Abstract
The challenge in the synthesis of protein-polymer conjugates for biological applications is to synthesize a stoichiometric (typically 1:1) conjugate of the protein with a monodisperse polymer, with good retention of protein activity, significantly improved pharmacokinetics and increased bioavailability, and hence improved in vivo efficacy. Here we demonstrate, using myoglobin as an example, a general route to grow a PEG-like polymer, poly(oligo(ethylene glycol) methyl ether methacrylate) [poly(OEGMA)], with low polydispersity and high yield, solely from the N-terminus of the protein by in situ atom transfer radical polymerization (ATRP) under aqueous conditions, to yield a site-specific (N-terminal) and stoichiometric conjugate (1:1). Notably, the myoglobin-poly(OEGMA) conjugate [hydrodynamic radius (Rh): 13 nm] showed a 41-fold increase in its blood exposure compared to the protein (Rh: 1.7 nm) after IV administration to mice, thereby demonstrating that comb polymers that present short oligo(ethylene glycol) side chains are a class of PEG-like polymers that can significantly improve the pharmacological properties of proteins. We believe that this approach to the synthesis of N-terminal protein conjugates of poly(OEGMA) may be applicable to a large subset of protein and peptide drugs, and thereby provide a general methodology for improvement of their pharmacological profiles.
Keywords: drug delivery, living radical polymerization, protein-polymer conjugate
Proteins and peptides are of interest as therapeutics by virtue of their high biological activity and specificity (1). However, the delivery of therapeutic proteins and peptides in their unmodified form has several limitations, which can include poor stability, low solubility, short in vivo circulation, and immunogenicity (2–5). Conjugating proteins and peptides with polymers is an effective strategy to overcome some of these limitations (2–5). Protein-polymer conjugates are typically prepared by reaction of semitelechelic polymers with the reactive side chains of protein residues. Modification of lysine or cysteine residues of proteins with poly(ethylene glycol) (PEG), generally known as PEGylation, is the most widely used polymer conjugation approach to improve the pharmacological profiles of proteins, and several PEGylated proteins are used clinically as therapeutics (2–5). Unfortunately, most proteins contain numerous chemically reactive residues that are promiscuously distributed on their surface, which can result in the formation of heterogeneous protein-polymer conjugates with unacceptably low biological activity (6–9).
An ideal method for the synthesis of protein-polymer conjugates for therapeutic applications must be carried out in aqueous conditions, and provide: 1) a stoichiometric conjugate (typically 1:1); 2) complete control of the site of conjugation; 3) high yield (> 50%); 4) good control of polymer dispersity; 5) retention of protein activity; and 6) a significant improvement in the pharmacokinetics and bioavailability of the conjugate compared to the unmodified protein. Recently, the direct growth of polymers from proteins has emerged as an alternative to postpolymerization conjugation (10–18), but these methods do not provide a general method for protein conjugation that meet all of these requirements. Furthermore, they have not been applied to synthesize polymers directly from a protein that improve the pharmacological profiles of proteins, nor have they been evaluated in vivo.
To devise a general strategy for the in situ growth of stoichiometric, site-specific polymer conjugates of proteins and peptides with improved pharmacological properties over the native molecules, we chose to grow a polymer chain directly from the N-terminus of a protein. We selected the N-terminus as the site of in situ growth, because the N-terminal amine has a sufficiently different pKa (≈7.8) from the amine group present on lysine side chains (pKa ≈ 10.5–12) that are typically distributed on the protein surface. As the different pKa of the N-terminal amine allows chemical reactions to be selectively performed at the N-terminus of a protein with no cross reaction at solvent-accessible lysine residues (19–24), we hypothesized that it should be possible to selectively attach a polymerization initiator to the N-terminus and subsequently grow a polymer chain solely from the N-terminus.
We chose poly(oligo(ethylene glycol) methyl ether methacrylate) [poly(OEGMA)] for in situ polymer growth because we had previously shown that poly(OEGMA) brushes grown on a planar surface confer exceptional protein and cell resistance to the surface of diverse materials (25–27), and we hypothesized that the corollary to protein resistance on surfaces is a long circulation half-life when the same polymer is grown from the surface of a protein. Furthermore, poly(OEGMA) can be synthesized by atom transfer radical polymerization (ATRP)—a living radical polymerization methodology—in aqueous medium (25–27) with good control of the polymer chain length (28), attributes that we believed would be invaluable for the growth of stoichiometric and functionally active polymer conjugates from the surface of a protein. In addition, poly(OEGMA) is likely to be biodegradable due to the enzyme-degradable and hydrolyzable ester linker (Fig. 1). The potential in vivo biodegradation of poly(OEGMA) may overcome the nonbiodegradable limitation of PEG for the design of the next generation of protein drugs (29).
