Abstract
Collagen Type I and fibrin are polymeric proteins commonly used in the field of regenerative medicine as the foundational matrix of engineered tissues. We examined the response of vascular smooth muscle cells (VSMC) to both two-dimensional (2D) substrates as well as three-dimensional (3D) matrices of these biopolymers. Pure collagen Type I, pure fibrin and composite matrices consisting of 1:1 mixtures of collagen and fibrin were studied. Relative gene expression of three ECM molecules (collagen Type I and III, and tropoelastin) and three integrin subunits (integrins α1, β1 and β3) was determined over 7 days in culture using quantitative RT-PCR. Expression of all of these marker genes was up-regulated in 3D matrices, relative to 2D substrates. Tropoelastin, integrin α1 and integrin β1 were highest in collagen matrices, while collagen III and integrin β3 expression were highest in pure fibrin, and collagen I expression was highest in the collagen-fibrin composite materials. Both the compositional and temporal expression patterns of these specific ECM-related genes were suggestive of a wound healing response. These results illuminate the short-term responses of VSMC to 2D and 3D biopolymer matrices, and have relevance to tissue engineering and cardiovascular biology.
Keywords: Biopolymers, collagen, fibrin, smooth muscle cells, extracellular matrix, wound healing
INTRODUCTION
The objective of this study was to examine how vascular smooth muscle cells (VSMC) interact with two-dimensional (2D) and three-dimensional (3D) protein matrices composed of the biopolymers collagen Type I and fibrin, as well as mixtures of these proteins. In particular, we were interested in the expression of genes associated with tissue remodeling as a function of time and matrix composition. In addition, this study was designed to add to the understanding of how cells interact differently with 3D matrices, as compared to the more commonly used 2D substrates. The use of 3D culture systems is gaining popularity due to their promise as improved models of tissue physiology, and because such systems can potentially be developed into engineered tissues for the treatment of disease. The field of tissue engineering, therefore, is in need of a better understanding of how cells interact with 3D matrices and how cell function can be controlled via cell-matrix interactions.
Collagen Type I and fibrin proteins are commonly used as biopolymers in tissue engineering due to their availability, scaffolding function, and bioactive qualities. Collagen is a major structural protein in a variety of tissues and is characterized by a high stiffness and tensile strength that imparts resistance to tensile and shear loads in soft tissues [1]. Collagen Type I in particular has been used in a wide variety of therapeutic applications, including drug delivery and tissue engineering (for a review, see, e.g., Ref. [2]). Fibrin biopolymer results from the thrombin-activated cleavage of the blood-borne protein fibrinogen, and it is the major fibrillar component of blood clots. In addition to its structural role, fibrin is a biochemical stimulant of the wound healing response. It is used clinically as a surgical sealant and also has been investigated as a scaffold material in tissue engineering (for a review, see, e.g., Ref. [3]). An attractive feature of using solubilized collagen and fibrin in tissue engineering is that they can be cast and reconstituted into essentially any desired geometry. In addition, it is possible to suspend living cells in the liquid polymer solution during molding and these cells subsequently become homogeneously embedded in the molded matrix upon gelation. The embedded cells can recognize, attach to and remodel the surrounding matrix, such that it becomes compacted to form a rudimentary tissue. In contrast, most synthetic polymers are polymerized under conditions that are toxic to cells and, therefore, the cellular component must be added onto the surface of the scaffold once it has been manufactured.
The extracellular environment is defined by a variety of biochemical and mechanical factors that affect cell function and behavior. The extracellular matrix (ECM) of most tissues is composed of many proteins and glycoproteins, each with specific structural and biological functions [4]. The amount of these proteins and their ratio relative to each other are not fixed. Indeed the ECM is dynamic and constantly evolving as a result of cell-mediated remodeling during development, injury and normal function. Cells interact with the ECM through specialized integrin receptors and the resulting signal initiation directs many cellular functions including proliferation, matrix synthesis [5], migration [6], differentiation [7] and apoptosis [8]. In addition, the ECM acts as a physical scaffold for cells and can, therefore, transmit mechanical signals, as well as sequester and release growth factors. In order to harness the full potential of natural biopolymers in tissue engineering and regenerative medicine, it is essential to understand how cells interact with them and remodel them.
The geometry of the matrix (i.e., 2D versus 3D) also plays a defining role in determining how a cell will respond to biochemical and mechanical cues, since in many native tissues cells are completely surrounded by ECM. Conventional 2D cell culture has provided important insight into how cells interact with their environment, but there is increasing interest in creating more physiologically relevant 3D cell culture models in order to study cell-matrix interactions and their effects on cell function. There is a growing literature that examines cell function in 3D protein matrices, particularly using the model of fibroblasts in a 3D collagen gel (see, e.g., Refs [9, 10]). These studies also are being extended to examine how matrix and mechanical factors interact to direct cell function [11, 12]. Such complex culture systems can be used as more advanced in vitro models of specific tissues to study how cell function is regulated, and VSMC also have been studied in 3D matrices [13, 14]. The insight gained from these culture systems can be applied to the creation of engineered tissues for transplantation into the body and understanding how matrix geometry affects cell-matrix interactions and remodeling in these scaffolds is important to controlling the functionality of engineered tissues.
