Abstract
Asparagine synthetase (ASNS) is an enzyme expressed ubiquitously in mammalian cells. Here, we discovered two 14-bp tandem repeat (2R, wild-type) sequences in the first intron of the gene. The 14-bp sequence is similar to the three GC-boxes (GC-I, -II, and -III) found in the promoter region of the ASNS gene, as well as, the binding site of transcription factor Sp-1. Approximately 75% of acute lymphoblastic leukemia (ALL) samples had the 2R sequence in both allele; however, 20% and 3% ALL samples had three (3R) and four (4R) 14-bp tandem repeats in one allele, respectively; the other allele had 2R. The tandem repeat sequence was not specific to the leukemia cells but represents a novel germline polymorphism. Interestingly, the 14-bp sequence functioned as a transcriptional enhancer element as shown by reporter analysis and formed a protein-DNA complex in vitro. Our data for the first time show that the ASNS gene has tandem repeated sequences as a polymorphism, and it can function as a transcriptional element; increased number of tandem repeat producing increased activity. Clinical significance in ALL requires further studies.
Keywords: asparagine synthetase gene, polymorphism, acute lymphoblastic leukemia, L-asparaginase
1. Introduction
Asparagine synthetase (ASNS) is an enzyme that produces asparagine from aspartic acid in an ATP-dependent manner [1]. Expression of ASNS is regulated by the nutritional status of cells [2]. Promoter analysis revealed three GC boxes consisting of GC-I (-148 to -139), GC-II (-128 to -119) and GC-III (-107 to -97), and two nutrient-sensing response elements, NSRE-1 (-68 to -60) and NSRE-2 (-48 to -43), which are known to be responsible for transcriptional activation [3, 4]. Transcription factors Sp1 and Sp3 are able to enhance the promoter via the GC boxes as shown in Sp-deficient Drosophila SL2 cells [5]; and ATF3, ATF4 and C/EBPβ are involved in gene expression via the NSRE-1 site as shown in human hepatoma HepG2 cells [6-9].
Methylation of CpG islands of genes is one of the epigenetic mechanisms to silence them; and aberrant methylation of CpG islands is observed in a cohort of genes in tumor cells [10, 11]. A CpG island in the region of the ASNS gene is reported to be methylated in the murine lymphoma cell line 6C3HED, as well as, the human leukemia cell lines 1873 and 1929; and these cells do not express ASNS. As a consequence, these cells are sensitive to l-asparaginase which is an effective drug for treatment with ALL [12-18]. The human ASNS gene has a CpG island located from -313 to +336 including 49 CpG sites. Previously, we found 74% of B-lineage ALL and 83% of T-ALL samples had methylation in the CpG island, but no methylation in breast and brain tumor samples [19]. During the analysis, we discovered two 14-bp tandem repeat (2R) sequences in the first intron of the gene. The 14-bp sequence (1R) is similar to the three GC-boxes (GC-I, -II, and -III) found in the promoter region of the ASNS gene, as well as, a binding site for the Sp-1 transcription factor. Here, we identified that this sequence cassette represents a novel polymorphism and investigated its function.
2. Materials and Methods
2.1. Genomic DNA samples and cell culture
Genomic DNA from diagnosis and remission B-ALL bone marrow samples was isolated from patients at the Institute of Human Genetics in Germany [19]. Genomic DNA of T-ALL was isolated from T-ALL patients at the University Hospital Benjamin Franklin in Germany. Genomic DNA was also examined from patients with childhood ALL collected at the Erasmus MC-Sophia Children's Hospital, Rotterdam, NL who were good, intermediate and poor responder to a therapeutic window with L-asparaginase as single agent given upfront of combination chemotherapy as described previously [20]. Genomic DNA of normal white blood cells (WBC) harvested from peripheral blood was isolated at Cedars-Sinai Medical Center. All samples were obtained after informed consent from the individuals.
Human leukemia cell lines Ball-1, HL-60, K562, Daudi, NCEB-1, SUDHL6 and SUDHL16 were maintained in RPMI 1640 (Invitrogen, Carlsbad, CA) medium supplemented with 10% fetal bovine serum (FBS, Invitrogen). Jeko1 and SP49 were cultured in RPMI 1640 medium with 20% FBS; and Ly4 and Ly10 were maintained in IMDM (Invitrogen) containing 10% FBS. Human embryonic kidney (HEK) 293T cells were cultured in DMEM (Invitrogen) containing 10% FBS.
2.2. Genomic PCR
Genomic DNA of various leukemia cell lines was isolated with DNeasy Tissue Kit (Qiagen, Valencia, CA). To detect tandem repeat sequences of the ASNS gene, diluted genomic DNA was used as a template, and PCR was performed with specific primers (sense primer: 5′- ATC CTC CAC CCC TTC CTT C-3′, antisense primer: 5′- ATC ACC CTG ACC TGC TTA CG-3′) that amplified from +34 to +150 of the ASNS gene. The PCR product was separated by 4% agarose gel electrophoresis containing 0.5 × TBE (44.5 mM Tris, 44.5 mM boric acid and 1mM EDTA) buffer.