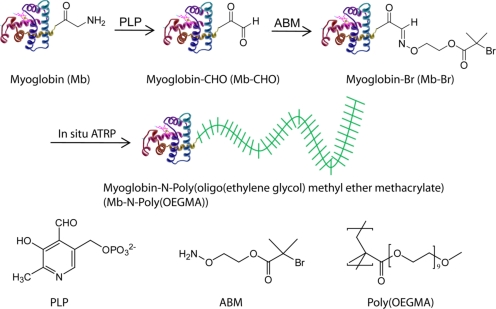
Fig. 1.
Schematic illustration of in situ growth of stoichiometric poly(oligo(ethylene glycol) methyl ether methacrylate) [poly(OEGMA)] at the N-terminus of myoglobin. First, the N-terminus (glycine) is transformed to an aldehyde through a biomimetic transamination reaction (Mb-CHO). Second, a hydroxylamine-functionalized ATRP initiator (ABM) is attached to the N-terminus, through a reaction between the aldehyde and the hydroxylamine, to form a macroinitiator (Mb-Br). Third, poly(OEGMA) is directly grown from the protein macroinitiator by atom transfer radical polymerization (ATRP).
Myoglobin (Mb) was selected as a model protein for proof of concept because: (1) it contains numerous (19) reactive lysine residues, most of which are distributed across its surface, so that it provides a stringent test-case of the selectivity of our strategy in growing a polymer solely from a protein's N-terminus; (2) Mb has a HEME prosthetic group with a characteristic Soret band at 409 nm, which is exquisitely sensitive to its local environment (24, 30), so that any deleterious changes to its tertiary structure upon attachment of the initiator or the growth of poly(OEGMA) would be reflected in changes in the absorbance of the Soret band; and (3) the renal clearance of hemoglobin and myoglobin based drug substitutes leads to nephrotoxicity, spurring many studies of modifications that prevent renal accumulation (1).
Results and Discussions
Attachment of an ATRP Intiator to the N-terminus of Mb.
To carry out ATRP from the N-terminus of Mb, a two-step procedure was used to attach the ATRP initiator solely to the N-terminal amine (Fig. 1). First, the protein was treated with pyridoxal-5-phosphate (PLP) in a phosphate buffer to yield an aldehyde at the N-terminus (Mb-CHO) through a transamination reaction (24). Two major peaks were observed by ESI-MS at 16,954 and 16,971 Da (Fig. S1B). Aldehydes can be catalyzed by acids to form hydrates, and the extent to which hydrates of aldehydes form gives a measure of the relative stability of aldehydes. We suggest this dynamic reaction led to the two peaks observed by ESI-MS. The peak at 16,971 Da is likely due to the hydrate form of Mb-CHO, Mb-C(H)(OH)(OH), as its experimentally observed molecular weight (MW) of 16,971 Da is consistent with the theoretical MW of 16,969 Da of this species. The second peak at 16,954 Da is likely to arise from two species, Mb-CHO (theoretical MW = 16,951 Da) and unmodified Mb, as the ESI-MS determined mass of control, native Mb (Fig. S1A) was 16,954 Da (theoretical MW = 16,952 Da). Note that N-terminal serine, cysteine, threonine, and tryptophan will be incompatible with this method due to known side reactions with aldehydes, and N-terminal proline will be expected to be unreactive (24).
Next, the mixture was treated with (2-(aminooxy)ethyl) 2-bromo-2-methylpropanoate (ABM). ESI-MS of the reaction mixture resulted in a single peak with a mass of 17,161 Da, which is in close agreement with the theoretical MW of 17,159 Da of the expected oxime product, myoglobin-Br (Mb-Br) (Fig. S1C). The overall yield of the N-terminal attachment of the ATRP initiator was ≈75% (Fig. S2), as seen by the area ratio of the peaks corresponding to Mb/Mb-CHO and Mb-Br. As a negative control, myoglobin was directly reacted with ABM without prior reaction with PLP and analyzed by ESI-MS. The MW of the control was determined to be 16,954 Da (Fig. S1D), which is identical to that of Mb (Fig. S1A). This means that Mb cannot react with ABM without prior reaction with PLP to form Mb-CHO.