Our laboratory has been developing collagen and fibrin biomaterials for use in cardiovascular tissue engineering [15]. The goal is to create mechanically and physiologically functional materials using natural biopolymers in a 3D configuration. In previous studies, we have used composite materials made of collagen and fibrin to modulate the biochemical [16] and mechanical [17] properties of the matrix. These composite biomaterials are of particular interest because they combine the structural features of collagen and the bioactive properties of fibrin. In addition, it is known that VSMC bind to these proteins via distinct integrins, which can act as centers of signal transduction. It recently has been shown that cell function can be altered by exposure to multiple matrix components, relative to pure protein substrates [18, 19]. Therefore, the rationale underlying our work is that by controlling the extracellular matrix that surrounds cells in an engineered tissue, we will be able to direct cell function and remodeling towards the creation of more robust and functional tissues. Achieving this goal requires improved knowledge of how cells interact with and remodel their 3D protein environment.
The present study was carried out to characterize the effects of collagen Type I and fibrin biopolymer materials specifically on matrix protein and integrin gene expression. Isolated vascular smooth muscle cells (VSMC) were cultured either on 2D substrates (i.e., traditional monolayer cultures atop ECM thin gels), or were embedded within free-floating 3D ECM hydrogels. The matrices were composed of collagen Type I (C), fibrin (F), or a 1:1 mixture of the two proteins (CF), and freely compacting gels were used because for tissue engineering applications it is desirable to obtain a dense and robust construct. We expected integrin expression to be altered by changing the geometry and composition of the matrix, and our interest was in characterizing how these changes affected cell function in terms of ECM expression. Our results show that ECM and integrin gene expression were markedly altered in 3D matrices, relative to 2D ECM cultures, and further thatthese changes were dependent on the composition of the matrix. Our goal is to apply a better understanding of the ECM cues that govern cell behavior in the 3D cellular environment to the development of improved biomaterials and therapies for a variety of diseases in which matrix accumulation is implicated, including atherosclerosis and hypertension. This knowledge also would be of benefit in the field of vascular tissue engineering, by enabling the controlled deposition of matrix proteins to enhance the mechanical robustness of tissues grown in vitro.
MATERIALS AND METHODS
Cell culture
Rat aortic smooth muscle cells (RASMC) were isolated from normal adult male Sprague-Dawley rats using a collagenase digestion method [4], and were used for experiments between passages 5 and 8. Cells and ECM constructs were maintained in Dulbecco’s Modified Eagle Medium (DMEM, Cellgro) supplemented with 10% fetal bovine serum (FBS, Cellgro), 2 mM l-glutamine (Cellgro) 100 U/ml penicillin G sodium (Cellgro), and 100 μg/ml streptomycin sulfate (Cellgro). The medium was changed every 2-3 days and plated cells were enzymatically removed from the flask using 0.05% trypsin (Cellgro).
Preparation of 2D collagen, fibrin and collagen-fibrin gels
Pure collagen substrates were made at 2.0 mg/ml protein content using bovine Type I collagen (MP Biomedicals). The lyophilized protein was dissolved in 0.02 M acetic acid to obtain a 2 × collagen solution (4.0 mg/ml). The ingredients for the final collagen substrates were combined: 2 × collagen solution (50%), FBS (10%), 0.1 M NaOH (10%), 5 × DMEM (14%) and DMEM (16%). This combination of constituents maintained the appropriate pH and osmolarity during gel formation, so that cell viability was preserved.
Pure fibrin gels were also made at 2.0 mg/ml protein content using bovine plasma Type IV fibrinogen (Sigma). The lyophilized protein was dissolved in cold DMEM supplemented with 4.0 mg/ml ε-amino caproic acid (ACA, Sigma), 2.0 mM l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin to obtain a 2 × fibrinogen solution. ACA is an inhibitor of plasmin activity that is used to prevent degradation of the fibrin polymer, such that the VSMC were in contact with intact fibrin fibers during the culture period. In addition, fibrin degradation products have been shown to affect VSMC function. The ingredients for the final fibrin solution were mixed: 2 × fibrinogen solution (50%), FBS (10%) and DMEM containing 0.1 U thrombin per mg fibrinogen (40%).
Collagen-fibrin gels were made using a 1:1 ratio of collagen to fibrin to obtain a final protein content of 2.0 mg/ml. ECM solutions and other reagents were prepared as described above.
Two-dimensional ECM substrates were formed by coating tissue culture plastic flasks with ECM solutions (2.0 mg/ml), and incubating them for 1 h at 37°C. The result was a thin “2D” film of gel that covered the TCP, onto which cells could be seeded. The matrix compositions of the 2D film gels matched the 3D counterparts. Each flask was rinsed with PBS before RASMC were plated at a cell density of 20 × 103 cells/cm2 of the culture surface. Cells were allowed to adhere undisturbed for at least 24 h, and culture medium was changed after 1, 2 and 6 days in culture.