2.3. Plasmid constructions and luciferase assay
The phRL-TK vector (Promega, Madison, WI) was digested with Bgl II and Hind III to isolate the HSV-TK promoter. The fragment was cloned into the Bgl II and Hind III sites of the pGL3-basic vector (Promega), which we then called the pGL3-TK vector. PCR products of the first intron from +34 to +150 (2R) from the ASNS gene either with or without additional 14-bp (3R) and 28-bp (4R) sequence, was cloned into the pCR 2.1 TOPO vector (Invitrogen) and sequenced. These plasmids were digested with Xho I and BamHI to isolate the inserts; and these fragments were cloned into Sal I and BamHI sites of the pGL3-TK vector. These pGL3-TK luciferase vectors carrying either two 14-bp tandem repeat (2R), three 14-bp tandem repeat (3R), or four 14-bp tandem repeat (4R) sequences are termed pGL3-TK-2R, -3R, or -4R, respectively.
HEK293T cells (4 × 105 cells) in a 6-cm dish were co-transfected with the phRL-TK and the pGL3-TK-2R, -3R, or -4R using LipofectAMIN 2000 (Invitrogen). Two days after transfection, cell lysates were prepared from these cells, and luciferase activity was assayed by the Dual-luciferase Reporter Assay System (Promega). Firefly luciferase expression levels were normalized to the level of Renilla luciferase expression. The assay was done in triplicate and repeated in three independent experiments.
2.4. Preparation of nuclear extract
Nuclear extract was isolated from the acute lymphoblastic leukemia cell line, Ball-1. Cells were resuspended in a buffer consisting of 20 mM HEPES (pH 7.9), 20% glycerol, 10 mM NaCl, 0.2 mM EDTA (pH 8.0) and 1.5 mM MgCl2 with a cocktail of proteinase inhibitors (Roche, Nutley, NJ). After incubation on ice for 15 min, samples were spun-down at 1,000 rpm for 10 min to pellet the nuclei. The pellet was resuspended at 2.5 × 107 nuclei/ml in the same buffer, and then 62.5μl of 5M NaCl was added to the sample with gentle vortexing. After incubation of the sample at 4°C for 30 min, it was centrifuged at 10,000 rpm. The supernatant was used as nuclear extract.
2.5. Electromobility shift assays
Oligonucleotides containing the 14-bp sequence (1R), the mutated 14-bp sequence (1R*), and the Sp-1 binding sequence were synthesized. Double-stranded probes were end-labeled with [γ-32P] ATP (Perkin Elmer, Waltham, MA) and purified by ProbeQuant G-50 Micro Columns (Amersham Biosciences, Piscataway, NJ). Probe sequences are shown in Figure 4A. For binding reaction, 10 μg of nuclear extract was incubated with radiolabeled probe, 2 μg of poly(dI-dC), and 300 μg/ml of BSA either with or without 200-fold excess of cold probe as a competitor. After incubation on ice for 40 min, the sample was subjected to electrophoresis in a non-denaturing 6% acrylamide gel at 100 V for 2h. The gel was dried and exposed to X-ray film.
