felsenstein J. Phylogeny inference package (version 3.2. Cladistics. 2005. pp. 164–166.
- Filchak KE, Roethele JB, Feder JL. Natural selection and sympatric divergence in the apple maggot Rhagoletis pomonella. Nature. 2000;407:739–742. doi: 10.1038/35037578. [DOI] [PubMed] [Google Scholar]
- Gavrilets S, Vose A. Dynamic patterns of adaptive radiation. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18040–18045. doi: 10.1073/pnas.0506330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A, Ferrand N, Nachman MW. Contrasting patterns of introgression at X-linked loci across the hybrid zone between subspecies of the European rabbit (Oryctolagus cuniculus). Genetics. 2006;173:919–933. doi: 10.1534/genetics.105.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DB, Linares AR, Cavallisforza LL, Feldman MW. Genetic absolute dating based on microsatellites and the origin of modern humans. Proc. Natl. Acad. Sci. U.S.A. 1995;92:6723–6727. doi: 10.1073/pnas.92.15.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross BL, Kane NC, Lexer C, Ludwig F, Rosenthal DM, Donovan LA, Rieseberg LH. Reconstructing the origin of Helianthus deserticola: Survival and selection on the desert floor. Am. Nat. 2004;164:145–156. doi: 10.1086/422223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross BL, Schwarzbach AE, Rieseberg LH. Origins of the diploid hybrid species Helianthus deserticola (Asteraceae). Am. J. Bot. 2003;90:1708–1719. doi: 10.3732/ajb.90.12.1708. [DOI] [PubMed] [Google Scholar]
- Heesacker A, Kishore VK, Gao W, Tang S, Kolkman JM, Gingle A, Matvienko M, Kozik A, Michelmore RM, Lai Z, Rieseberg LH, Knapp SJ. SSRs and INDELs mined from the sunflower EST database: Abundance, polymorphisms, and cross-taxa utility of sunflower EST-SSRs. Theor. Appl. Genet. 2008;117:1021–1029. doi: 10.1007/s00122-008-0841-0. [DOI] [PubMed] [Google Scholar]
- Heiser CB. Hybridization between the sunflower species Helianthus annuus and petiolaris. Evolution. 1947;1:249–262. [Google Scholar]
- Heiser CB. Studies in the evolution of the sunflower species Helianthus annuus and H. bollanderi. Univ. Calif. Publ. Bot. 1949;23:157–196. [Google Scholar]
- Heiser CB. Hybridization in the annual sunflowers: Helianthus annuus x H. debilis var. cucumerifolius. Evolution. 1951;5:42–51. [Google Scholar]
- Heiser CB, Smith DM, Clegenger S, Martin WC. The North American sunflowers (Helianthus). Mem. Torrey Bot. Club. 1969;22:1–218. [Google Scholar]
- Kane NC, Rieseberg LH. Selective sweeps reveal candidate genes for adaptation to drought and salt tolerance in common sunflower, Helianthus annuus. Genetics. 2007;175:1823–1824. doi: 10.1534/genetics.106.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane NC, Rieseberg LH. Genetics and the evolution of weediness in Helianthus annuus. Mol. Ecol. 2008;17:384–394. doi: 10.1111/j.1365-294X.2007.03467.x. [DOI] [PubMed] [Google Scholar]
- Lai Z, Nakazato T, Salmaso M, Burke JM, Tang S, Knapp SJ, Rieseberg LH. Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species. Genetics. 2005;171:291–303. doi: 10.1534/genetics.105.042242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Gross BL, Zou Y, Andrews J, Rieseberg LH. Microarray analysis reveals differential gene expression in hybrid sunflower species. Mol. Ecol. 2006;15:1213–1227. doi: 10.1111/j.1365-294X.2006.02775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexer C, Fay MF, Joseph JA, Nica MS, Heinze B. Barrier to gene flow between two ecologically divergent Populus species, P. alba (white poplar. and P. tremula (European aspen): The role of ecology and life history in gene introgression. Mol. Ecol. 2005;14:1045–1057. doi: 10.1111/j.1365-294X.2005.02469.x. [DOI] [PubMed] [Google Scholar]
- Llopart A, Lachaise D, Coyne JA. Multilocus analysis of introgression between two sympatric sister species of Drosophila: Drosophila yakuba and D. santomea. Genetics. 2005;171:197–210. doi: 10.1534/genetics.104.033597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhok OP, Walker RB. Magnesium nutrition of two species of sunflower. Plant Physiol. 1969;44:1016–1022. doi: 10.1104/pp.44.7.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Martinsen GD, Whitham TG, Turek RJ, Keim P. Hybrid populations selectively filter gene introgression between species. Evolution. 2001;55:1325–1335. doi: 10.1111/j.0014-3820.2001.tb00655.x. [DOI] [PubMed] [Google Scholar]
- Mayr E. Animal species and evolution. Harvard Univ. Press; Cambridge, MA: 1963. [Google Scholar]