Preparation of 3D collagen, fibrin and collagen-fibrin gels
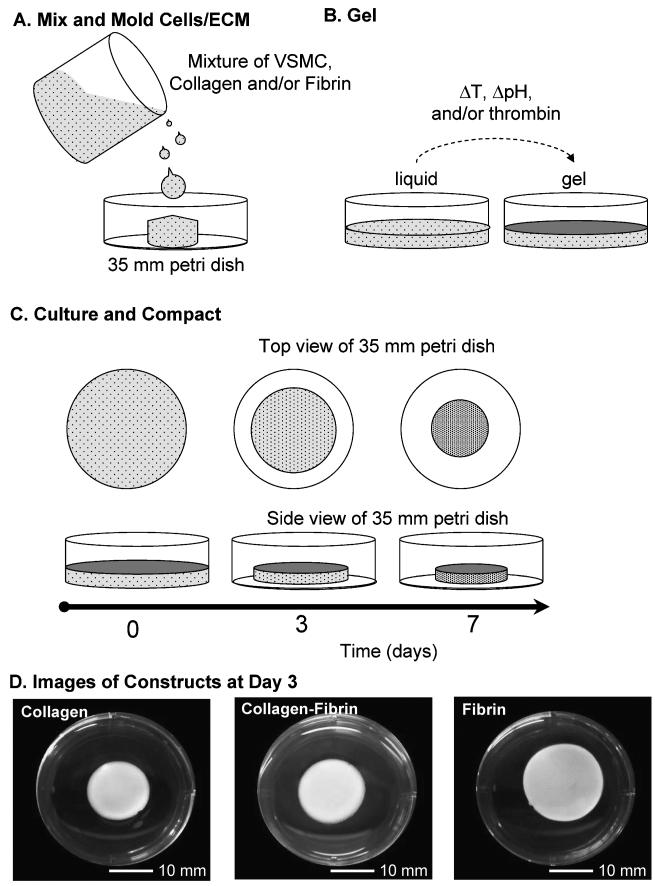
The matrix materials and compositions used to create 3D hydrogels were the same as used to make 2D substrates. Figure 1 shows a schematic representation of ECM gel fabrication and the phenomona of cell-mediated compaction over time. Three-dimensional constructs using the three matrix materials were prepared by mixing the ECM solutions (concentrations given above) with cells at a final concentration of 1.0 × 106 cells/ml. The cell-ECM suspension was then gently pipetted into a 6-well plate (Fig. 1A), which was subsequently incubated at 37.0°C for 30 min to allow for gelation (Fig. 1B). After gelation, the edges of constructs were freed using the tip of a Pasteur pipette to allow for unconstrained cell-mediated gel compaction (Fig. 1C). After 1, 3 and 7 days in culture, samples were collected and frozen. Constructs were rinsed with PBS and rapidly frozen using liquid nitrogen and stored at -80°C.
Figure 1.
Fabrication of 3D protein hydrogel constructs. Solubilized ECM is mixed with VSMC and the mixture is then poured into a mold, in this case a 35 mm diameter Petri dish (A). The liquid suspension is exposed to conditions that initiate gelation, such as pH or temperature change or addition of an enzyme (B). Over time in culture the gelled material is compacted by cell-mediated remodeling of the protein matrix (C), resulting in rudimentary 3D tissue constructs in which the VSMC are surrounded by matrix proteins (D).
Figure 1D shows representative images of constructs after 3 days in culture. Compaction of the constructs over time was assessed by taking two diameter measurements at 90 degrees to each other and then averaging. The thickness of the constructs was estimated by assuming that the degree of compaction was the same in all dimensions. These dimensions could then be used to determine a percent compaction relative to the original construct volume.
Total RNA extraction from cells and constructs
All samples were lysed using cold Trizol (Invitrogen) according to the manufacturer’s instructions. For harvesting of cells and RNA extraction, 2D cell samples were scraped off the flask bottom using a plastic scraper. Frozen gel constructs were carefully homogenized by hand using a tissue grinder. Samples were mixed with Trizol reagent, followed by phase separation using chloroform and collection of the aqueous phase. RNA was precipitated using isopropyl alcohol and the RNA pellet was collected and washed. Extracted samples were treated with DNase (Invitrogen) according to the manufacture. The concentration of RNA was determined by measuring the absorbance at 260 nm in a spectrophotometer (Bio-Tek). Only RNA samples with A260/A280 ≥ 1.65 were used.
Reverse transcriptase and real-time polymerase chain reaction
DNA was transcribed with You-Prime-Ready-to-Go-Beads (Amersham) using the manufacturer’s instructions. 0.5 μg of DNase-treated total RNA and random hexamer primers (Bio-Rad) were used. Samples were stored at -20°C for future use.
Real-time quantitative PCR (qRT-PCR) was performed in the Bio-Rad iCycler 4 (Bio-Rad) using the SYBR green fluorescent dye detection kit (Bio-Rad). Primer sequences are shown in Table 1. Quantification was based on the relative expression of the target genes versus the expression of the commonly used housekeeping gene GAPDH as a reference (endogenous control). GAPDH expression stability was verified for the experimental conditions of this study via comparison of Ct values (data not shown). Relative quantification was performed as instructed by the manufacturer.
Table 1.