Figure 4. Electromobility shift assays using the 14-bp sequence.
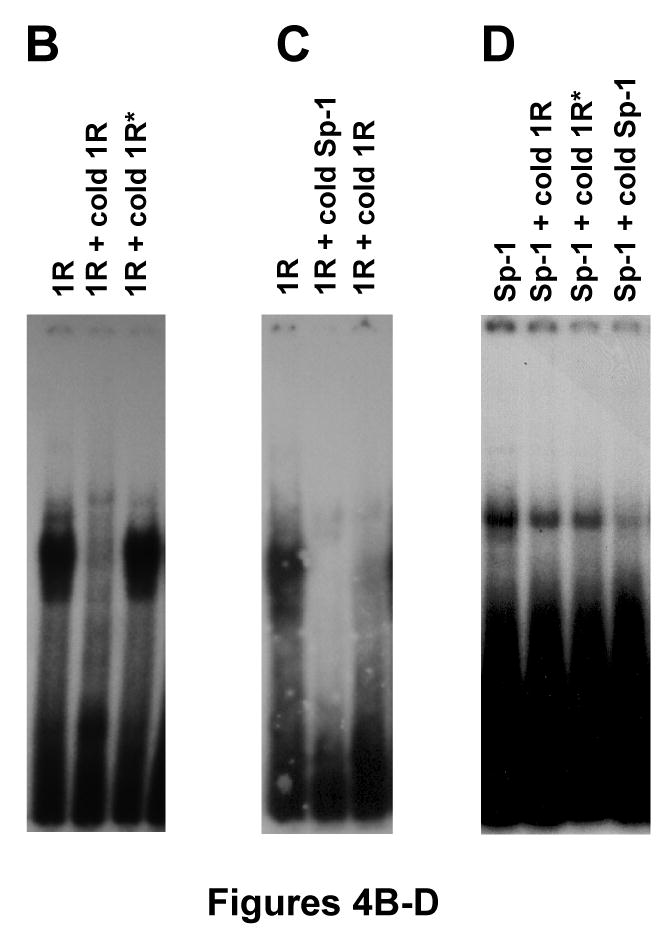
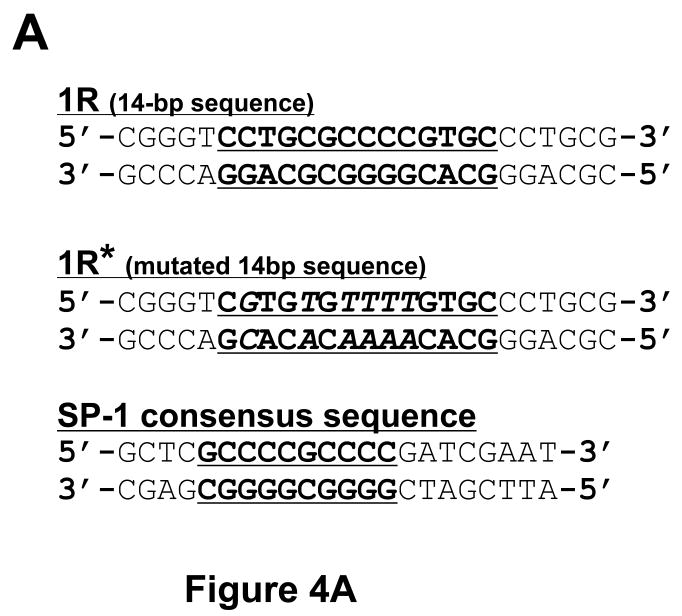
(A) Wild-type (1R) and mutated 14-bp (1R*) oligonucleotide sequences, as well as, the Sp-1 binding site. Bold characters indicate wild-type consensus sequences, and italic letters are mutated sequences as described previously. (B) Radioisotope labeled 1R oligonucleotide sequences were incubated with nuclear extract from Ball1 ALL cells either with or without 200-fold cold 1R oligonucleotide or 1R* oligonucleotide sequences. (C) Radioisotope labeled 1R oligonucleotide sequences were incubated with nuclear extract from Ball1 ALL cells either with or without 200-fold cold Sp-1 binding site or cold 1R oligonucleotide sequences. (D) Radioisotope labeled Sp-1 binding site sequences were incubated with nuclear extract from Ball1 ALL cells either with or without 200-fold cold 1R, 1R*, or Sp-1 binding site sequences. Abbreviations: 1R; 14-bp oligonucleotide sequences, 1R*; mutated 14-bp oligonucleotide sequences, Sp-1; Sp-1 binding site sequences.
3. Results
3.1. Insertional sequence in the asparagine synthetase gene in ALL
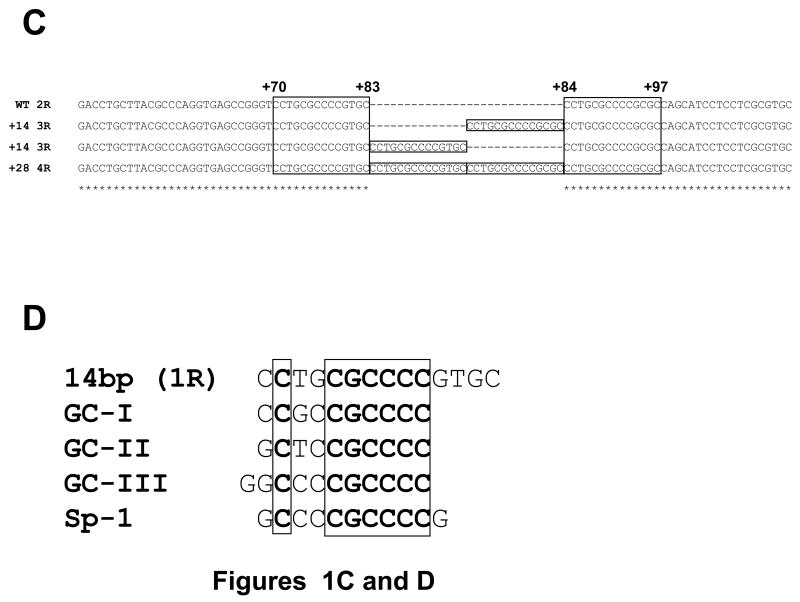
The CpG island of asparagine synthetase gene is located from -313 to +336 in its promoter region. During methylation analysis of the CpG island as described before [19], we discovered two 14-bp tandem repeat (2R, wild-type) sequence between +70 and +97 located in the first intron of the gene (Figures 1A and C). The 14-bp sequence was [CCTGCGCCCCG (C/T) GC] (1R), and the sequence is repeated twice. To analyze the sequence in detail, we amplified a 117-bp region from +34 to +150 by PCR using genomic DNA from ALL bone marrow, and the PCR products were separated by agarose gel electrophoresis. As shown in Figure 1B, most of the ALL bone marrow samples showed a single band of 117-bp; however, four samples (patient Nos. KS-10, KK-25, A-42 and S-68) displayed a band other than the 117-bp band. The upper band from patient A-42 contained one additional 14-bp sequence and thus formed three-tandem-repeats (3R); and an even higher band from patient KS-10 contained two additional 14-bp sequence forming four-tandem-repeats (4R) (Figure 1C). These results showed that the 14-bp or 28-bp insertions were found in the first intron of the ASNS gene in the ALL samples. Interestingly, the 14-bp sequence (1R) is similar to both the three GC-boxes (GC-I, -II, and -III) found in the promoter region of the ASNS gene, as well as, the binding site for the Sp-1 transcription factor (Figure 1D).
Figure 1. Identification of tandem repeat sequences in the ASNS gene.
