Abstract
Following recent success in genome-wide association studies, a critical focus of human genetics is to understand how genetic variation at implicated loci influences cellular and disease processes. Crohn’s disease (CD) is associated with SNPs around IRGM1,2, but coding-sequence variation has been excluded as a source of this association2. We identified a common, 20-kb deletion polymorphism, immediately upstream of IRGM and in perfect linkage disequilibrium (r2 = 1.0) with the most strongly CD-associated SNP, that causes IRGM to segregate in the population with two distinct upstream sequences. The deletion (CD risk) and reference (CD protective) haplotypes of IRGM showed distinct expression patterns. Manipulation of IRGM expression levels modulated cellular autophagy of internalized bacteria, a process implicated in CD. These results suggest that the CD association at IRGM arises from an alteration in IRGM regulation that affects the efficacy of autophagy and identify a common deletion polymorphism as a likely causal variant.
Recently, SNP rs13361189 was found to be strongly associated (P = 2.1 × 10−10) with Crohn’s disease in a genome-wide association scan and independent replication study1,2. rs13361189 lies immediately upstream of IRGM, a gene previously shown to be essential for autophagy3. Because the most strongly associated SNPs span the 5′ end of IRGM, and because CD risk is also associated with the autophagy gene ATG16L1 (refs. 4,5), the association signal seems to arise from IRGM2. However, coding-sequence variation in IRGM has been excluded as the source of the association signal: resequencing IRGM exons in 248 individuals revealed only three coding-sequence variants, of which two were uncorrelated with CD risk and the third was a synonymous exonic SNP that did not affect IRGM protein sequence or splice sites2.
HapMap SNPs upstream of IRGM showed a pattern of assay failure (multiple SNPs yielding null genotypes in the same 34 samples) that is characteristic of structural polymorphisms6. To directly assess whether structural polymorphisms reside near IRGM, we analyzed experimental data in which DNA from the 270 HapMap samples were analyzed using a hybrid array of SNP and copy-number probes (SNP6.0 array; S.A.M., F.G. Kuruvilla, J.M. Korn, M.J.D. and D.A., unpublished data) (Fig. 1a). Six copy-number probes spanning the 13-kb region from 150.186 Mb to 150.199 Mb (spanning the failing HapMap SNPs) showed a correlated variation in intensity across samples, suggesting the existence of a common copy-number polymorphism upstream of IRGM (Fig. 1a).
Figure 1.
A common, 20-kb deletion polymorphism upstream of IRGM.
(a) Hybridization of DNA from 90 HapMap YRI samples to the Affymetrix SNP 6.0 array reveals a correlated pattern of variation in intensity across six copy-number probes spanning the region upstream of IRGM, suggesting the existence of a common copy-number polymorphism. (Darker shades of red represent reduced hybridization intensity.) Quantitative PCR indicated that the copy-number-variable region was an insertion/deletion (Supplementary Fig. 1). (b) Mapping of the deletion breakpoints by microarray and PCR. Red indicates evidence for deletion; black indicates evidence to the contrary; blue arrows indicate locations of flanking PCR primers able to generate a PCR product in individuals with at least one deletion allele. (c) Sequence of the reference and deletion alleles. The CD-associated SNP rs13361189 is indicated in boldface red type. Physical coordinates are on the reference human genome (build 35/36); allele frequencies are for extended HapMap analysis panels, including 540 samples. (d) CD association of typed and imputed polymorphisms in the IRGM region in the NIDDK IBDGC genome scan5,8. Blue trace indicates recombination map. The deletion polymorphism and rs13361189 are indicated by red arrows. rs13361189 (which is in perfect linkage disequilibrium with the deletion, r2 = 1.0) was also the most strongly associated SNP in the combined WTCCC genome scan and replication study2.
Quantitative PCR assays across the identified region revealed that individuals have 0, 1 or 2 copies of the region per diploid genome, indicating that the structural polymorphism is an insertion/deletion (Supplementary Fig. 1 online). The insertion/deletion was in perfect linkage disequilibrium with rs13361189 (r2 = 1.0) in all HapMap analysis panels, indicating that it is an ancestral mutation and making it a candidate to explain the association signal at rs13361189.