The site specificity of the modification at the N-terminus was confirmed by subjecting Mb, Mb-CHO and Mb-Br to a proteolytic digest with trypsin. Myoglobin was also directly reacted with ABM without prior reaction with PLP and then digested with trypsin, as a negative control. A N-terminal fragment with an aldehyde was observed only for Mb-CHO (Fig. 2A) by analysis of the resulting peptide fragments by liquid chromatography/mass spectrometry/mass spectrometry (LC-MS/MS), which indicated that only the N-terminus of Mb was modified after reaction with PLP, leaving the 19 other amine groups in the lysine side chains unaffected. Furthermore, a N-terminal peptide fragment with a bromine was only observed for Mb-Br (Fig. 2A), which confirmed the successful modification of the aldehyde with ABM. The experimental isotopic distribution of the fragment (Fig. 2B) was comparable to the theoretical Br isotopic distribution (Fig. 2C), which further confirmed the incorporation of Br into this N-terminal fragment. The peptide fragments from the control sample were experimentally indistinguishable from those observed for unmodified Mb, indicating a lack of reaction between ABM and Mb without prior treatment of Mb with PLP. These data confirm that the ATRP initiator was solely attached to the N-terminal amine of the protein to form a N-terminal brominated macroinitiator (Mb-Br) despite the presence of 19 lysine residues in the protein. After reaction, residual PLP and ABM were removed by centrifugal ultrafiltration with a cut off MW of 3,000 Da.
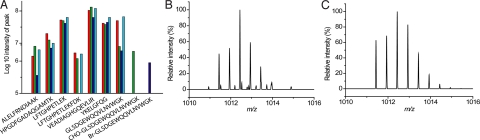
Fig. 2.
Analysis of peptide fragments by LC-MS/MS after trypsin digest. (A) Log10 intensity of myoglobin peptide fragments. Red column: Mb; Green column: Mb-CHO; Blue column: Mb-Br; Cyan column: control. Note: Log10 = 5 is the approximate S/N limit. (B) Measured isotope distribution of [*Br-GLSDGEWQQVLNVWGK]2+, m/z = 1011.4377 Da, and mass measurement error is 3.46 ppm. (C) Theoretical isotope distribution of [*Br- GLSDGEWQQVLNVWGK]2+, m/z = 1011.4332 Da.
In Situ Growth of Poly(OEGMA) at the N-Terminus of Mb.
In situ ATRP of OEGMA was next carried out from Mb-Br in buffer. The progress of ATRP was monitored by size exclusion chromatography (SEC) as a function of polymerization time (Fig. 3 A–C), and by SDS polyacrylamide gel electrophoresis (SDS/PAGE) analysis (Fig. S3). Refractive index (RI) and UV-visible absorbance detection at 280 nm (mainly protein) and 409 nm (HEME absorbance) allowed the protein (a mixture of unmodified Mb, residual Mb-CHO, and Mb-Br), to be unequivocally discriminated from poly(OEGMA) and the Mb-N-poly(OEGMA) conjugate in the polymerization mixture. The SEC traces for both RI (Fig. 3A) and absorbance at 280 nm and 409 nm (Fig. 3 B and C) clearly showed the intriguing appearance of a peak with a lower overall elution time than the peak corresponding to Mb; furthermore, this peak shifted to lower elution times with increasing polymerization times. The lower overall elution time of this peak is consistent with the in situ formation of a higher MW Mb-N-poly(OEGMA) conjugate and the shift of this peak to lower elution time with increasing ATRP time suggests increasing MW of the polymer in the conjugate, as expected. The absorbance traces at 280 nm and 409 nm in Fig. 3 B and C (blue traces) show that the polymerization mixture was composed primarily of the protein-polymer conjugate (≈70% by molar fraction) and a smaller (≈30%) fraction of residual protein (Mb, MB-CHO and Mb-Br). Because the 70% conversion efficiency of the protein to conjugate was close to that observed for the cumulative efficiency of conversion of the Mb to Mb-Br, it indicated that the initiation efficiency of the in situ ATRP from Mb-Br was nearly quantitative. SDS/PAGE analysis also clearly showed that the bands for Mb-N-poly(OEGMA) conjugates had a larger MW than for Mb-Br (Fig. S3) and increased with increasing polymerization times, which confirmed both the polymerization of OEGMA from Mb-Br and the increase in the MW of the polymer with increasing polymerization time.
Fig. 3.
In situ ATRP of OEGMA from the ATRP-initiator modified N terminus of myoglobin as a function of polymerization time. The polymerization mixtures were directly analyzed by SEC with a refractive index detector (RID) and a UV detector (UVD). (A–C) The SEC traces detected by RID and UVD at wavelengths of 280 and 409 nm: Mb-Br (black), 5 min ATRP time (red), 15 min ATRP time (green), and 60 min ATRP time (blue).
In a control experiment, in which native Mb was used as an initiator for ATRP under the same polymerization conditions as were used for Mb-Br, the SEC peak traces of the protein did not shift to lower elution times even after 1.5 h (Fig. S4), clearly indicating that ATRP did not occur without an ATRP initiation site on the protein. Note that the third peak with the longest elution time of 10.5 min in Fig. 3A corresponds to phosphate, sodium chloride or other small molecules, which have a different refractive index from the mobile phase. The absence of a peak corresponding to free polymer indicated that removal for residual ABM from the polymerization mixture was successful and ATRP was carried out in a controlled manner, so that there was no free polymer generated in these ATRP experiments.
