Abstract
MSX1 has a critical role in craniofacial development, as indicated by expression assays and transgenic mouse phenotypes. Previously, MSX1 mutations have been identified in three families with autosomal-dominant tooth agenesis. To test the hypothesis that MSX1 mutations are a common cause of congenital tooth agenesis, we screened 92 affected individuals, representing 82 nuclear families, for mutations, using single-strand conformation analysis. A Met61Lys substitution was found in two siblings from a large family with autosomal-dominant tooth agenesis. Complete concordance of the mutation with tooth agenesis was observed in the extended family. The siblings have a pattern of severe tooth agenesis similar that in to previous reports, suggesting that mutations in MSX1 are responsible for a specific pattern of inherited tooth agenesis. Supporting this theory, no mutations were found in more common cases of incisor or premolar agenesis, indicating that these have a different etiology.
Keywords: MSX1, homeobox, odontogenesis, dental patterning
Introduction
Congenital agenesis of one or more permanent teeth, also known as hypodontia, is among the most well-recognized morphologic anomalies in humans, and yet the etiology is largely unknown (Vastardis, 2000). Oligodontia has been defined as agenesis of more than 6 permanent teeth (Stockton et al., 2000). In Caucasians, tooth agenesis most commonly involves third molars, with from 10 to 25% of the population affected. Reports on the overall incidence of missing permanent teeth, excluding third molars, vary substantially, from 2% to 10%. In Caucasians, approximately 80% of tooth agenesis cases involve only one or two teeth.
Evidence supporting a genetic etiology for tooth agenesis is well-established (reviewed by (Vastardis, 2000). Tooth agenesis usually presents as an isolated anomaly. However, it is known to occur in association with syndromes or inherited disorders, many of which have known genetic defects (Gorlin, 1990).
It has been proposed that tooth shape and position are specified by multiple homeobox genes expressed in neural-crest-derived mesenchyme in the mandibular and maxillary processes of the first branchial arch (Sharpe, 1995; Thomas and Sharpe, 1998). These expression patterns and the dental abnormalities observed in transgenic mice of these genes support such a theory (Satokata and Maas, 1994; Qiu et al., 1997; Winograd et al., 1997; Satokata et al., 2000).
MSX1, a non-clustered homeobox protein, has considerable evidence suggesting that it plays a role in dental development. Homozygous Msx1-deficient mice have complete secondary cleft palate, complete failure of incisor development, and bud-stage arrest of molar development (Satokata and Maas, 1994). An Arg196Pro missense mutation in the homeodomain of MSX1 was described in a large family with a severe form of autosomal-dominant tooth agenesis (Vastardis et al., 1996). Another large family with a similar pattern of tooth agenesis was found to have a MSX1 Ser105Stop mutation (van den Boogaard et al., 2000). Interestingly, some affected individuals also had cleft lip or cleft palate, extending the phenotypes associated with MSX1 mutations in humans and supporting previous associations reported between MSX1 and non-syndromic cleft lip and cleft palate (Lidral et al., 1998; Beaty et al., 2001; Blanco et al., 2001). More recently, a MSX1 Ser202Stop mutation was reported to be associated with the Witkop syndrome, which includes tooth agenesis and nail dysgenesis (Jumlongras et al., 2001).
Thus, Msx1 expression is essential for murine dental development, and mutations in the homeobox of human MSX1 are responsible for some instances of tooth agenesis in humans. The purpose of this study is to test the hypothesis that mutations in MSX1 are common causes of tooth agenesis of the human permanent dentition.
Materials & Methods
Patient and Control Samples
A total of 92 individuals, constituting 82 nuclear families, affected with congenital tooth agenesis was recruited from The Ohio State University College of Dentistry and Children's Hospital in Columbus, OH. Written consent was obtained, and the study was approved by the Ohio State University IRB (Protocol No. 97H0047). The inclusion criterion was congenital agenesis of at least one secondary tooth, not including third molars, as verified by radiographs and dental history. Instances of tooth agenesis adjacent to a cleft site were not included, because the absence of such teeth is likely the consequence of local developmental anomalies at the cleft site. Third molar agenesis was not characterized in all subjects, since some were too young for this trait to be determined. The Universal Tooth Numbering System was used to designate which teeth were missing (Parreidt, 1882). Medical, birth defect, and family histories were gathered to identify possible associated anomalies. In addition, 40 Caucasian controls, from Ohio, who were not affected with tooth agenesis (excluding third molars) were also recruited.