To determine the physical extent and molecular nature of the deletion polymorphism, we used PCR assays to map its breakpoints (Fig. 1b). PCR capture and sequencing of the deletion breakpoints revealed that the deletion removes 20,103 nucleotides, replacing them with 7 nucleotides (Fig. 1c). Identical lesions were identified in 6/6 individuals tested, reinforcing the linkage disequilibrium data in suggesting that this insertion/deletion represents a single ancestral mutation. The 20-kb affected sequence was observed at the same genomic location in the chimpanzee genome, indicating that the insertion allele is the ancestral state. The right breakpoint of the deletion was 123 bp from the CD-associated SNP rs13361189 (Fig. 1c) and 2.7 kb before the reported IRGM transcription start7.
We then sought to determine whether this deletion polymorphism showed CD association consistent with it being the causal allele at this locus. First, to directly confirm that the deletion was associated with risk of inflammatory bowel disease and CD, we typed the polymorphism in a North American case-control collection of 685 individuals. Relative to its frequency in unaffected individuals (10%), the deletion allele showed an elevated frequency in individuals with inflammatory bowel disease (15%, odds ratio (OR) = 1.5, P < 0.01), including association to CD (allele frequency 15%, OR = 1.6, P < 0.01) and ulcerative colitis (allele frequency 14%, OR = 1.4, P < 0.05). These data contained 150 copies of the deletion allele and showed a perfect (r2 = 1.0) correlation between the deletion and the CD-associated SNP rs13361189, further indicating that rs13361189 is a proxy for the deletion. We further confirmed this relationship by evaluating regional probe-intensity information and rs13361189 genotypes from newly generated data on 990 additional extended HapMap samples run on the SNP 6.0 array (Supplementary Fig. 1). In total, combining IBD, HapMap and extended HapMap data, we observed perfect correlation of rs13361189 and the deletion polymorphism across 933 instances of the minor allele of each in a sample comprising individuals of various ancestries. These results indicate equivalence of rs13361189 and the structural polymorphism for the purposes of association.
To compare the association signal at rs13361189 and the deletion to other SNPs in the region, we used additional SNP data from the National Institute of Diabetes and Digestive and Kidney Diseases Inflammatory Bowel Disease Genetics Consortium (NIDDK IBDGC) genome scan5,8. As in the combined Wellcome Trust Case Control Consortium (WTCCC) and replication study2, rs13361189 and its perfectly correlated neighbors showed the strongest CD association (P = 3.0 × 10−4) of all SNPs in the region (Fig. 1d). A second set of SNPs at IRGM (rs4958847 and its perfectly correlated neighbors), also reported in the WTCCC replication study2 and partially correlated with rs13361189 and the deletion, was more modestly associated with CD (P = 0.003). In combination with the WTCCC results, these SNPs showed association more than two orders of magnitude less significant than rs13361189 (3.8 × 10−10 versus 2.1 × 10−12) and therefore did not seem to explain the association. Because the earlier HapMap CEU data suggested the existence of a large block of linkage disequilibrium that extended across the nearby gene ZNF300, we also examined linkage disequilibrium and CD association in the genes near IRGM. The extended HapMap sample and IBDGC CD cohort indicated that SNPs in other genes were only partially correlated with rs13361189. Notably, in the CD cohort, rs13361189 remained associated (P < 0.05) conditional on genotypes at all SNPs beyond the boundaries of IRGM, but no SNPs showed association conditional on genotype at rs13361189. Thus, rs13361189 and its strongly correlated neighbors at the 5′ end of IRGM, including the 20-kb deletion polymorphism, are the primary polymorphisms that can explain the CD association in this region.