Purification and Characterization of Mb-N-Poly(OEGMA).
To further characterize the polydispersity and size of the conjugate, a Mb-N-poly(OEGMA) conjugate that was synthesized by in situ ATRP for 1.0 h was separated from the residual protein by size exclusion chromatography (SEC). The SEC trace for absorbance detection at 280 nm of the purified conjugate (red trace, Fig. 4A) showed only one peak corresponding to an elution time of 6 min and the absence of the peak corresponding to the unreacted Mb that was observed in the unpurified protein polymer conjugate (Fig. 4A: black trace, peak elution time ≈9.3 min), showing that the unreacted protein had been completely removed from Mb-N-poly(OEGMA). These results were confirmed by absorbance detection at 409 nm (Fig. S5B) and RI detection as well (Fig. S5A). SDS/PAGE analysis provided visual confirmation of these results, as the band corresponding to Mb with a mass of ≈17,000 Da disappeared, and a smear of the Mb-poly(OEGMA) conjugate with a higher mass of approximately 150 kDa was exclusively seen after purification by SEC (Fig. S6).
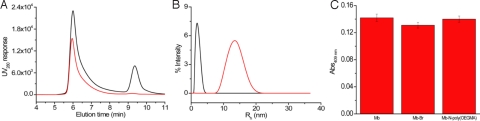
Fig. 4.
Purification and characterization of Mb-N-poly(OEGMA). The polymerization mixtures were purified by SEC. (A) The SEC traces from UVD at 280 nm, wherein black and red curves correspond to samples of 60 min ATRP solution and purified Mb-N-poly(OEGMA), respectively. (B) DLS data of Mb (black curve) and Mb-N-Poly(OEGMA) (red curve) in phosphate buffer solution. (C) Peroxidase activity of Mb, Mb-Br and Mb-N-poly(OEGMA).
The number-average molecular weight of the purified Mb-N-poly(OEGMA) conjugate with a polymerization time of 1.5 h was approximately 120 kDa, as determined by SEC calibrated with PEG standards, and the polydispersity of the conjugate was 1.4. The polydispersity may have some impact on the in vivo performance of the conjugate, although we note that it could potentially be reduced by careful optimization of the reaction conditions, or by the use of alternative polymerization methods such as reversible addition fragmentation chain transfer (RAFT) polymerizations, that are believed to provide better control over polymer dispersity. The efficient purification of the conjugate from the protein was also confirmed by dynamic light scattering (DLS) which revealed only one distribution of particles with a hydrodynamic radius (Rh) of 13 nm with a polydispersity of 11.2% after purification, and the absence of a species corresponding to myoglobin, which has a Rh of 1.7 nm as shown in Fig. 4B. 1H NMR analysis of the conjugate confirmed the synthesis of poly(OEGMA) from the protein (Fig. S7). As expected, characteristic signals of poly(OEGMA) were clearly observed at 1, 2, 3.4, 3.6–3.8, and 4.2 ppm that correspond to CCH3, CH2C, OCH3, OCH2CH2, and C(O)OCH2, respectively. Note that the protein signals are not visible in the NMR spectrum under the conditions used for NMR analysis (16). Elemental analysis of the Mb-N-poly(OEGMA) conjugate indicated the absence of copper in the conjugate. We also measured the absorbance of the HEME prosthetic group at 409 nm of the conjugate relative to the native protein. The absorbance of the HEME group at 409 nm was almost unchanged over the course of the reaction (Fig. S8), which indicated that the site specific polymer conjugation of Mb at the N-terminus through in situ ATRP did not perturb the tertiary structure of the protein. We further confirmed the retention of the functional activity of the conjugate by quantifying the peroxidase-like activity of Mb (31). The absorbance at 409 nm was monitored by UV-visible spectrophotometry after treatment of Mb and modified derivatives with 2,2'-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS) following reaction with hydrogen peroxide (Fig. 4C). Mb, before and after the attachment of the ATRP initiator, showed almost the same activity, which was determined by comparing the absorbance at 409 nm of Mb at the same concentration of protein (1 μM). The original activity of Mb was also retained after the in situ synthesis of the poly(OEGMA) conjugate. These data clearly indicated the peroxidase activity of Mb was retained after in situ growth of a stoichiometric (1:1) poly(OEGMA) conjugate at the N-terminal site.
In Vivo Pharmacological Evaluation.