MSX1 Mutation Screen and Sequencing
DNA was extracted from all 92 subjects (Richards et al., 1993), and 8 overlapping primer pairs spanning the two MSX1 exons were amplified and screened for single-strand conformation polymorphisms (SSCPs) (Lidral et al., 1998). Positive controls consisting of known MSX1 variants (Lidral et al., 1998) were also loaded on the gel for comparison with observed shifts in the tooth agenesis samples. Shifted bands were excised from the gel, re-amplified, and re-analyzed by single-strand conformation analysis (SSCA). This revealed that the shifted bands were true SSCPs and not heteroduplexes. Both the genomic DNA and the excised band shifts were sequenced in both directions by means of the ABI Prism BigDye terminator cycle sequencing kit on an ABI 310 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Linkage Analysis
Nine additional family members of the affected siblings with the Met61Lys mutation were recruited, for a total of seven affected and four unaffected individuals. The T to A change at nucleotide 620 obliterates a NlaIII restriction site. The entire family was genotyped for the Met61Lys mutation by NlaIII digestion of the X1.2 PCR products. We performed parametric linkage analysis using GENEHUNTER (Kruglyak et al., 1996), with the penetrance set at 95% for an autosomal-dominant inheritance pattern, 5% phenocopy rate, and a 2.4% disease allele frequency.
Results
Tooth Agenesis Frequency
A total of 92 individuals constituted the sample, representing 82 unrelated families. Eight were non-Caucasian: two Asian, three African, one Iranian, one Latin-American, and one Indian. The age range was from 5 to 57 yrs old. Thirty-one had an additional first-degree relative reported to have tooth agenesis. Fifteen are known to be affected with another congenital abnormality or systemic disease; eight are affected with cleft lip, cleft palate, or submucous cleft palate, of which one also has subaortic stenosis, three with heart defects, one with heart, kidney, and ear anomalies, two siblings with hypospadias and undescended testicles, and one with mitral prolapse, essential hypertension, and thyroid disease. Sixty-nine individuals in the sample are Caucasians unaffected by other congenital abnormalities or systemic diseases. Excluding third molar agenesis, 74% are missing only one or two teeth, while only 10% are missing 5 or more (Fig. 1). The most commonly missing teeth are the mandibular second premolars, followed by the maxillary lateral incisors and subsequently the maxillary second premolars (Fig. 2). These findings are in agreement with previous reports on dental agenesis in Caucasian populations.
Figure 1.
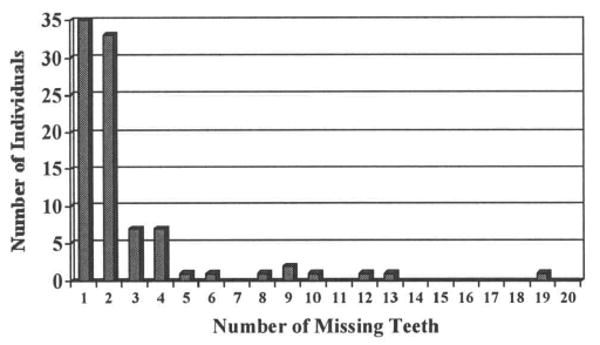
Number of missing teeth per individual, not including 3rd molar agenesis.
Figure 2.
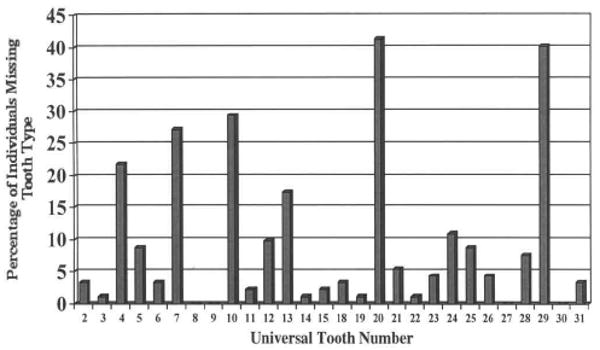
Frequency of individuals missing each tooth type.