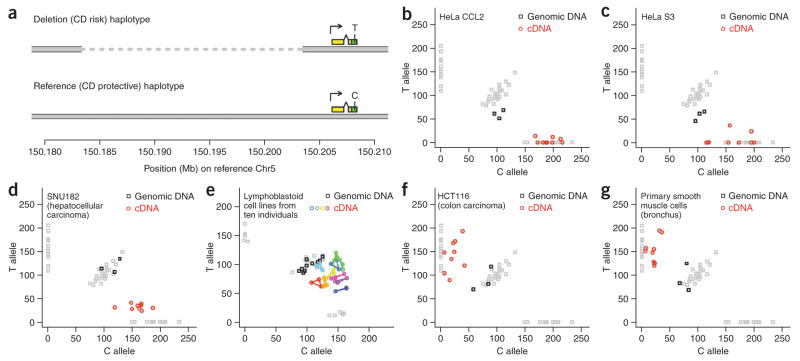
Given the nature and location of these potential causal polymorphisms, we next assessed whether the IRGM haplotypes differ in their regulation of IRGM expression and whether IRGM expression levels have physiological consequence. To assess whether the deletion (CD risk) and reference (CD protective) haplotypes of IRGM differ in their ability to activate IRGM expression, we measured the relative abundance of IRGM transcripts derived from the two haplotypes in cell lines that were heterozygous for the two haplotypes. Comparing the relative expression of two alleles in heterozygous cells allows the analysis of cis-acting variation in a way that controls for trans effects and environmental influences9,10. This approach was facilitated by the existence of an exonic synonymous SNP (rs10065172) in IRGM that was in strong linkage disequilibrium (r2 = 1.0 in samples tested) with both rs13361189 and the deletion polymorphism, such that transcripts arising from the risk (deletion) haplotype carry the T allele of rs10065172, and transcripts arising from the protective (reference) haplotype carry the C allele (Fig. 2a).
Figure 2.
Differential expression of IRGM from the deletion (CD risk) and reference (CD protective) haplotypes. (a) The exonic SNP rs10065172, which is in strong linkage disequilibrium with the 20-kb insertion/deletion polymorphism upstream of IRGM, can be used to distinguish IRGM transcripts that arise from the deletion haplotype from IRGM transcripts that arise from the reference haplotype. This makes it possible to measure the relative expression of the two haplotypes in heterozygotes by measuring the relative abundance of the two rs10065172 alleles in cDNA. (Yellow and green rectangles indicate transcribed sequence; green rectangles indicate protein-coding sequence.) (b–g) Differential expression of IRGM from the two structural haplotypes in heterozygotes. The relative abundance of the C and T alleles of rs10065172 in cDNA (colored circles) and genomic DNA (black squares) from human tissues and cell lines were measured. Gray squares indicate control measurements for genomic DNA from 48 HapMap YRI individuals, identifying the three reference genotype classes CC, CT and TT. Genomic DNA from all other samples was heterozygous for the C and T alleles (black squares); HeLa genomic DNA (b,c) showed a 2:1 allelic ratio (CCT) reflecting HeLa’s triploid 5q karyotypic status28. In e, cDNA and genomic DNA from lymphoblastoid cell lines from ten different heterozygous individuals were analyzed: the ten different colors represent the ten individuals; technical replicates from the same cell line are connected by line segments. HeLa, SNU182 and the lymphoblastoid cell lines expressed the C allele of rs10065172 more strongly than the T allele (b–e); HCT116 and primary bronchial cells expressed the T allele much more strongly than the C allele (f,g).
The two IRGM haplotypes showed different patterns of expression across a panel of heterozygous cell lines (Fig. 2). cDNA from HeLa cells, whose genomic DNA was heterozygous for the two IRGM haplotypes, almost exclusively contained the C allele arising from the protective (reference) haplotype; this result was consistent across multiple HeLa isolates (Fig. 2b,c). Similarly, the hepatocellular carcinoma cell line SNU182 expressed the C allele 4–6 times more strongly than the T allele (Fig. 2d), and lymphoblastoid cell lines from ten heterozygous individuals all expressed the C allele more strongly than the T allele (Fig. 2e). In cells derived from some other tissues, however, we observed much stronger expression of IRGM from the deletion haplotype: both the colon carcinoma cell line HCT116 and primary smooth muscle cells from human bronchus expressed the T allele approximately six times more strongly than the C allele (Fig. 2f,g). These results indicate that the CD risk (deletion) and CD protective (reference) haplotypes activate IRGM expression in different cellular contexts.