- Morjan CL, Rieseberg LH. How species evolve collectively: Implications of gene flow and selection for the spread of advantageous alleles. Mol. Ecol. 2004;13:1341–1356. doi: 10.1111/j.1365-294X.2004.02164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MAF, Grams KL, Bertucci LA, Reiland J. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. U.S.A. 2001;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Kindt R, Legendre P, O'Hara RB. Vegan: Community ecology package version 1.8−6. 2007 Available at: http://cran.r-project.org/
- Pamilo P, Nei M. Relationships between gene trees and species trees. Mol. Biol. Evol. 1988;5:568–583. doi: 10.1093/oxfordjournals.molbev.a040517. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J, Liang F, Holt I, Pertea G, Upton J. The TIGR gene indices: Reconstruction and representation of expressed gene sequences. Nucl. Acids Res. 2000;28:141–145. doi: 10.1093/nar/28.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2006. ISBN 3−900051−07−0. Available at: http://www.R-project.org. [Google Scholar]
- Rannala B, Yang ZH. Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics. 2003;164:1645–1656. doi: 10.1093/genetics/164.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al. The Arabidopsis information resource (TAIR): A model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhymer JM, Simberloff D. Extinction by hybridization and introgression. Annu. Rev. Ecol. Syst. 1996;27:83–109. [Google Scholar]
- Rieseberg LH. Homoploid reticulate evolution in Helianthus (Asteraceae): Evidence from ribosomal genes. Am. J. Bot. 1991;78:1218–1237. [Google Scholar]
- Rieseberg LH. Hybrid origins of plant species. Annu. Rev. Ecol. Syst. 1997;28:359–389. [Google Scholar]
- Rieseberg LH. Hybrid speciation in wild sunflowers. Ann. Mo. Bot. Gard. 2006;93:34–48. [Google Scholar]
- Rieseberg LH, Burke JM. The biological reality of species: Gene flow, selection, and collective evolution. Taxon. 2001;50:47–67. [Google Scholar]
- Rieseberg LH, Wendel J. Introgression and its consequences in plants. In: Harrison R, editor. Hybrid zones and the evolutionary process. Oxford Univ. Press; New York: 1993. pp. 70–1114. [Google Scholar]
- Rieseberg LH, Willis JH. Plant speciation. Science. 2007;317:910–914. doi: 10.1126/science.1137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Beckstrom-Sternberg S, Doan K. Helianthus annuus ssp. texanus has chloroplast DNA and nuclear ribosomal RNA genes of Helianthus debilis ssp. cucumerifolius. Proc. Natl. Acad. Sci. U.S.A. 1990;87:593–597. doi: 10.1073/pnas.87.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Choi HC, Ham D. Differential cytoplasmic versus nuclear introgression in Helianthus. J. Hered. 1991;82:489–493. [Google Scholar]
- Rieseberg LH, Linder CR, Seiler GJ. Chromosomal and genic barriers to introgression in Helianthus. Genetics. 1995;141:1163–1171. doi: 10.1093/genetics/141.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Baird SJE, Desrochers AM. Patterns of mating in wild sunflower hybrid zones. Evolution. 1998;52:713–726. doi: 10.1111/j.1558-5646.1998.tb03696.x. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Kim MJ, Seiler GJ. Introgression between the cultivated sunflower and a sympatric wild relative, Helianthus petiolaris (Asteraceae). Int. J. Plant Sci. 1999;160:102–108. [Google Scholar]
- Rosenthal DM, Schwarzbach AE, Donovan LA, Raymond O, Rieseberg LH. Phenotypic differentiation between three ancient hybrid taxa and their parental species. Int. J. Plant Sci. 2002;163:387–398. [Google Scholar]
- Rosenthal DM, Rieseberg LH, Donovan LA. Re-creating ancient hybrid species’ complex phenotypes from early-generation synthetic hybrids: Three examples using wild sunflowers. Am. Nat. 2005;166:26–41. doi: 10.1086/430527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundle HD, Nagel L, Boughman JW, Schluter D. Natural selection and parallel speciation in sympatric sticklebacks. Science. 2000;287:306–308. doi: 10.1126/science.287.5451.306. [DOI] [PubMed] [Google Scholar]
- Sambatti JBM, Rice KJ. Functional ecology of ecotypic differentiation in the Californian serpentine sunflower Helianthus exilis. New Phytol. 2007;175:107–119. doi: 10.1111/j.1469-8137.2007.02068.x. [DOI] [PubMed] [Google Scholar]
- Sambatti JBM, Ortiz-Barrientos D, Baack EJ, Rieseberg LH. Ecological selection maintains cytonuclear incompatibilities in hybridizing sunflowers. Ecol. Lett. 2008;11:1082–1091. doi: 10.1111/j.1461-0248.2008.01224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling EE, Heiser CB. Infrageneric classification of Helianthus (Compositae). Taxon. 1981;30:393–403. [Google Scholar]