MSX1 Mutation Screen and Sequencing
No new variants were observed in 90 subjects in any portion of the coding region of MSX1. Novel SSCPs were observed in two affected siblings by means of the MSX1 X1.2 primers (Fig. 3). These siblings are from a large pedigree of 16 individuals affected with tooth agenesis segregating in an autosomal-dominant manner. Sequencing confirmed a T to A mutation for nucleotide 620 (Hewitt et al., 1991), resulting in a Met61Lys substitution (Fig. 3). NlaIII restriction digest and agarose gel electrophoresis of the 11 available family members verified the presence of the mutation in all seven affecteds, but not in the four unaffecteds or controls (data not shown). Linkage analysis yielded a LOD score of 1.68 (theta = 0.0). Furthermore, the mutation was not found in 80 Caucasian control chromosomes or in a previous study of 24 CL/P and 69 patients with isolated cleft palate (Lidral et al., 1998).
Figure 3.
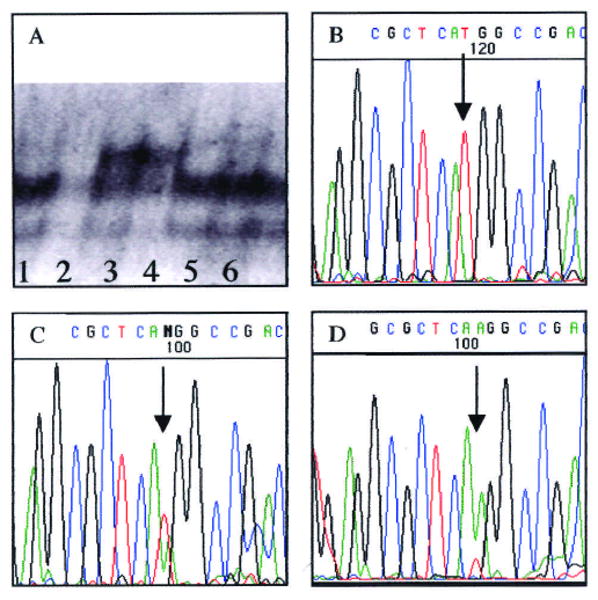
MSX1 mutation and sequence. (A) Lanes 3 and 4 show the band shift indicating SSCPs in the two initially identified siblings. Lanes 1, 2, 5, and 6 show unrelated individuals. (B) MSX1 sequence of unaffected individual; arrow indicates nucleotide 620, which matches published sequence (Hewitt et al., 1991). (C) MSX1 sequence of an affected heterozygous individual, with arrow indicating location of overlapping A and T peaks. (D) MSX1 sequence of DNA isolated from band shift. Arrows indicate mutated nucleotide.
Discussion
This study reveals a Met61Lys mutation in a large kindred with hereditary tooth agenesis, resulting in a LOD score of 1.68, which is the maximum possible LOD score for this family. The 100% concordance in this large pedigree and the fact that we did not find the mutation on 80 normal control chromosomes confirm the linkage of MSX1 mutations and hereditary tooth agenesis. This is in contrast to two studies that did not find MSX1 linkage or mutations in families or subjects with tooth agenesis (Nieminen et al., 1995; Scarel et al., 2000). Currently, PAX9 is the only other gene having a mutation associated with isolated tooth agenesis (Stockton et al., 2000).
In the first report linking tooth agenesis with MSX1 mutation, tooth agenesis was reported to be associated with mild maxillary anterior-posterior hypoplasia (Vastardis et al., 1996). Others have postulated that congenital absence of teeth may result in decreased mesenchymal tissues required for normal growth of the maxilla (Ferguson, 1994). However, cephalometric analysis of the initially identified two affected siblings in this family revealed normal facial proportions (data not shown).
The fact that only one instance of MSX1 mutation was found in our sample indicates that other factors are responsible for the more commonly occurring cases of incisor or premolar agenesis. Eight individuals in this study had cleft lip, cleft palate, or submucous cleft palate in addition to tooth agenesis. However, no MSX1 mutations were found in these individuals, suggesting that the clefts were due to other causes. All of the individuals in this study who also had an orofacial cleft were missing only 1 or 2 teeth. Given the high frequency of tooth agenesis in the general population, it may be coincidence that both phenotypes were observed.