Primer sequences used in RT-PCR analysis
| Gene (Accession No.) | Forward | Reverse |
|---|---|---|
| Collagen Type I α-chain (Z78279) |
GGAGAGTACTGGATCGACCCTAAC | CTGACCTGTCTCCATGTTGCA |
| Collagen Type III α-chain (X70369) |
GAAAAAACCCTGCTCGGAATT | GGATCAACCCAGTATTCTCCACTCT |
| Tropoelastin (M86376) |
CCTAGGAGCCAGGCCATTC | TGGGCTGGGTAGATA |
| Integrin α1 (X52140) |
GGATTTAATGACGTCGTGA | CCACTGCCATGATAAATGTACACA |
| Integrin β1 (U12309) |
GGAGAAAACTGTGATGCCATACAT | TGGGCTGGTACAGTTTTGTTCA |
| Integrin β3 (S58529) |
GGAAGGCTGGCAGGCATT | TGGTTGTCCCGGCCAA |
SEM imaging
Samples were washed three times in phosphate buffered saline solution (PBS, 0.01 M Na2HPO4, 0.15 M NaC, pH 7.4), fixed for 1 h in 4% EM grade glutaraldehyde (Electron Microscopy Sciences) in PBS and then washed three times in PBS. This was followed by dehydration in a series of sequentially more concentrated ethanol solutions: 25%, 35%, 50%, 70%, 80%, 90%, 95% and three incubations in 100%. Dehydrated gels were transferred into anhydrous 100% ethanol in the specimen chamber of a critical-point dryer (CPD, Tousimis). After the chamber was sealed, the ethanol was exchanged for liquid CO2 by alternately filling the chamber with the high pressure fluid and waiting several minutes for equilibration, and venting and replacing with fresh CO2. The dried samples were mounted on aluminum stubs, sputter-coated with platinum, and viewed by scanning electron microscopy (LEO 1550VP Field Emission SEM, LEO Electron Microscopy) operated at 3-5 kV accelerating voltage.
Data analysis
For all time points (days 1, 3 and 7), ECM geometries (2D and 3D), and ECM compositions (collagen, collagen-fibrin and fibrin), the normalized values from four independent experiments were averaged and expressed as mean ± standard error (SE). Data sets were statistically analyzed for significance using unpaired, two-tailed Student’s t-tests. Significance of correlations was determined using the Fisher Transformation.
RESULTS
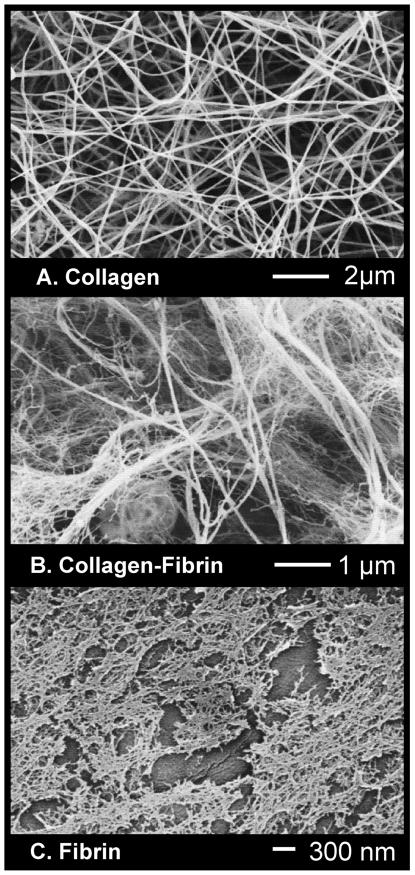
Figure 2 shows microscale (D-F) images of the biopolymer constructs. Collagen constructs consistently compacted to the greatest degree (to approx. 40% of their original volume at day 3), followed by collagen-fibrin (to approx. 45%) and fibrin (to approx. 60%) hydrogels. The SEM images in panels A-C reveal that collagen matrices (A) were composed of long thin filaments, whereas fibrin matrices (C) had a fine mesh-like appearance. Mixing these two biopolymers into a collagen-fibrin composite (B) produced an interpenetrating network with a morphology that incorporated both long thing filaments and a fine mesh. VSMC (not shown in these images) were incorporated directly into these matrices, such that most cells were entirely surrounded by protein fibers.
Figure 2.
SEM images of the microstructure of the protein matrix in 3D constructs, revealing distinct fiber morphologies: collagen long and filamentous (A), collagen-fibrin interpenetrated network of filaments and mesh (B), fibrin fine mesh-like structure (C).