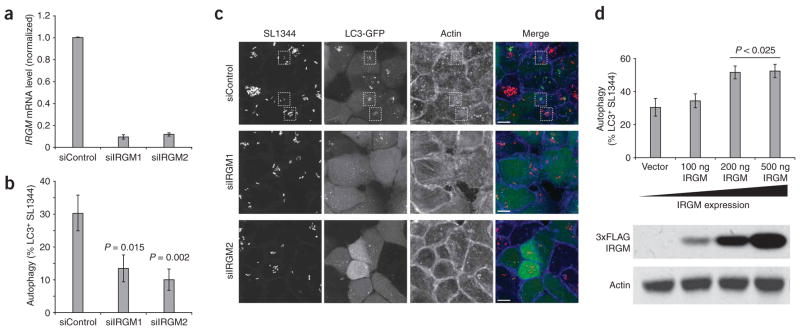
We then sought to assess whether a relationship between IRGM expression and cellular autophagy existed in a manner that could plausibly be linked to CD. To address an emerging connection between CD and autophagic processing of internalized bacteria5, we manipulated IRGM expression in HeLa cells infected with Salmonella typhimurium, and then assayed the ability of the infected cells to form autophagic vesicles around the infecting bacteria. Reductions in IRGM expression, using siRNA constructs that reduced IRGM mRNA expression by six- to eightfold (Fig. 3a), significantly compromised the efficiency of anti-bacterial autophagy (Fig. 3b,c). Together with existing data on cellular control of Mycobacterium tuberculosis3, these data support a role for IRGM in anti-bacterial autophagy. To test a further hypothesis that expression of IRGM, a GTPase with putative signaling function, can actually regulate rates of autophagy, we next overexpressed IRGM in HeLa cells. Modest overexpression of IRGM enhanced autophagy of Salmonella (Fig. 3d), indicating that endogenous cellular levels of IRGM limit autophagic efficiency. These results indicate that the expression level of IRGM can regulate the efficiency of the anti-bacterial autophagic response.
Figure 3.
IRGM expression levels affect the autophagy of Salmonella typhimurium in human epithelial cells. (a) siRNA constructs directed at IRGM reduced endogenous IRGM transcripts by six- to eightfold in HeLa cells. Cells were transfected with control (siControl) or IRGM-targeted (siIRGM1, siIRGM2) siRNA duplexes and assayed 48 h later by quantitative RT-PCR. Error bars, s.d. (b) IRGM knockdown reduces the efficiency of anti-bacterial autophagy. HeLa cells stably expressing LC3-GFP (a marker for autophagic vesicles) were transfected with control or IRGM-directed siRNA oligos, then infected with S. typhimurium after 48 h. The percentage of bacteria encapsulated in LC3-positive vesicles was determined by microscopy. Reductions in IRGM expression resulted in a significant loss of anti-bacterial autophagy compared to control cells. (c) Microscopic examination reveals a decreased number of autophagically encapsulated bacteria in IRGM knockdown cells. Control siRNA-treated (siControl) and IRGM siRNA-treated (siIRGM1, siIRGM2) cells were infected with S. typhimurium SL1344 and imaged by confocal microscopy. Control cells show numerous bacteria (red in merged image) surrounded by LC3-GFP membranes (green in merged image). Such bacteria-containing autophagosomes (indicated by dashed boxes) were almost completely absent in IRGM-deficient cells. The actin cytoskeleton was visualized using phalloidin (blue in merge). Images are flat projections of confocal z-stacks. Scale bars, 10 μm. (d) HeLa cells stably expressing LC3-GFP were transfected with increasing amounts of an IRGM expression construct and infected 24 h later with S. typhimurium. The percentage of bacteria found within LC3-positive vesicles increased with increasing amounts of exogenous IRGM. Protein expression levels were confirmed by blotting lysates for Flag-IRGM expression, using an antibody to Flag to detect exogenous IRGM and actin as a loading control (lower panel). Error bars in b and d, s.e.m.
Together, these results establish that the risk and protective alleles of IRGM differ strongly in the extent to which they are expressed in different cell types, and that expression levels of IRGM regulate the efficiency of anti-bacterial autophagy; they also identify a large deletion polymorphism upstream of IRGM resulting in population segregation of IRGM with two distinct upstream sequences, which we propose as a candidate explanation for the observed difference in expression patterns and association to CD.