The Met61Lys mutation may alter normal MSX1 function by a variety of mechanisms. MSX and DLX proteins have been shown to form dimeric complexes (Zhang et al., 1997). DLX proteins have also been shown to be important in dental development (Qiu et al., 1997). The overlapping expression patterns of MSX and DLX genes and their involvement in epithelial-mesenchymal signaling cascades of murine odontogenesis suggest that MSX and DLX proteins form heterodimeric complexes in vivo which provide a mechanism for transcriptional regulation via functional antagonism. It is possible that the Met61Lys may interfere with dimerization of MSX1 with DLX proteins. MSX1 has also been shown to interact with TBP and other components of the core transcriptional complex (Catron et al., 1995; Zhang et al., 1996). However, both the interactions with DLX proteins and TBP have been shown to be mediated by the homeodomain, specifically amino acids in the N-terminal region, which are located some distance from the Met61Lys mutation, suggesting that interference with these interactions may not be a likely explanation. Alternatively, a likely explanation may involve a highly conserved 10- to 12-amino-acid region (53-LPFSVEALMA-62) in MSX1, MSX2, and MSX3 across many species (Ekker et al., 1997). This region, which is quite hydrophobic, has also been suggested to be similar to the engrailed homology (EH-1) repression domain (Smith and Jaynes, 1996). Repression by the EH-1 domain is mediated by Groucho, a basic-helix-loop-helix protein, that interacts with a variety of motifs in other transcription repression proteins (Jimenez et al., 1999). Thus, this conserved MSX1 region may also have repression activity and interact with Groucho. Previous studies did not identify this region as being involved in MSX1 repression (Catron et al., 1995, 1996). However, the Met61Lys mutation may be affecting an uncharacterized repression domain in the protein.
In the family reported here, the second premolars and the third molars were most often missing in the five affected individuals in whom the identity of the affected teeth could be verified (Table). Thus, there is a similar pattern of tooth agenesis among the four families reported to have MSX1 mutations (Vastardis et al., 1996; van den Boogaard et al., 2000; Jumlongras et al., 2001). The lower second premolars are the most commonly affected, followed by upper second premolars, upper first premolars, and upper lateral incisors. Third molar agenesis, while difficult to document due to the young age in some subjects, also is a common feature in these families. The most distal tooth of each tooth type is most often affected, and, as the severity worsens, there appears to be a pattern of anterior progression of agenesis for each tooth type. Another striking feature of MSX1 mutations is the large number of teeth missing in the affected individuals, with an average of 11.0/person (Vastardis et al., 1996), 8.4/person (van den Boogaard et al., 2000), 16.4/person (Jumlongras et al., 2001), and 12.2/person in this report. MSX1 phenotypes are in contrast to PAX9 phenotypes, in that molar agenesis was observed in the families with PAX9 mutations (Stockton et al., 2000).
Table.
Summary of Congenitally Missing Teeth in Family with Met61Lys Mutation
| Right | Left | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID Number | 8a | 7 | 6 | 5 | 4 | 3 | 2 | 1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| III-3 Maxillary | Wearing maxillary denture; therefore unable to identify missing teeth. | |||||||||||||||
| Mandibular | * | * | * | * | * | * | * | |||||||||
| III-5 Maxillary | * | * | * | * | * | * | ||||||||||
| Mandibular | * | * | * | * | ||||||||||||
| IV-4 Maxillary | * | * | * | * | * | * | ||||||||||
| Mandibular | * | * | * | |||||||||||||
| IV-8 Maxillary | * | * | * | * | * | * | * | * | ||||||||
| Mandibular | * | * | * | * | * | * | * | * | * | |||||||
| IV-9 Maxillary | * | * | * | * | * | * | ||||||||||
| Mandibular | * | * | * | * | * | * | ||||||||||
1 = central incisor; 2 = lateral incisor; 3 = canine; 4 and 5 = first and second premolars, respectively; 6, 7, and 8 = first, second, and third molars, respectively.
Interestingly, the existing teeth of the two affected siblings in this study, for whom orthodontic study models were available, are slightly smaller than average (data not shown). The maxillary second molars do not have a distal lingual cusp, and the mandibular first molars have only 4 cusps, missing the distal buccal cusp. Hence, there appears to be a transformation to a morphology of a more posterior tooth type. These findings suggest that MSX1 may play a role in both the patterning and morphogenesis of the dentition. MSX1 is expressed in the developing tooth bud during morphogenesis (MacKenzie et al., 1991), supporting this theory.