The pharmacokinetics of Mb and the purified Mb-N-poly(OEGMA) conjugate with a hydrodynamic radius of 13 nm was determined by intravenously administering 125I labeled Mb and conjugate to nude mice (BALB/c nu/nu), and collecting blood samples at various time intervals after administration (Fig. 5). A two-compartment model was used to fit the data. Distribution and elimination data were represented by the following parameters: area under the curve (AUC), total body clearance (CL), and blood half-life for the distribution and elimination phase (t1/2α, t1/2β) (Table 1). All pharmacokinetic parameters revealed a biphasic behavior (Fig. 5). After bolus administration, the concentration of unmodified Mb rapidly decreased in blood, with a short distribution phase (t1/2α = 3.0 min) and a rapid terminal elimination phase (t1/2β = 2 h). In contrast, the distribution phase of poly(OEGMA) conjugation increased by 40 times (t1/2α = 2 h), and the terminal elimination phase was prolonged to 18 h. There was also significant difference in the clearance rate between Mb (1.43 mL/h) and Mb-N-polyOEGMA (0.035 mL/h) respectively. These differences in the pharmacokinetics resulted in a 41-fold greater area under the curve (AUC) for the Mb-N-poly(OEGMA) conjugate (2.9 ×107 h*cpm/mL) respectively as compared to myoglobin (7.0 × 105 h*cpm/mL, respectively), indicating N-terminal site-specific conjugation with poly(OEGMA) significantly enhanced the cumulative exposure of Mb in the blood. This dramatic improvement of blood pharmacokinetic parameters demonstrates that this is an exciting protein conjugation method for prolonging the circulation of protein and peptide therapeutics with potential applications ranging from blood substitutes to targeting solid tumors.
Fig. 5.
Blood concentration as a function of time postinjection. After i.v. administration of 125I labeled Mb (green triangle) and Mb-N-poly(OEGMA) (black square), blood was sampled from the tail of mouse at given time points and quantified for radioactivity.
Table 1.
Pharmacokinetic parameters calculated from a two-compartment model
| Protein | T1/2α, h | T1/2β, h | AUC, h·CPM/mL | CL, mL/h |
|---|---|---|---|---|
| Mb | 0.05 | 3.0 | 7.0 × 105 | 1.43 |
| Mb-N-poly(OEGMA) | 2.0 | 18.0 | 2.9 × 107 | 0.035 |
The data in Fig. 5 were fit to a two-compartment model shown as a solid line, to quantify the elimination and distribution response of Mb and Mb-N-poly(OEGMA).
Conclusions
We report a general approach to directly grow stoichiometric (1:1) polymer conjugates from a defined and ubiquitous location on a protein scaffold—the N-terminus—via a living radical polymerization under aqueous conditions with high yield, with no free polymer byproduct, and complete retention of protein activity. This approach of in situ polymerization from a protein's N-terminus also yields two findings that are relevant to the field of biological therapeutics: first, we report a biodegradable, PEG-like polymer conjugate with a notably different polymer architecture—a PEG-like comb polymer—as compared to the predominantly linear and lightly branched PEGs that are currently used in the biopharmaceutical industry; second, we report exciting in vivo data for this class of polymer conjugates, and show that a poly(OEGMA) conjugate significantly prolongs the half-life of a relatively short-lived protein after IV administration to mice; third, together, the demonstration of this methodology to grow a polymer from a protein's N-terminus, allied with the observation that a pharmaceutically relevant—poly(OEGMA)—conjugate has a dramatic effect on increasing the in vivo half-life of a protein, provides a synthetic methodology and a pharmaceutically relevant polymer for the development of peptide and protein therapeutics to a field that has relied on the chemical attachment of PEGs to proteins for far too long, in its longstanding efforts to improve the pharmaceutical profiles of protein and peptide drugs. From a broader perspective, our results demonstrate that the importance of protein resistance in medicine and biotechnology spans length scales from the macroscopic to the molecular: modification of the macroscale surfaces with protein-resistant polymers to improve their performance as diagnostics (32) has a suitable corollary in the modification of the molecular surfaces of protein pharmaceutics with the same class of polymers to improve their pharmacokinetic profiles.
Methods
Synthesis of ATRP Initiator.
The ATRP initiator (2-(aminooxy)ethyl) 2-bromo-2-methylpropanoate·HCl (ABM) was synthesized as described in the SI Text.
Conjugation of ATRP Initiator to the N-Terminus of Myoglobin.
A 50-mL Eppendorf tube was charged with a solution of Mb (25 mL of a 100 μM solution in 25 mM phosphate buffer, pH 6.5) and a solution of pyridoxal 5′-phosphate (PLP) (25 mL of a 20 mM solution in 25 mM phosphate buffer, pH adjusted to 6.5 with 2 M NaOH). The mixture was briefly agitated to ensure proper mixing, and was incubated without further agitation at 37 °C for 36 h. The PLP was removed from the reaction mixture via ultracentrifugation (Amicon Ultra-15 centrifugal filter; 3,000 MWCO). The purified mixture (25 mL of a 50 μM solution in 50 mM phosphate buffer, pH 5.5) was treated with the ATRP initiator, ABM (25 mL of a 2 mM solution in 50 mM phosphate buffer, pH 5.5) and allowed to sit without agitation for 36 h. The reaction solution was similarly purified by ultracentrifugation.