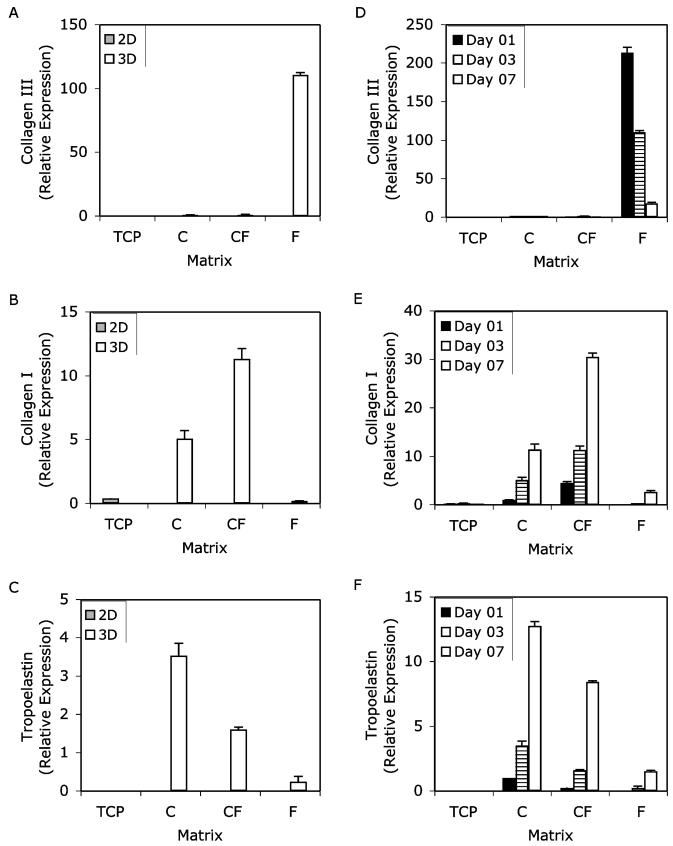
Both matrix composition and geometry affected ECM protein and integrin gene expression by VSMC in the materials studied. Overall, 3D cultures stimulated expression of these genes to a greater extent than monolayer culture. Figures 3 and 4 show the effects of matrix geometry and time in culture on the ECM and integrin genes of interest in this study. Collagen Type III, collagen Type I and tropoelastin are shown in Fig. 3, and integrins β3, α1 and β3 are shown in Fig. 4. In both figures, the designation “2D” represents VSMC cultured on top of biopolymer substrates, with tissue culture plastic (TCP) used as a control, whereas “3D” represents VSMC cultured inside biopolymer matrices. Panels A-C show a comparison of 2D and 3D matrices at day 3 in culture, whereas panels D-F show the time-course of gene expression in 3D matrices only.
Figure 3.
Gene expression of collagen Type III α-chain (A and D), collagen Type I α-chain (B and E) and tropoelastin (C and F) over 7 days in culture. Results are normalized relative to 3D collagen on day 1. Two matrix geometries were used: two-dimensional monolayer culture (2D) and three-dimensional matrices (3D). The matrix materials used were tissue-culture plastic (TCP), collagen (C), collagen-fibrin (CF) and fibrin (F). Panels A-C depict the effects of matrix geometry on ECM gene expression at day 3, which is representative of the temporal data. Panels D-F show the effects of matrix composition in 3D systems over time in culture. RT-PCR results were normalized to GAPDH expression at each time point and for each matrix condition. Results are expressed as the mean ± SEM.
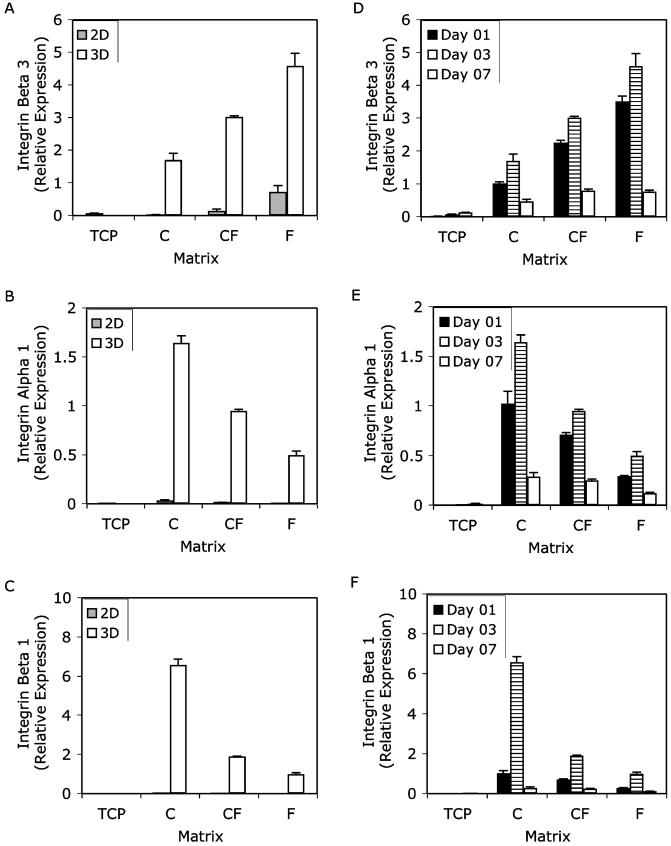
Figure 4.
Gene expression of integrins α1 (A and D), β1 (B and E) and β3 (C and F) over the course of a 7-day culture period. RT-PCR results were normalized to GAPDH expression for each geometry and matrix condition, and are shown relative to 3D collagen day 1. Data from day 3 only are shown in panels A-C, while panels D-F show gene expression over time. Two matrix geometries were used two-dimensional monolayer (2D) and three-dimensional matrices (3D). The matrix materials used were tissue-culture plastic (TCP), collagen (C), collagen-fibrin (CF) and fibrin (F). The effects of matrix geometry are shown in panels A-C and the effects of composition and time in TCP and 3D systems are shown in panels D-F. Results are expressed as the mean ± SEM.