The study of autophagy has to date relied upon knockout or siRNA ablation of gene products, revealing little of how the regulation and signaling involved in initiation of autophagy are affected by expression levels. Although components of the autophagic core apparatus may be required in only catalytic amounts11, it is likely that the signaling molecules that initiate autophagy are required to exceed an initiation threshold before initiation takes place12; in addition, active signaling molecules may be quickly sequestered by the local autophagy machinery. The hypothesis that the degree of expression of such signaling molecules limits rates of autophagy is supported by our data indicating that IRGM overexpression enhances the anti-bacterial autophagic efficiency of HeLa cells (Fig. 3).
The CD risk and protective haplotypes, which carry different genomic sequences upstream of IRGM (Fig. 1), showed different patterns of tissue-specific expression of IRGM (Fig. 2). The extent to which human gene expression variation is tissue specific is not yet known, as large-scale surveys of the genetic basis of human gene expression variation have primarily used a single cell type (lymphoblastoid cell lines). A recent study of allelic imbalance in the liver, spleen and brain of F1 mice suggests that tissue specificity of expression variation is common: one-third (11/33) of genes with allelic imbalance showed differences in allelic imbalance between tissues, and several (3/33) showed strongly opposite allelic effects in different tissues10, analogous to our observations for IRGM. The replacement of the upstream sequence of a gene by genomic polymorphism may also increase the prior likelihood of a complex pattern of expression differences such as that observed at IRGM.
IRGM seems to have arrived at its primate genomic location as a small translocation or retroposition of an ancestral gene that was encoded elsewhere in the genome; the genomic region upstream of IRGM at this new locus has subsequently undergone intense evolutionary change, with heavy modification by retroposons along the primate lineage (Supplementary Note online). Although the reproducible cellular phenotype of multiple IRGM siRNAs3 (Fig. 3b,c) indicates that IRGM is expressed at a level sufficient to be functional, we found no conserved transcription factor binding sequences at IRGM, and its expression in most tested cell lines was low. One or more unknown genomic feature(s) seem to be able to activate the transcription of IRGM at a low but functionally relevant level. The most likely candidates may be among the 33 subfamilies of retroposon sequences that have populated the region upstream of IRGM during primate evolution (Supplementary Note); such sequences are increasingly observed to have tissue-specific enhancer properties, although the association of specific sequences with specific expression patterns is at an early stage13–18.
The extent to which linkage disequilibrium–based approaches will be able to identify associations between genome structural polymorphisms and disease risk is the subject of intense debate6,19–23. Here we have identified such an association by combining SNP association data with linkage disequilibrium analysis of a common structural polymorphism we found in the associated region. Genome-wide maps of human structural polymorphisms and the SNP haplotypes on which they segregate, together with data from genome-wide SNP association studies, could in principle enable large-scale investigation of the relationships between structural polymorphisms and human disease.
METHODS
Genotyping of insertion/deletion polymorphism
Initial genotyping of the insertion/deletion polymorphism in the 270 HapMap DNA samples and in human cell-line and tissue samples was done using a two-color TaqMan assay in which the affected locus and a control, two-copy locus were simultaneously amplified and detected using TaqMan probes (primer and probe sequences are listed in Supplementary Table 1 online). Samples were typed in three replicates; delta-Ct values were summarized by median polish and then clustered (Supplementary Fig. 1a). To address the need for a robust genotyping assay to generate data approaching 100% completeness and accuracy across a range of clinical DNA qualities and experimental conditions, we developed and extensively validated a breakpoint-based genotyping assay (Supplementary Methods and Supplementary Fig. 1b online) for use in clinical cohorts.
Genotyping deletion polymorphism in individuals with inflammatory bowel disease
The study cohort consisted of 688 individuals (344 control individuals and 344 with inflammatory bowel disease, including 172 with CD and 171 with ulcerative colitis). Affected individuals and geographically matched controls were ascertained through the Cedars-Sinai Medical Center, Johns Hopkins University, University of Chicago, University of Montreal, University of Pittsburgh and the University of Toronto Genetics Research Centers. Informed consent was obtained from all participants, and protocols were approved by the local institutional review board in all participating institutions. The NIDDK genome scan (from which genotype data were also used here) has been described earlier5,8; the MACH software (see URLs section below) was used to impute additional SNPs. The association plot (Fig. 1d) was made using an R script developed by P. deBakker.