In Situ ATRP from the N-Terminus of Myglobin.
A deoxygenated solution of 1 mL PBS, 1.05 mmol poly(ethylene glycol) methyl ether methacrylate (OEGMA) (MW = 475, Sigma-Aldrich), 0.02 mmol CuCl, 0.044 mmol CuCl2, and 0.087 mmol 1,1,4,7,10,10-hexamethyltriethylenetetramine (HMTETA) was transferred into a deoxygenated myoglobin-Br (Mb-Br) solution in PBS. The polymerization was allowed to proceed for a given time under argon and then exposed to air. The polymerization mixture was further purified with HPLC (AKTA, GE Life Science) using a size exclusion column.
Trypsin Digestion of Myoglobin.
Protein samples were anayzed using a miniBradford assay to determine protein concentration (Bio-Rad, Inc.) followed by concentration normalization, and brief digestion with trypsin (Promega) by incubation of protein with trypsin at a 25:1 (wt/wt) ratio for 1 h at 37 °C in a solution of 50 mM ammonium bicarbonate. Samples were then acidified in 0.1% formic acid to stop proteolysis. Approximately 2.5 pmol of myoglobin equivalent (by Bradford assay) was analyzed by LC-MS/MS using a Waters nanoAcquity and Thermo LTQ-Orbitrap XL mass spectrometer (Thermo Corporation). Peptides were separated on a 75 μm × 250 mm BEH C18 column using a gradient of 5 to 40% vol/vol acetonitrile with 0.1% formic acid in 90 min, with a flow rate of 0.3 μL/min and column temperature of 45 °C. Electrospray ionization was used to introduce the sample into the Orbitrap mass spectrometer in real time, with precursor ion scanning from 400 to 2,000 m/z with 60,000 resolution. The top two precursor ions above a threshold of 10,000 counts were selected for MS/MS analysis in a data dependent fashion, and read out in the Orbitrap with 7,500 resolution. Both precursor and product ion spectra are collected in a high-resolution, accurate-mass mode, allowing database searching to be performed at 10 ppm parent ion and 0.02 Da fragment ion mass tolerance.
Tandem mass spectra (MS/MS) were extracted, and their charge state was deconvoluted and deisotoped using Rosetta Elucidator software, (version 3.2, Rosetta Bioinformatics). All MS/MS spectra were analyzed using Mascot (Matrix Science; version 2.2.0). Mascot was set up to search SwissProt (version 55.6, mammalian taxonomy, 390,696 sequences) with trypsin as the proteolytic enzyme. Mascot was searched with a fragment ion mass tolerance of 0.02 Da and a parent ion tolerance of 10 ppm. Oxidation of methionine was specified in Mascot as a variable modification. All raw data were imported into Rosetta Elucidator and the four LC-MS/MS runs (one per sample) were aligned and features extracted using the PeakTeller algorithm. Database searching (described above) was used to assign peptide identifications to the quantified features. All peptides reported were unambiguously identified using MS/MS database searching, with the exception of the brominated N-terminal peptide, whose identity was confirmed by accurate mass and isotope distribution modeling. Relative quantitation of each of the Mb peptides in the four different samples was performed by comparing the extracted ion intensities between the samples.
Evaluation of Peroxidase Activity of Myoglobin.
Briefly, hydrogen peroxide (5 μL, 40 mM) was added to 1 mL of a 1 μM solution of Mb or modified Mb in 100 mM phosphate buffer at pH 7.0. After 5 min incubation at 20 °C, excess catalase from bovine liver (2 mg/mL, 5 μL) was added, and the solution was further incubated for 2 min to completely decompose the excessive hydrogen peroxide. 2, 2′-azino-di-(3-ethyl)benzthiazoline-6-sulfonic acid (ABTS) (10 mM, 5 μL) was added to the solution. The oxidation reaction was monitored on a Cary-300 UV-visible spectrophotometer at 409 nm. Protein concentrations were determined by the BCA protein assay (Pierce).
Attachment of Radiohalogens to Myoglobin.
Iodination of myoglobin with 125I was performed with a Pierce IODO-Gen Precoated tube and purified by desalting on a dextran column (Pierce). Briefly, 100 μL of 27 μM Mb (or conjugate) was added to 1 mCi 125INa loaded in IODO-Gen precoated tubes. The tubes were shaken every 30 s for 15 min. The iodinated protein or conjugate was purified by gel-filtration chromatography on a dextran column. The radioactivity was counted by a gamma-counter (LKB-Wallac) and the labeling efficiency was calculated by the equation: % Efficiency = (Radioactivity of collected conjugate/Total loaded radioactivity) × 100.