Collagen III α1-chain mRNA was markedly up-regulated in 3D fibrin culture compared with both its 2D analog and other the two collagen containing 3D matrices (Fig. 3A). This up-regulation in 3D fibrin persisted across all time-points with the highest observed levels recorded after 1 day in culture, after which they tapered off to the lowest level observed on day 7 (Fig. 3D). Collagen III expression was 213-fold higher at day 1, but this increase diminished with time to 111-fold at day 3 and 18-fold at day 7. Like collagen III, collagen I α1-chain mRNA was elevated in 3D matrices relative to their 2D counterparts and tissue culture plastic; however, unlike collagen III maximum expression of collagen Type I was measured in 3D collagen-fibrin matrices (Fig. 3B). Over the course of this study, collagen I gene expression in each of the 3D matrices increased steadily with the highest observed levels on day 7 (Fig. 3E). Over all time points, collagen I expression was highest in 3D collagen-fibrin. Analysis of tropoelastin gene expression revealed that pure collagen matrices elicited a greater response than fibrin-containing matrices (Fig. 3C). Like collagen I gene expression, tropoelastin expression was lowest on the first day of culture and highest on the seventh day of culture (Fig. 3F) in all 3D matrices.
Gene expression of the three integrins evaluated in this study also was sensitive to both matrix geometry and composition. More specifically, expression of integrins α1 and β1 was very similar to each other, and different from expression of integrin β3. There were two clear trends: one pertaining to the effects of matrix geometry, the other to composition. First, all three integrin subunits were markedly up-regulated in 3D culture relative to 2D (Fig. 4A-C). Second, in 3D matrices, the same temporal pattern of integrin gene expression persisted across the various protein compositions: increasing between days 1 and 3, then decreasing between days 3 and 7 (Fig. 4D-F). The most notable effect of matrix composition on integrin expression was that in fibrin-containing 3D matrices integrin β3 was more up-regulated compared with pure collagen matrices (Fig. 4D). However, for integrins α1 and β1 the reverse was observed, with collagen-containing matrices having higher levels of expression than pure fibrin matrices (Fig. 4E and 4F).
DISCUSSION
Understanding ECM expression, synthesis and remodeling in response to different biopolymer compositions and geometries is of importance to bioengineers who wish to use these materials for tissue engineering and as models to study biology. Our results show that both ECM protein and integrin gene expression were up-regulated in 3D matrices, relative to 2D substrates. We focused on vascular cells and three ECM genes with relevance to the vasculature, in combination with three integrin genes associated with the biopolymer matrices used in this study. Collagen Type I and fibrin polymers have distinct biochemical structures, and the fibers formed by polymerization were also distinct in their morphology. Culture in these 3D matrices led to increased expression of all genes relative to 2D; however, the expression of ECM genes demonstrated a temporal pattern that resembled the sequence of events in wound healing.
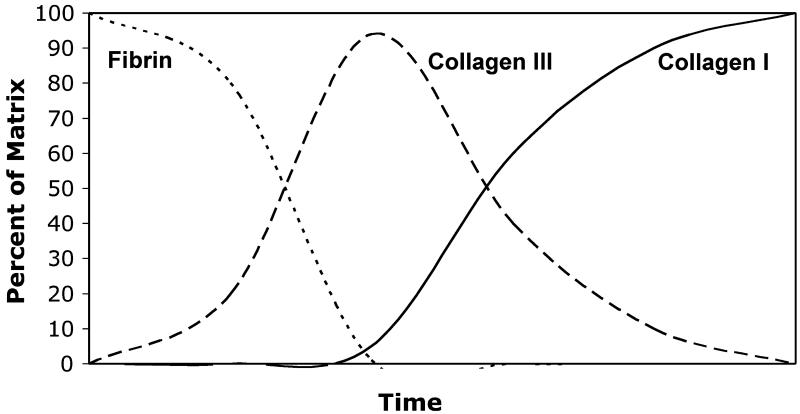
The ECM of a wound bed evolves over the course of time from a fibrin clot to a collagen scar (for a review, see, e.g., Ref. [20]), as depicted schematically in Fig. 5. In our system, fibrin gels were most similar to the ECM content of the initial blood clot. The collagen-fibrin composite material is a rough representation of the evolving granulation tissue, which contains fibrin and a collagen provisional matrix, although in native tissues the provisional matrix is primarily established with collagen Type III. Finally, the pure collagen gels simulate the later stages of wound healing, where a predominantly collagen Type I scar has replaced the provisional matrix. In this study, collagen Type III expression was highly elevated in 3D fibrin matrices, but was only modestly up-regulated in collagen-containing matrices, relative to 2D controls. In contrast, collagen I and tropoelastin expression were increased in pure collagen and collagen-fibrin composites. In addition, the temporal expression data showed that collagen III decreased over time, while collagen I and tropoelastin increased over 1 week in 3D culture.
Figure 5.
Schematic diagram showing the progression of the ECM during adult wound healing over time. Over a period of days and weeks the fibrin clot is replaced by a provisional matrix, which is composed mainly of collagen Type III. The provisional matrix is then degraded and replaced by a collagen Type I scar.