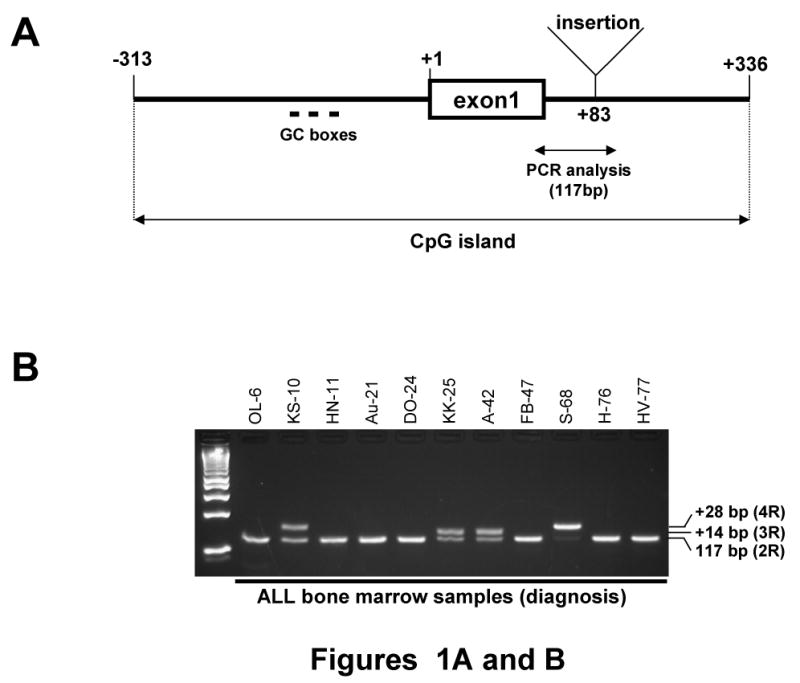
(A) Structure of promoter region of the ASNS gene. The CpG island of the ASNS gene is located from -313 to +336. Sequences of either 14-bp or 28-bp were discovered between +83 and +84 located in the first intron of the gene in some ALL samples. GC boxes (GC-I, -II and -III) occur in the promoter region. (B) Identification of insertional fragment by PCR. To distinguish between the dominant allele (2R) and an allele with an insert (3R or 4R), intron of the ASNS gene (+34 to +150) was analyzed by PCR and gel electrophoresis. The 117-bp PCR product had 2R. KK-25 and A-42 had shifted band which contain 3R, and KS-10 and S-68 had a greater shifted band that has 4R. (C) Nucleotide sequence of the insertional fragment. Two-tandem-repeat (2R) sequences were found from +70 to +97 in the dominant allele; each repeat sequence was 14-bp [CCTGCGCCCCG (C/T) GC] (1R). The 14-bp (1R) inserted allele contained an additional element, and thus formed three-tandem-repeats (3R), and the 28-bp insert formed four-tandem-repeats (4R). (D) Sequence alignment of the 14-bp (1R), GC-I, -II, -III and Sp-1 binding site. Each was highly homologous in nucleotide sequences.
3.2. Germline polymorphism of ASNS gene
These findings prompted us to examine whether the insertional sequence was specific for leukemic cells. First, we analyzed for the presence of the insertion in matched diagnosis and remission samples. Genomic PCR showed that the insertion was found in both the diagnosis and the remission samples (Figure 2A), suggesting that the insertion was a germline polymorphism in the ALL patients. One sample gave a surprising result. We detected that both alleles had 4R in the diagnosis sample from patient S-68, but the remission sample revealed that one allele had 4R and the other was 2R. This result probably reflects that one allele (2R allele) was deleted; and the second allele (4R allele) was duplicated in the ALL sample.
Figure 2. Germline polymorphism of ASNS gene.
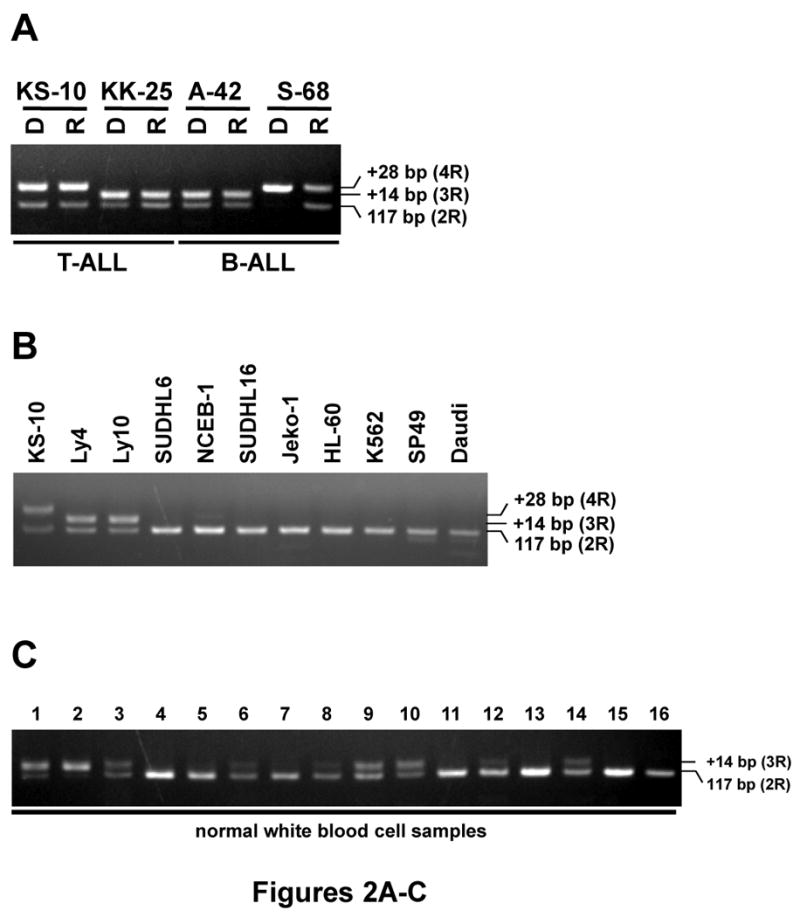
(A) PCR products of the first intron from +34 to +150 were amplified from genomic DNA of ALL samples at diagnosis (D) and at remission (R) from the bone marrow from children (two samples with T-ALL and two samples with B-ALL). (B) Several leukemia cell lines. (C) Peripheral blood lymphocytes from normal individuals. The PCR products were separated in 4% agarose gel. The 117-bp band is from the dominant allele (2R), and the upper bands originate from either 3R- or 4R-containing allele.