The hypodontia pattern observed with MSX1 mutations suggests that a threshold level of MSX1 function is vital in the development of only selected teeth and that MSX1 functions to pattern the dentition. This corroborates the hypothesized odontogenic homeobox code proposed by Sharpe (1995). The variation observed suggests that other factors modulate the effects of MSX1 mutations. Thus, we conclude that MSX1 mutations result in a specific pattern of inherited tooth agenesis, and the cause for the more common cases of tooth agenesis, where only one or two teeth are missing, is not explained by MSX1 mutations.
Acknowledgments
The invaluable cooperation of the participating patients is greatly appreciated. In addition, the efforts by the following individuals in identifying patients is recognized: Drs. Heather Abrahams, Mark Bentele, Alex Cassinelli, Dale Anne Featheringham, Jim Eimer, Brian Hockenberger, Ceil Markham, Ana Mercado, Elizabeth Peruchini, Robert Pham, Arnold Riesmeijer, Michelle Renick, Brenda Wilhelm, Roger Zody. Karen Neal, Valarie Jones, and Sam King, and Larry Druhan provided invaluable technical assistance. Insightful comments were provided by Drs. Lina Moreno and Peter Jezewski. The efforts of Drs. Wayne Mahar, Jr., Joseph Moriello, and Stanley Vermilyea to verify the identity of missing teeth in the extended family are greatly appreciated. This project was supported by start-up funds provided by the College of Dentistry, Ohio State University.
References
- Beaty TH, Wang H, Hetmanski JB, Fan YT, Zeiger JS, Liang KY, et al. A case-control study of nonsyndromic oral clefts in Maryland. Ann Epidemiol. 2001;11:434–442. doi: 10.1016/s1047-2797(01)00222-8. [DOI] [PubMed] [Google Scholar]
- Blanco R, Chakraborty R, Barton SA, Carreno H, Paredes M, Jara L, et al. Evidence of a sex-dependent association between the MSX1 locus and nonsyndromic cleft lip with or without cleft palate in the Chilean population. Hum Biol. 2001;73(1):81–89. doi: 10.1353/hub.2001.0002. [DOI] [PubMed] [Google Scholar]
- Catron KM, Zhang H, Marshall SC, Inostroza JA, Wilson JM, Abate C. Transcriptional repression by Msx-1 does not require homeodomain DNA-binding sites. Mol Cell Biol. 1995;15:861–871. doi: 10.1128/mcb.15.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catron KM, Wang H, Hu G, Shen MM, Abate-Shen C. Comparison of MSX-1 and MSX-2 suggests a molecular basis for functional redundancy. Mech Dev. 1996;55:185–199. doi: 10.1016/0925-4773(96)00503-5. [DOI] [PubMed] [Google Scholar]
- Ekker M, Akimenko M, Allende M, Smith R, Drouin G, Langille R, et al. Relationships among msx gene structure and function in zebrafish and other vertebrates. Mol Biol Evol. 1997;14:1008–1022. doi: 10.1093/oxfordjournals.molbev.a025707. [DOI] [PubMed] [Google Scholar]
- Ferguson MWJ. Craniofacial malformations: towards a molecular understanding. Nat Genet. 1994;6:329–330. doi: 10.1038/ng0494-329. [DOI] [PubMed] [Google Scholar]
- Gorlin RJ. Syndromes of the head and neck. New York: Oxford University Press; 1990. [Google Scholar]
- Hewitt JE, Clark LN, Ivens A, Williamson R. Structure and sequence of the human homeobox gene HOX7. Genomics. 1991;11:670–678. doi: 10.1016/0888-7543(91)90074-o. [DOI] [PubMed] [Google Scholar]
- Jimenez G, Verrijzer CP, Ish-Horowicz D. A conserved motif in goosecoid mediates groucho-dependent repression in Drosophila embryos. Mol Cell Biol. 1999;19:2080–2087. doi: 10.1128/mcb.19.3.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumlongras D, Bei M, Stimson JM, Wang WF, DePalma SR, Seidman CE, et al. A nonsense mutation in MSX1 causes Witkop syndrome. Am J Hum Genet. 2001;69:67–74. doi: 10.1086/321271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- Lidral AC, Romitti PA, Basart AM, Doetschman T, Leysens NJ, Daack-Hirsch S, et al. Association of MSX1 and TGFB3 with nonsyndromic clefting in humans. Am J Hum Genet. 1998;63:557–568. doi: 10.1086/301956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie A, Leeming GL, Jowett AK, Ferguson MWJ, Sharpe PT. The homeobox gene Hox 7.1 has specific regional and temporal expression patterns during early murine craniofacial embryogenesis, especially tooth development in vivo and in vitro. Development. 1991;111:269–285. doi: 10.1242/dev.111.2.269. [DOI] [PubMed] [Google Scholar]