Pharmacokinetics.
Seven nude mice bearing mouse breast carcinoma xenograft (4T1) were randomly divided into two groups. Group 1 (three mice) and 2 (four mice) received IV bolus injections of a given amount of PBS solution of 125I labeled Mb or Mb-N-poly(OEGMA) conjugate, respectively. Each animal was intravenously dosed with ≈0.3 μg Mb (component) with ≈5 μCi of 125I. Blood samples (20 μL) was collected from the tail vein of the mice at 40 s, 15, 30 and 60 min, 2, 4, 8, 24, 48 and 72 h after injection. The whole blood sample was analyzed by gamma-counting. The blood concentration time course was analyzed with a standard two-compartment pharmacokinetic model to approximate both distribution and elimination of the samples. All animal experiments were performed in accordance with the Duke University Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments.
We thank David Gooden at the Duke University Small Molecule Synthesis Facility for synthesis of the ATRP initiator ABM; George R Dubay at the Department of Chemistry of Duke University for MALDI-MS, ESI-MS, and NMR measurements; and J. Will Thompson, Erik J. Soderblom, and Laura G. Dubois at Duke Institute for Genome Sciences & Policy for protein sequencing.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904378106/DCSupplemental.
References
- 1.Lee VHL. Peptide and protein drug delivery. New York: Mercel Dekker; 1991. [Google Scholar]
- 2.Nucci ML, Shorr R, Abuchowski A. The therapeutic value of poly(ethylene glycol)- modified proteins. Adv Drug Deliv Rev. 1991;6:133–151. [Google Scholar]
- 3.Harris JM, Martin NE, Modi M. PEGylation: A novel process for modifying pharmacokinetics. Clin Pharmacokinet. 2001;40:539–551. doi: 10.2165/00003088-200140070-00005. [DOI] [PubMed] [Google Scholar]
- 4.Caliceti P, Veronese FM. Pharmacokinetic and biodistribution properties of poly(ethylene glycol)-protein conjugates. Adv Drug Deliv Rev. 2003;55:1261–1277. doi: 10.1016/s0169-409x(03)00108-x. [DOI] [PubMed] [Google Scholar]
- 5.Parveen S, Sahoo SK. Nanomedicine: Clinical applications of polyethylene glycol conjugated proteins and drugs. Clin Pharmacokinet. 2006;45:965–988. doi: 10.2165/00003088-200645100-00002. [DOI] [PubMed] [Google Scholar]
- 6.Francis GE, et al. PEGylation of cytokines and other therapeutic proteins and peptides: The importance of biological optimization of coupling techniques. Int J Hematol. 1998;68:1–18. doi: 10.1016/s0925-5710(98)00039-5. [DOI] [PubMed] [Google Scholar]
- 7.Veronese FM. Peptide and protein PEGylation: A review of problems and solutions. Biomaterials. 2001;22:405–417. doi: 10.1016/s0142-9612(00)00193-9. [DOI] [PubMed] [Google Scholar]
- 8.Roberts MJ, Bentley MD, Harris JM. Chemistry for peptide and protein PEGylation. Adv Drug Deliv Rev. 2002;54:459–476. doi: 10.1016/s0169-409x(02)00022-4. [DOI] [PubMed] [Google Scholar]
- 9.Basu A, et al. Structure-function engineering of interferon-1b for improving stability, solubility, potency, immunogenicity, and pharmacokinetic properties by site-selective mono-PEGylation. Bioconjug Chem. 2006;17:618–630. doi: 10.1021/bc050322y. [DOI] [PubMed] [Google Scholar]
- 10.Lele BS, Murata H, Matyjaszewski K, Russell AJ. Synthesis of uniform protein-polymer conjugates. Biomacromolecules. 2005;6:3380–3387. doi: 10.1021/bm050428w. [DOI] [PubMed] [Google Scholar]
- 11.Bontempo D, Maynard HD. Streptavidin as a macroinitiator for polymerization: In-situ protein-polymer conjugate formation. J Am Chem Soc. 2005;127:6508–6509. doi: 10.1021/ja042230+. [DOI] [PubMed] [Google Scholar]
- 12.Heredia KL, et al. In situ preparation of protein-“smart” polymer conjugates with retention of bioactivity. J Am Chem Soc. 2005;127:16955–16960. doi: 10.1021/ja054482w. [DOI] [PubMed] [Google Scholar]
- 13.Nicolas J, et al. Fluorescently tagged polymer bioconjugates from protein derived macroinitiators. Chem Commun. 2006;45:4697–4699. doi: 10.1039/b609935a. [DOI] [PubMed] [Google Scholar]