We also examined for the presence of an insertion in various leukemia cell lines including Ly4, Ly10, SUDHL6, SUDHL16, NCEB-1, Jeko-1, HL-60, K562, SP49 and Daudi. Only Ly4 and Ly10 showed one allele with 3R; and other 8 cell lines had 2R (Figure 2B). Of interest, only Ball-1 cells had 4R in both alleles (data not shown).
Next, we examined whether normal white blood cells (WBC) isolated from peripheral blood contained the insertion. As shown in Figure 2C, some normal WBC samples had 3R in one or both allele. To calculate the frequency of the insertion, 73 WBC samples from normal individuals and 92 ALL (B- and T-ALL) bone marrow samples at diagnosis were analyzed by genomic PCR (Table 1). A total of 48 of 73 (66%) normal WBC samples and 69 of 92 (75%) ALL bone marrow samples had 2R in both alleles (referred to as 2R/2R), 21 (29%) normal WBC samples and 18 (20%) ALL bone marrow samples contained 3R in one allele (referred to as 2R/3R), and 4 (5%) normal WBC samples and 1 (1%) ALL bone marrow sample contained 3R in both alleles (referred to as 3R/3R, Table 1). Interestingly, 3 (3%) ALL samples had 4R in one allele (referred to as 2R/4R), 1 (1%) ALL sample (S68) showed only 4R allele (referred to as 4R/4R), and no normal WBC sample had 2R/4R nor 4R/4R. These results showed that the insertion is not specific for leukemia samples, but a novel germline polymorphism of the ASNS gene.
Table 1.
Frequency of insertional sequences in the ASNS gene in genomic DNA samples from normal WBCs and ALL cells.
| Samples | Normal WBC (n=73) | ALL (n=92) | ||||||
|---|---|---|---|---|---|---|---|---|
| Allele type | 2R/2R | 3R/3R | 2R/3R | 2R/2R | 3R/3R | 2R/3R | 2R/4R | 4R/4R |
| Number of samples | 48 | 4 | 21 | 69 | 1 | 18 | 3 | 1 |
| Ratio | 66% | 5% | 29% | 75% | 1% | 20% | 3% | 1% |
Genomic DNA from 73 white blood cell (WBC) samples from normal individuals and 92 ALL bone marrow samples (both B- and T-ALL) at diagnosis were analyzed by PCR to determine the frequencies of the insertional sequences in the ASNS gene. Abbreviations: 2R/2R; both allele have 2R, 3R/3R; both alleles have 3R, 2R/3R; one allele is 2R and the other allele is 3R, 2R/4R; one allele is 2R and the other allele is 4R, 4R/4R; both alleles have 4R.
3.3. Transcriptional regulation activity of the 14-bp sequence
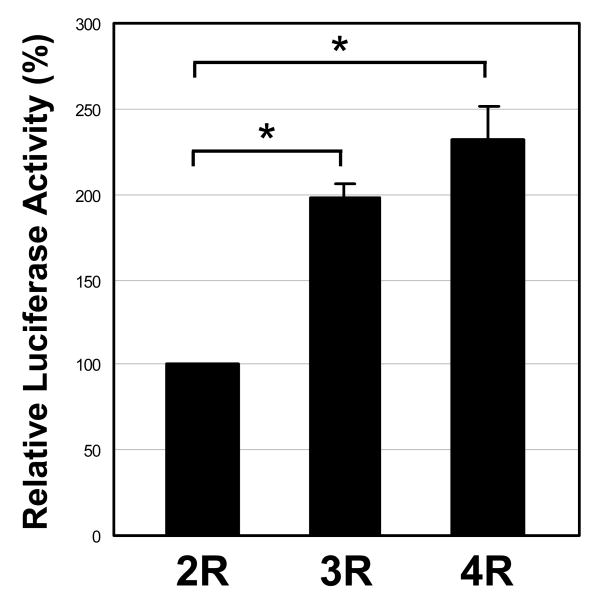
Next, we explored the functional significance of the insertional sequence using a luciferase reporter assay, since the 14-bp sequence is similar to three GC-boxes that are present in the promoter region of the ASNS gene (Figure 1D). A luciferase reporter plasmid having the HSV TK promoter, as well as, the first intron from +34 to +150 (2R) from the ASNS gene either with or without the additional 14-bp (3R) or 28-bp (4R) sequence (pGL3-TK-2R, -3R, or -4R) were transfected into HEK293T cells, and luciferase activity was examined (Figure 3). The luciferase activity of pGL3-TK-3R and -4R showed about a 2-fold increased activity compared with the 2R reporter plasmid (Figure 3), suggesting that the insertional sequence can function as a transcriptional enhancer.