- Nieminen P, Arte S, Pirinen S, Peltonen L, Thesleff I. Gene defect in hypodontia: exclusion of MSX1 and MSX2 as candidate genes. Human Genet. 1995;96:305–308. doi: 10.1007/BF00210412. [DOI] [PubMed] [Google Scholar]
- Parreidt J. Zählung der Zähne und Benennung der verschiedenen Zahnsorten. In: Felix A, editor. Zahnärzliche Mitteilungen aus der chirurgischen Universitätspoliklinik zu Leipzig. 1882. pp. 10–15. [Google Scholar]
- Qiu M, Bulfone A, Ghattas I, Meneses JJ, Christensen L, Sharpe PT, et al. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev Biol. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- Richards B, Skoletsky J, Shuber AP, Balfour R, Stern RC, Dorkin HL, et al. Multiplex PCR amplification from the CFTR gene using DNA prepared from buccal brushes/swabs. Hum Molecular Genet. 1993;2:159–163. doi: 10.1093/hmg/2.2.159. [DOI] [PubMed] [Google Scholar]
- Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, et al. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat Genet. 2000;24:391–395. doi: 10.1038/74231. [DOI] [PubMed] [Google Scholar]
- Scarel RM, Trevilatto PC, Di Hipolito O, Jr, Camargo LE, Line SR. Absence of mutations in the homeodomain of the MSX1 gene in patients with hypodontia. Am J Med Genet. 2000;92:346–349. doi: 10.1002/1096-8628(20000619)92:5<346::aid-ajmg10>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Sharpe PT. Homeobox genes and orofacial development. Connect Tissue Res. 1995;32(14):17–25. doi: 10.3109/03008209509013701. [DOI] [PubMed] [Google Scholar]
- Smith S, Jaynes J. A conserved region of engrailed, shared among all en-, gsc-, Nk1-, Nk2- and msh-class homeoproteins, mediates active transcriptional repression in vivo. Development. 1996;122:3141–3150. doi: 10.1242/dev.122.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockton DW, Das P, Goldenberg M, D'Souza RN, Patel PI. Mutation of PAX9 is associated with oligodontia. Nat Genet. 2000;24:18–19. doi: 10.1038/71634. [DOI] [PubMed] [Google Scholar]
- Thomas BL, Sharpe PT. Patterning of the murine dentition by homeobox genes. Eur J Oral Sci. 1998;106(Suppl 1):48–54. doi: 10.1111/j.1600-0722.1998.tb02153.x. [DOI] [PubMed] [Google Scholar]
- van den Boogaard MJ, Dorland M, Beemer FA, van Amstel HK. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat Genet. 2000;24:342–343. doi: 10.1038/74155. published erratum appears in. [DOI] [PubMed] [Google Scholar]; Nat Genet. 2000 May;25(1):125. doi: 10.1038/75532. [DOI] [PubMed] [Google Scholar]
- Vastardis H. The genetics of human tooth agenesis: new discoveries for understanding dental anomalies. Am J Orthod Dentofac Orthop. 2000;117:650–656. [PubMed] [Google Scholar]
- Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet. 1996;13:417–421. doi: 10.1038/ng0896-417. see comments. [DOI] [PubMed] [Google Scholar]
- Winograd J, Reilly MP, Roe R, Lutz J, Laughner E, Xu X, et al. Perinatal lethality and multiple craniofacial malformations in MSX2 transgenic mice. Hum Mol Genet. 1997;6:369–379. doi: 10.1093/hmg/6.3.369. [DOI] [PubMed] [Google Scholar]
- Zhang H, Catron KM, Abate-Shen C. A role for the Msx-1 homeodomain in transcriptional regulation: residues in the N-terminal arm mediate TATA binding protein interaction and transcriptional repression. Proc Natl Acad Sci USA. 1996;93:1764–1769. doi: 10.1073/pnas.93.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hu G, Wang H, Sciavolino P, Iler N, Shen M, et al. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol Cell Biol. 1997;17:2920–2932. doi: 10.1128/mcb.17.5.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]