- Schilling EE, Linder CR, Noyes RD, Rieseberg LH. Phylogenetic relationships in Helianthus (Asteraceae) based on nuclear ribosomal DNA internal transcribed spacer region sequence data. Syst. Bot. 1998;23:177–187. [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Arlequin: A software for population genetics data analysis. Ver 2.000. Genetics and Biometry Lab, Dept. of Anthropology, University of Geneva; 2000. [Google Scholar]
- Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000;18:233–234. doi: 10.1038/72708. [DOI] [PubMed] [Google Scholar]
- Schwarzbach AE, Rieseberg LH. Likely multiple origins of a diploid hybrid sunflower species. Mol. Ecol. 2002;11:1703–1715. doi: 10.1046/j.1365-294x.2002.01557.x. [DOI] [PubMed] [Google Scholar]
- Scotti-Saintagne C, Mariette S, Porth I, Goicoechea PG, Barreneche T, Bodenes C, Burg K, Kremer A. Genome scanning for interspecific differentiation between two closely related oak species (Quercus robur L. and Q petraea [Matt. Liebl.]). Genetics. 2004b;168:1615–1626. doi: 10.1534/genetics.104.026849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. The rate of spread of an advantageous allele in a subdivided population. In: Karlin S, Nevo E, editors. Population genetics and ecology. Academic Press; New York: 1976. pp. 767–780. [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL, Daly K. Changes in the variation pattern of a hybrid population of Helianthus over an eight-year period. Evolution. 1961;15:60–71. [Google Scholar]
- Strasburg JL, Rieseberg LH. Molecular demographic history of the annual sunflowers Helianthus annuus and H. petiolaris— large effective population sizes and rates of long-term gene flow. Evolution. 2008;62:1936–1950. doi: 10.1111/j.1558-5646.2008.00415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump AD, Fitzpatrick MC, Lobo NF, Traore S, Sagnon N, Costantini C, Collins FH, Besansky NJ. Centromere-proximal differentiation and speciation in Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15930–15935. doi: 10.1073/pnas.0508161102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates; Sunderland, MA: 2003. [Google Scholar]
- Takahata N. Gene geneology in three related populations: Consistency probability between gene and population trees. Genetics. 1989;122:957–966. doi: 10.1093/genetics/122.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme RE, Simpson BB, Linder RC. High-resolution phylogeny for Helianthus (Asteraceae) using the 18S-26S ribosomal DNA external transcribed spacer. Am. J. Bot. 2007;94:1837–1852. doi: 10.3732/ajb.94.11.1837. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. Plos Biol. 2005;3:1572–1578. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer MC, Baird SJE, Pan J, Rieseberg LH. Rapid hybrid speciation in wild sunflowers. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11757–11762. doi: 10.1073/pnas.95.20.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S, Bouck AC, Skillman S. Reproductive isolation between divergent races of pea aphids on two hosts. II. Selection against migrants and hybrids in the parental environments. Evolution. 2000;54:1626–1637. doi: 10.1111/j.0014-3820.2000.tb00707.x. [DOI] [PubMed] [Google Scholar]
- Walker DR, Koonin EV. SEALS: A system for easy analysis of lots of sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1997;5:333–339. [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Welch ME, Rieseberg LH. Patterns of genetic variation suggest a single, ancient origin for the diploid hybrid species Helianthus paradoxus. Evolution. 2002;56:2126–2137. doi: 10.1111/j.0014-3820.2002.tb00138.x. [DOI] [PubMed] [Google Scholar]
- Wernersson R, Pedersen AG. RevTrans: Multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC, McCauley DE. Some population genetic consequences of colony formation and extinction: Genetic correlations within founding groups. Evolution. 1990;44:1717–1724. doi: 10.1111/j.1558-5646.1990.tb05243.x. [DOI] [PubMed] [Google Scholar]
- Whitmore AT, Schaal BA. Interspecific gene flow in sympatric oaks. Proc. Natl. Acad. Sci. U.S.A. 1991;88:2540–2544. doi: 10.1073/pnas.88.6.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI. The genic view of the process of speciation. J. Evol. Biol. 2001;14:851–865. [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. BioSci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: A program package for phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yatabe Y, Kane NC, Scotti-Saintagne C, Rieseberg LH. Rampant gene exchange across a strong reproductive barrier between the annual sunflowers, Helianthus annuus and H. petiolaris. Genetics. 2007;175:1883–1893. doi: 10.1534/genetics.106.064469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Collection locations for populations used.
Table S2. Names, locations, and other information for microsatellite markers used.