- 14.Zeng Q, et al. Chemoselective derivatization of a bionanoparticle by click reaction and ATRP reaction. Chem Commun. 2007;14:1453–1455. doi: 10.1039/b617534a. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, et al. In situ formation of protein-polymer conjugates through reversible fragmentation chain transfer polymerization. Angew Chem Int Ed. 2007;46:3099–3103. doi: 10.1002/anie.200604922. [DOI] [PubMed] [Google Scholar]
- 16.Boyer C, et al. Well-defined protein-polymer conjugates via in situ RAFT polymerization. J Am Chem Soc. 2007;129:7145–7154. doi: 10.1021/ja070956a. [DOI] [PubMed] [Google Scholar]
- 17.Droumaguet BL, Velonia K. In situ ATRP-mediated synthesis of hierarchical formation of giant amphiphile bionanoreactors. Angew Chem Int Ed. 2008;47:6263–6266. doi: 10.1002/anie.200801007. [DOI] [PubMed] [Google Scholar]
- 18.De P, et al. Temperature-regulated activity of responsive polymer-protein conjugates prepared by grafting-from via RAFT polymerization. J Am Chem Soc. 2008;130:11288–11289. doi: 10.1021/ja804495v. [DOI] [PubMed] [Google Scholar]
- 19.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 20.Tam JP, Yu Q, Miao Z. Orthogonal Ligation Strategies for Peptide and Protein. Biopolymers. 1999;51:311–332. doi: 10.1002/(SICI)1097-0282(1999)51:5<311::AID-BIP2>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 21.Kinstler O, et al. Mono-N-terminal poly(ethylene glycol)-protein conjugates. Adv Drug Deliv Rev. 2002;54:477–485. doi: 10.1016/s0169-409x(02)00023-6. [DOI] [PubMed] [Google Scholar]
- 22.Lee H, et al. N-terminal site-specific mono-PEGylation of epidermal growth factor. Pharm Res. 2003;20:818–825. doi: 10.1023/a:1023402123119. [DOI] [PubMed] [Google Scholar]
- 23.Baker DP, et al. N-terminally PEGylated human interferon-α 1a with improved pharmacokinetic properties and in vivo efficacy in a melanoma angiogenesis model. Bioconj Chem. 2006;17:179–188. doi: 10.1021/bc050237q. [DOI] [PubMed] [Google Scholar]
- 24.Gilmore JM, Scheck RA, Esser-Kahn AP, Joshi NS, Francis MB. N-terminal protein modification through a biomimetic transamination reaction. Angew Chem Int Ed. 2006;45:5307–5311. doi: 10.1002/anie.200600368. [DOI] [PubMed] [Google Scholar]
- 25.Ma HW, Hyun JH, Stiller P, Chilkoti A. “Non-fouling” oligo(ethylene glycol)-functionalized polymer brushes synthesized by surface-initiated atom transfer radical polymerization. Adv Mater. 2004;16:338–341. [Google Scholar]
- 26.Ma HW, Wells M, Beebe TP, Chilkoti A. Surface-initiated atom transfer radical polymerization of oligo(ethylene glycol) methyl methacrylate from a mixed self-assembled monolayer on gold. Adv Funct Mater. 2006;16:640–648. [Google Scholar]
- 27.Ma HW, Li DJ, Sheng X, Zhao B, Chilkoti A. Protein-resistant polymer coatings on silicon oxide by surface-initiated atom transfer radical polymerization. Langmuir. 2006;22:3751–3756. doi: 10.1021/la052796r. [DOI] [PubMed] [Google Scholar]
- 28.Feng W, Chen R, Brash JL, Zhu S. Surface-initiated atom transfer radical polymerization of oligo(ethylene glycol) methacrylate: Effect of solvent on graft density. Macromol Rapid Commun. 2005;26:1383–1388. [Google Scholar]
- 29.Gaberc-Porekar V, Zore I, Podobnic B, Menart V. Obstacles and pitfalls in the PEGylation of therapeutic proteins. Curr Opin Drug Discov Devel. 2008;11:242–250. [PubMed] [Google Scholar]
- 30.Kawamura-konishi Y, Kihara H, Suzuki H. Reconstitution of myoglobin from apoprotein and heme, monitored by stopped-flow absorption, fluorescence and circular dichroism. Eur J Biochem. 1988;170:589–595. doi: 10.1111/j.1432-1033.1988.tb13738.x. [DOI] [PubMed] [Google Scholar]
- 31.Hayahsi T, et al. Peroxidase activity of myoglobin is enhanced by chemical mutation of HEME-propionates. J Am Chem Soc. 1999;121:7747–7750. [Google Scholar]
- 32.Hucknall A, Kim DH, Rangarajan S, Hill RT, Reichert WM, Chilkoti A. Simple fabrication of antibody microarrays on nonfouling polymer brushes with femtomolar sensitivity for protein analytes in serum and blood. Adv Mater. 2009;21:1968–1971. doi: 10.1002/adma.200803125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.