Figure 3. Insertional sequences stimulate reporter gene expression.
HEK293T cells were transiently transfected with luciferase reporter plasmid having the HSV TK promoter with the first intron from +34 to +150 (2R) of the ASNS gene either with or without additional 3R and 4R insertional sequences. Firefly luciferase expression levels were normalized to the level of Renilla luciferase expression. Assays were done in triplicate and repeated in three independent experiments. Abbreviations: 2R; pGL3-TK-2R plasmid, 3R; pGL3-TK-3R plasmid, 4R; pGL3-TK-4R plasmid, and *; p < 0.01.
3.4. The 14-bp sequence forms a complex with a protein(s)
A previous report showed that GC-II and GC-III formed a complex with the transcription factors Sp-1 and Sp-3 [5]. The 14-bp sequence (1R) is similar to GC-II and GC-III, as well as, the binding site for the Sp-1 transcription factor. Using the electromobility shift assay (EMSA), we explored the possibility of the 14-bp sequence forming a complex with a protein(s). As shown in Figure 4B, the 14-bp oligonucleotide (1R) formed a complex with a protein(s) in vitro. Unlabeled competitor (200 ×) inhibited formation of the complex, but it was not competed by an excess of unlabeled oligonucleotide that contained the mutated 14-bp sequences (1R*) which was shown in Figure 4A. Interestingly, the complex was also competed by cold Sp-1 consensus oligonucleotide, as well as, a cold 1R oligonucleotide (Figure 4C). Reciprocally, the Sp-1 consensus oligonucleotide was competed by its cold oligonucleotide; however, the complex was not inhibited by either a cold 1R or a 1R* oligonucleotides (Figure 4D), suggesting that the unknown factor, which binds to the 14-bp sequence, is not Sp-1; but another molecule which also binds to the Sp-1 consensus oligonucleotide. Further studies are required to clarify the unknown molecule that binds to the 14-bp sequence.
3.5. Relationship between resistance to L-asparaginase in ALL patients and tandem repeat sequences in the ALL cells
L-asparaginase is an effective drug for the treatment of ALL, but some patients show resistance to it. We compared the relationship between the tandem repeat sequences and clinical response to L-asparaginase in ALL patients. Among 22 childhood ALL patients, 15, 5 and 2 patients were good, intermediate and poor responders to a therapeutic window with L-asparaginase given upfront of combination chemotherapy [20]. As shown in Table 2, the genotypes 2R/2R:2R/3R:3R/3R were 11:4:0 in the good responders; 4:1:0 in the intermediate responders; and 1:0:1 in the poor responders. Interestingly, 3R/3R was found only in the poor responder. Our population size is clearly too small to make statistical statements, but these results suggest that resistance to L-asparaginase and tandem repeat sequences may be independent of each other.
Table 2.
Relationship between L-asparaginase resistance in patients and tandem repeat sequences in their ALL cells
| Response | Genotype | Total | ||
|---|---|---|---|---|
| 2R/2R | 2R/3R | 3R/3R | ||
| Good | 11 | 4 | 0 | 15 |
| Intermediate | 4 | 1 | 0 | 5 |
| Poor | 1 | 0 | 1 | 2 |
| Total | 16 | 5 | 1 | 22 |
A good clinical response to L-asparaginase for the child with ALL is defined by less than 1 × 109/blasts/L, an intermediate response by 1 − 10 × 109/blasts/L, and a poor response by more than 10 × 109/blasts/L in the blood at day 5 after treatment with L-asparaginase.
4. Discussion
In the present study, we found that either a 14-bp (3R) or 28-bp (4R) insertional nucleotide sequence are novel polymorphisms in the first intron of ASNS, and demonstrated that the 14-bp sequence functions as a transcriptional regulatory element. Furthermore, the sequence formed a protein-DNA complex in vitro, and the complex was competed with a Sp-1 consensus oligonucleotide sequence.
The 14-bp sequence is not found in any other genomic DNA by homology search (data not shown), but this sequence is very similar to the binding site of the transcription factor Sp-1, as well as, three GC-boxes that are present in the promoter region of the ASNS gene. These three GC-boxes are known to be required to maintain basal transcription of the ASNS gene; and they acts as modulators to allow maximal activation via the NSRE sites following nutrient deprivation [5]. Interestingly, our data show increased enhancer activity of the 3R or 4R sequences, suggesting that the insertional sequences might be involved in gene expression.
Two of the GC-boxes, GC-II and GC-III, are known to form protein-DNA complexes with either Sp1 or Sp3; and Sp3 has been demonstrated to enhance activation of the ASNS promoter [5]. Our EMSA analysis revealed that the 14-bp nucleotide sequence (1R) formed a protein-DNA complex, and the complex disappeared in the presence of unlabeled Sp-1 consensus oligonucleotide. The Sp-1 consensus oligonucleotide was competed by its cold oligonucleotide; however, the complex was not inhibited by either a cold 1R or a 1R* oligonucleotides, suggesting that the unknown factor, which binds to the 14-bp sequence, is not Sp-1; but another molecule which also binds to the Sp-1 consensus oligonucleotide. Further studies are required to clarify the unknown molecule that binds to the 14-bp sequence.