Some aspects of ECM gene expression by cells embedded within 3D protein lattices have previously been examined. Collagen Type I gene expression has produced conflicting results: while some studies observed a decrease in collagen synthesis [5], others have observed an increase as a result of 3D culture [21]. Another study showed both effects, and attributed the increase in gene expression and subsequent decrease in protein expression to posttranslational modifications of the nascent protein [22]. It also has been shown that the mechanical environment to which a collagen gel is exposed (e.g., constrained versus unconstrained compaction) can affect cell function in 3D matrices [23, 24], and cyclic stretching increased gene expression of collagens Type I and III by vascular cells in fibrin matrices [25]. In addition, it is known that the effect of growth factors on matrix synthesis is modulated by the presence of a pre-existing protein lattice [26], and different cell types will respond differently to similar matrices [27]. In contrast to the findings we report, fibrin gels have been shown to stimulate more tropoelastin gene expression by VSMC than collagen lattices [28], though these studies were carried our over a period of 5 weeks using neonatal VSMC. This highlights an important aspect of this study: we examined gene expression over short-term culture periods because our study sought to evaluate the immediate reaction of VSMC to matrix environment. The subsequent translation and processing of proteins, which were not characterized in the present work, also may be affected by the presence of a 3D matrix.
The expression of integrin subunits in this study suggests that cells mediated their own adhesion by expressing the integrins needed to anchor to their environment. In fibrin cultures, the β3-subunit, which binds fibrin when coupled with the αV subunit, was expressed at higher levels in fibrin cultures than the collagen-binding integrin subunits α1 and β1. The inverse was true for collagen-containing cultures. These results suggest that VSMC transcribe integrin genes specific to their environments to adhere to their matrix, and that specific integrin signaling also is involved in directing the remodeling of the matrix. For example, VSMC have been shown to modulate their integrin expression in response to the type of ECM presented. VSMC tend to bind to native collagen using α1β1 integrins [29] but also can bind via α2β1 integrins, and it has been suggested that collagen reorganization is specifically mediated through α2β1 integrins [30]. When fibroblasts in 3D collagen gels adhere to the matrix via α1β1, collagen gene expression is up-regulated; however, adhesion via α2β1 integrins leads to up-regulation of MMP-1 [31]. ECM also has been shown to modulate PDGF regulation of integrin gene expression in fibroblasts [32]. In an osteogenic cell line normally expressing only α1β1 integrin receptors, α2β1 integrin expression can be induced through culture in 3D collagen [33]. Similarly, microvascular endothelial cells are known to more potently up-regulate integrin αVβ3 expression in 3D fibrin matrices than in 3D collagen matrices [34]. Mesenchymal progenitor cells also regulate ECM and integrin expression via integrin binding [35]. Taken together, these results suggest that matrix-mediated expression of specific integrin types can be used to control cell function, though few practical applications of this knowledge have emerged to date.
Understanding and guiding cell-matrix interactions and matrix remodeling is a goal of vascular tissue engineering that also has important implications in vascular biology. Composite materials composed of collagen and fibrin are attractive because they capitalize on the structural integrity of collagen as well as the wound healing properties of fibrin. The absolute amount and ratio of collagen and fibrin potentially can be varied in order to tailor the biochemical and mechanical properties of these composite biomaterials. This study demonstrates that such 3D contextual cues are necessary to stimulate the evolution of the ECM in a process that to some extent mimics wound healing. The presence of 3D ECM led to ECM gene expression that suggested a directed development of the matrix not observed in 2D. These results can be used to help direct the dynamic cellular remodeling response of VSMC in defined protein matrices, with the goal of creating more robust and functional engineered tissues.
Acknowledgements
This manuscript was submitted in honor of Prof. Buddy Ratner’s 60th birthday, to help acknowledge his many contributions to biomaterials science in general, and hydrogel characterization in particular. It is aligned with the symposium titled “Biomaterials from 2D to 3D to Larger than Life”, held to celebrate Dr. Ratner’s birthday in Maui, Hawaii in December 2006. This work was supported in part by the American Heart Association through grant SDG-043511N and the National Institute of Biomedical Imaging and Bioengineering through grant R21-EB003978.