In addition, 4R allele was found only in 4 ALL samples and an ALL cell line, Ball1 (Table 1, and data not shown). Many more normal and ALL samples need to be studied to determine if an association exists between this germline polymorphism and development of ALL or responsiveness to L-asparaginase. The ASNS gene is located on chromosome 7q21.3. Monosomy 7, deletion of 7q, and uniparental disomy (UPD) of 7q were found in 21%, 21% and 1% of childhood ALL [21, 26]. Of interest, ALL cells of patient S-68 probably had either monosomy 7, 7q deletion or UPD resulting in deletion of the dominant allele (2R) since this patient contained a 4R/4R type at presentation of ALL. The frequency of 3R and 4R from patients with ALL is lower (25%, 23/92) than that of 3R in the normal population (34%, 25/73); however, the difference is not significant (p=0.228, Fisher's exact test).
Thymidylate synthase catalyzes the conversion of deoxyuridylate into thymidylate, and becomes a target for several anti-tumor drugs including methotrexate and 5-fluorouracil. A tandem repeat sequence is present in the promoter region of the gene; and this sequence is polymorphic containing either two (2r) or three (3r) 28-bp repeat [22]. The 3r is associated with higher expression of the thymidylate synthase mRNA than the 2r. Importantly, children with ALL having homozygous 3r (3r/3r) are associated with a worse prognosis than those with 2r/2r and 2r/3r [23]. Here, we compared the tandem repeat sequences and resistance to L-asparaginase, an effective drug for the treatment of ALL; and we found that they were independent. The promoter region of the ASNS gene is often methylated in ALL samples, suggesting that this epigenetic change is a major mechanism to silence the gene. Nevertheless, studies showed that resistance to L-asparaginase is associated with high expression of ASNS mRNA in TEL-AML1-negative, but not TEL-AML1-positive ALL cells [20, 24]. Interestingly, a recent study suggested a novel mechanism of L-asparaginase resistance. The investigators reported that bone marrow-derived mesenchymal stem cells (MSCs) expressed high-level of ASNS mRNA, and MSCs protected ALL cells from L-asparaginase cytotoxicity by providing asparagine to the local environment [25]. Taken together, several factors are probably involved in L-asparaginase resistance.
Further studies including the relationship between the insertional polymorphism in the ASNS gene and prognosis of ALL patients will provide novel insights concerning this novel polymorphism both in ALL and normal cells.
Acknowledgments
We are grateful to Drs. Kent Taylor and Talin Haritunians for their discussion about statistical analysis. We are also thankful to members of our laboratory for helpful discussions. This work was supported by NIH grants as well as Parker Hughes Fund. H. P. K. is the holder of the Mark Goodson endowed Chair in Oncology Research and is a member of the Jonsson Cancer and Molecular Biology Institute, UCLA. The study is dedicated to David Golde, a mentor and friend.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contribution: TA performed research and wrote the manuscript; DY and NK were technically involved in cell culture; CRB, WKH and MLD provided genomic DNA of ALL cells; JHS and CWM provided genomic DNA of normal white blood cell and ALL cells; and HPK directed the overall study.
Conflict-of-interest statements: The authors declare no competing financial interests.
References
- 1.Richards NG, Schuster SM. Mechanistic issues in asparagine synthetase catalysis. Adv Enzymol Relat Areas Mol Biol. 1998;72:145–198. doi: 10.1002/9780470123188.ch5. [DOI] [PubMed] [Google Scholar]
- 2.Kilberg MS, Barbosa-Tessmann IP. Genomic sequences necessary for transcriptional activation by amino acid deprivation of mammalian cells. J Nutr. 2002;132:1801–1804. doi: 10.1093/jn/132.7.1801. [DOI] [PubMed] [Google Scholar]
- 3.Barbosa-Tessmann IP, Chen C, Zhong C, Schuster SM, Nick HS, Kilberg MS. Activation of the unfolded protein response pathway induces human asparagine synthetase gene expression. J Biol Chem. 1999;274:31139–31144. doi: 10.1074/jbc.274.44.31139. [DOI] [PubMed] [Google Scholar]
- 4.Barbosa-Tessmann IP, Chen C, Zhong C, Siu F, Schuster SM, Nick HS, Kilberg MS. Activation of the human asparagine synthetase gene by the amino acid response and the endoplasmic reticulum stress response pathways occurs by common genomic elements. J Biol Chem. 2000;275:26976–26985. doi: 10.1074/jbc.M000004200. [DOI] [PubMed] [Google Scholar]
- 5.Leung-Pineda V, Kilberg MS. Role of Sp1 and Sp3 in the nutrient-regulated expression of the human asparagine synthetase gene. J Biol Chem. 2002;277:16585–16591. doi: 10.1074/jbc.M110972200. [DOI] [PubMed] [Google Scholar]
- 6.Siu F, Bain PJ, LeBlanc-Chaffin R, Chen H, Kilberg MS. ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J Biol Chem. 2002;277:24120–24127. doi: 10.1074/jbc.M201959200. [DOI] [PubMed] [Google Scholar]