REFERENCES
- 1.Gentleman E, Lay AN, Dickerson DA, Nauman EA, Livesay GA, Dee KC. Biomaterials. 2003;24:3805. doi: 10.1016/s0142-9612(03)00206-0. [DOI] [PubMed] [Google Scholar]
- 2.Wallace DG, Rosenblatt J. Adv. Drug Deliv. Rev. 2003;55:1631. doi: 10.1016/j.addr.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Hubbell JA. Curr. Opin. Biotechnol. 2003;14:551. doi: 10.1016/j.copbio.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Aumailley M, Gayraud B. J. Mol. Med. 1998;76:253. doi: 10.1007/s001090050215. [DOI] [PubMed] [Google Scholar]
- 5.Thie M, Schlumberger W, Semich R, Rauterberg J, Robenek H. Eur. J. Cell Biol. 1991;55:295. [PubMed] [Google Scholar]
- 6.Naito M, Stirk CM, Smith EB, Thompson WB. Thromb. Res. 2000;98:165. doi: 10.1016/s0049-3848(99)00202-9. [DOI] [PubMed] [Google Scholar]
- 7.Engler AJ, Griffin MA, Sen S, Bonnemann CG, Sweeney HL, Discher DE. J. Cell Biol. 2004;166:877. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engbers-Buijtenhuijs P, Buttafoco L, Poot AA, Geelkerken RH, Feijen J, Vermes I. Tissue Eng. 2005;11:1631. doi: 10.1089/ten.2005.11.1631. [DOI] [PubMed] [Google Scholar]
- 9.Xu J, Clark RA. J. Cell Biol. 1997;136:473. doi: 10.1083/jcb.136.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grinnell F. Trends Cell Biol. 2003;13:264. doi: 10.1016/s0962-8924(03)00057-6. [DOI] [PubMed] [Google Scholar]
- 11.Rhee S, Grinnell F. Adv. Drug Deliv. Rev. 2007;59:1299. doi: 10.1016/j.addr.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green JA, Yamada KM. Adv. Drug Deliv. Rev. 2007;59:1293. doi: 10.1016/j.addr.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S, Moon JJ, Miao H, Jin G, Chen BP, Yuan S, Hu Y, Usami S, Chien S. J. Vasc. Res. 2003;40:378. doi: 10.1159/000072702. [DOI] [PubMed] [Google Scholar]
- 14.Stegemann JP, Nerem RM. Exp. Cell Res. 2003;15:283. doi: 10.1016/s0014-4827(02)00041-1. [DOI] [PubMed] [Google Scholar]
- 15.Cummings CL, Gawlitta D, Nerem RM, Stegemann JP. Biomaterials. 2004;25:3699. doi: 10.1016/j.biomaterials.2003.10.073. [DOI] [PubMed] [Google Scholar]
- 16.Hong H, McCullough CM, Stegemann JP. Biomaterials. 2007;28:3824. doi: 10.1016/j.biomaterials.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rowe SL, Stegemann JP. Biomacromolecules. 2006;7:2942. doi: 10.1021/bm0602233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alavi A, Stupack DG. Methods Enzymol. 2007;426:85. doi: 10.1016/S0076-6879(07)26005-7. [DOI] [PubMed] [Google Scholar]
- 19.Feng Z, Chian KS, Ong WF, Mhaisalka PS, Chan V, Ratner BD. J. Biomed. Mater. Res. A. 2007;82:788. doi: 10.1002/jbm.a.31123. [DOI] [PubMed] [Google Scholar]
- 20.Martin P. Science. 1997;276:75. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 21.Li S, Lao J, Chen BP, Li YS, Zhao Y, Chu J, Chen KD, Tsou TC, Peck K, Chien S. FASEB J. 2003;17:97. doi: 10.1096/fj.02-0256fje. [DOI] [PubMed] [Google Scholar]
- 22.Redecker-Beuke B, Thie M, Rauterberg J, Robenek H. Arterioscler. Thromb. 1993;13:1572. doi: 10.1161/01.atv.13.11.1572. [DOI] [PubMed] [Google Scholar]
- 23.Fringer J, Grinnell F. J. Biol. Chem. 2001;276:31047. doi: 10.1074/jbc.M101898200. [DOI] [PubMed] [Google Scholar]
- 24.Stegemann JP, Nerem RM. Ann. Biomed. Eng. 2003;31:391. doi: 10.1114/1.1558031. [DOI] [PubMed] [Google Scholar]
- 25.Flanagan TC, Cornelissen C, Koch S, Tschoeke B, Sachweh JS, Schmitz-Rode T, Jockenhoevel S. Biomaterials. 2007;28:3388. doi: 10.1016/j.biomaterials.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 26.Schlumberger W, Thie M, Rauterberg J, Robenek H. Arterioscler. Thromb. 1991;11:1660. doi: 10.1161/01.atv.11.6.1660. [DOI] [PubMed] [Google Scholar]
- 27.Williams C, Johnson SL, Robinson PS, Tranquillo RT. Tissue Eng. 2006;12:1489. doi: 10.1089/ten.2006.12.1489. [DOI] [PubMed] [Google Scholar]
- 28.Ross JJ, Tranquillo RT. Matrix Biol. 2003;22:477. doi: 10.1016/s0945-053x(03)00078-7. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto M, Yamato M, Aoyagi M, Yamamoto K. Exp. Cell Res. 1995;219:249. doi: 10.1006/excr.1995.1225. [DOI] [PubMed] [Google Scholar]
- 30.Lee RT, Berditchevski F, Cheng GC, Hemler ME. Circ. Res. 1995;76:209. doi: 10.1161/01.res.76.2.209. [DOI] [PubMed] [Google Scholar]
- 31.Langholz O, Rockel D, Mauch C, Kozlowska E, Bank I, Krieg T, Eckes B. J. Cell Biol. 1995;131:1903. doi: 10.1083/jcb.131.6.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu J, Clark RA. J. Cell Biol. 1996;132:239. doi: 10.1083/jcb.132.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riikonen T, Westermarck J, Koivisto L, Broberg A, Kahari VM, Heino J. J. Biol. Chem. 1995;270:13548. doi: 10.1074/jbc.270.22.13548. [DOI] [PubMed] [Google Scholar]
- 34.Feng X, Clark RA, Galanakis D, Tonnesen MG. J. Invest. Dermatol. 1999;113:913. doi: 10.1046/j.1523-1747.1999.00786.x. [DOI] [PubMed] [Google Scholar]
- 35.Heckmann L, Fiedler J, Mattes T, Brenner RE. Cells Tissues Organs. 2006;182:143. doi: 10.1159/000093964. [DOI] [PubMed] [Google Scholar]