- 7.Pan Y, Chen H, Siu F, Kilberg MS. Amino acid deprivation and endoplasmic reticulum stress induce expression of multiple activating transcription factor-3 mRNA species that, when overexpressed in HepG2 cells, modulate transcription by the human asparagine synthetase promoter. J Biol Chem. 2003;278:38402–38412. doi: 10.1074/jbc.M304574200. [DOI] [PubMed] [Google Scholar]
- 8.Siu F, Chen C, Zhong C, Kilberg MS. CCAAT/enhancer-binding protein-beta is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J Biol Chem. 2001;276:48100–48107. doi: 10.1074/jbc.M109533200. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Pan YX, Dudenhausen EE, Kilberg MS. Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive basic region/leucine zipper transcription factors as well as localized histone acetylation. J Biol Chem. 2004;279:50829–50839. doi: 10.1074/jbc.M409173200. [DOI] [PubMed] [Google Scholar]
- 10.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 11.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 12.Cooney DA, Handschumacher RE. l-asparaginase and l-asparagine metabolism. Annu Rev Pharmacol. 1970;10:421–440. doi: 10.1146/annurev.pa.10.040170.002225. [DOI] [PubMed] [Google Scholar]
- 13.Wriston JC, Jr, Yellin TO. l-asparaginase. Adv Enzymol Relat Areas Mol Biol. 1973;39:185–248. doi: 10.1002/9780470122846.ch3. [DOI] [PubMed] [Google Scholar]
- 14.Muller HJ, Boos J. Use of l-asparaginase in childhood ALL. Crit Rev Oncol Hematol. 1998;28:97–113. doi: 10.1016/s1040-8428(98)00015-8. [DOI] [PubMed] [Google Scholar]
- 15.Pinheiro JP, Boos J. The best way to use asparaginase in childhood acute lymphatic leukaemia-still to be defined? Br J Haematol. 2004;125:117–127. doi: 10.1111/j.1365-2141.2004.04863.x. [DOI] [PubMed] [Google Scholar]
- 16.Broome JD. Studies on the mechanism of tumor inhibition by l-asparaginase. Effects of the enzyme on asparagine levels in the blood, normal tissues, and 6C3HED lymphomas of mice: differences in asparagine formation and utilization in asparaginase-sensitive and -resistant lymphoma cells J Exp Med. 1968;127:1055–1072. doi: 10.1084/jem.127.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng H, Shen N, Qian L, Sun XL, Koduru P, Goodwin LO, Issa JP, Broome JD. Hypermethylation of CpG islands in the mouse asparagine synthetase gene: relationship to asparaginase sensitivity in lymphoma cells. Partial methylation in normal cells. Br J Cancer. 2001;85:930–935. doi: 10.1054/bjoc.2001.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren Y, Roy S, Ding Y, Iqbal J, Broome JD. Methylation of the asparagine synthetase promoter in human leukemic cell lines is associated with a specific methyl binding protein. Oncogene. 2004;23:3953–3961. doi: 10.1038/sj.onc.1207498. [DOI] [PubMed] [Google Scholar]
- 19.Akagi T, Yin D, Kawamata N, Bartram CR, Hofmann WK, Wolf I, Miller CW, Koeffler HP. Methylation analysis of asparagine synthetase gene in acute lymphoblastic leukemia cells. Leukemia. 2006;20:1303–1306. doi: 10.1038/sj.leu.2404216. [DOI] [PubMed] [Google Scholar]
- 20.Appel IM, den Boer ML, Meijerink JP, Veerman AJ, Reniers NC, Pieters R. Up-regulation of asparagine synthetase expression is not linked to the clinical response L-asparaginase in pediatric acute lymphoblastic leukemia. Blood. 2006;107:4244–4249. doi: 10.1182/blood-2005-06-2597. [DOI] [PubMed] [Google Scholar]
- 21.Kawamata N, Ogawa S, Zimmermann M, et al. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high-resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood. 2008;111:776–784. doi: 10.1182/blood-2007-05-088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horie N, Aiba H, Oguro K, Hojo H, Takeishi K. Functional analysis and DNA polymorphism of the tandemly repeated sequences in the 5′-terminal regulatory region of the human gene for thymidylate synthase. Cell Struct Funct. 1995;20:191–197. doi: 10.1247/csf.20.191. [DOI] [PubMed] [Google Scholar]
- 23.Krajinovic M, Costea I, Chiasson S. Polymorphism of the thymidylate synthase gene and outcome of acute lymphoblastic leukaemia. Lancet. 2002;359:1033–1034. doi: 10.1016/S0140-6736(02)08065-0. [DOI] [PubMed] [Google Scholar]
- 24.Stams WA, den Boer ML, Beverloo HB, Meijerink JP, Stigter RL, van Wering ER, Janka-Schaub GE, Slater R, Pieters R. Sensitivity to l-asparaginase is not associated with expression levels of asparagine synthetase in t(12;21)+ pediatric ALL. Blood. 2003;101:2743–2747. doi: 10.1182/blood-2002-08-2446. [DOI] [PubMed] [Google Scholar]
- 25.Iwamoto S, Mihara K, Downing JR, Pui CH, Campana D. Mesenchymal cells regulate the response of acute lymphoblastic leukemia cells to asparaginase. J Clin Invest. 2007;117:1049–1057. doi: 10.1172/JCI30235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heerema NA, Nachman JB, Sather HN, La MK, Hutchinson R, Lange BJ, Bostrom B, Steinherz PG, Gaynon PS, Uckun FM; Children's Cancer Group Deletion of 7p or monosomy 7 in pediatric acute lymphoblastic leukemia is an adverse prognostic factor: a report from the Children's Cancer Group. Leukemia. 2004;18:939–947. doi: 10.1038/sj.leu.2403327. [DOI] [PubMed] [Google Scholar]