Extended HapMap samples and SNP 6.0 SNP/CNP genotyping array
We previously developed an array platform and set of algorithms for finding and genotyping CNPs alongside SNPs (S.A.M., F.G. Kuruvilla, J.M. Korn, D.A. and M.J.D., unpublished data). Briefly, to genotype the IRGM deletion polymorphism, we summarized intensity measurements for the six copy-number probes across the deleted region into a single measurement for each sample using median polish; these measurements were then clustered across samples, allowing each individual sample to be assigned to one of three discrete copy-number classes (Supplementary Fig. 1c), corresponding to individuals with 0, 1 or 2 copies of the locus per diploid genome. Concordance of the deletion genotypes from the SNP 6.0 array with the PCR breakpoint assay (described above) was 100% across 270 samples. Array data on the 1,260 ‘extended HapMap’ samples have been deposited to the International HapMap Consortium and will be available on the HapMap web site; these samples represent individuals with ancestry from Europe, East Asia, West Africa, East Africa, India and North America.
Genotyping human cell lines for insertion/deletion polymorphism and SNPs
For genotyping of the insertion/deletion polymorphism and tightly linked SNPs in human cell lines, we used either pure genomic DNA or (where necessary) genomic DNA that was present in initial preparations of extracted RNA. Genotypes for the deletion polymorphism, rs13361189 and rs10065172, were perfectly correlated (r2 = 1.0) in all cell lines tested, defining the haplotypes shown in Figure 2. In addition to the heterozygous cell lines for which data are shown in Figure 2b–g, the following cell lines were genotyped and found to be homozygous for the protective (insertion) haplotype: CaCo2, Sw480, HT29, HEK293, Jurkat, Daudi, U937, MOLT4, MonoMac6, HepG2, A549 and K562. The THP1 cell line was found to be homozygous for the risk (deletion) haplotype.
Cell culture and RNA extraction
Cell lines were maintained under normal conditions (37 °C, 5% CO2) in standard culture media (DMEM containing 10% FCS + Fe2+ and 50 μg/ml gentamicin for adherent cell lines; IMDM containing 10% FCS + Fe2+, 100 μM β-mercaptoethanol and 50 μg/ml gentamicin for suspension cell lines). We extracted RNA from 5 × 106 cells using RNeasy spin columns according to the manufacturer’s instructions (Qiagen).
cDNA synthesis
To remove genomic DNA from RNA samples before cDNA synthesis, we incubated 2 μg RNA with 1U DNase at 37 °C for 40 min; we then added EDTA (to 100 nM) to protect RNA from degradation and further incubated the samples at 75 °C for 10 min to denature the DNase. cDNA was synthesized using the SuperScript III First-Strand Synthesis Kit (Invitrogen). We assessed the presence of contaminating genomic DNA using a TaqMan assay interrogating the presence of a nontranscribed genomic locus.
Measurement of allelic ratios in cDNA and genomic DNA
To assess the relative expression of the two alleles of IRGM, we carried out SNP genotyping analysis using a cDNA template and used the quantitative allele-specific measurements that are generated during SNP genotyping. In order to assess the reproducibility of any findings, we used two different SNP genotyping platforms: a mass-extension platform (Sequenom) and a quantitative PCR platform (TaqMan). The mass-extension platform works by mass spectrometric analysis of a primer (designed to genomic sequence next to the SNP site) that is extended across the SNP site in the presence of a partial mixture of dNTPs; the quantitative PCR (TaqMan) platform uses probes that distinguish between the two SNP alleles. Each experimental plate included cDNA samples, genomic DNA from the same cell lines, and control genomic DNA from 48 samples (HapMap YRI) with different rs10065172 genotypes; the control genomic DNA samples (with known genotypes) allowed all measurements for new samples to be calibrated against the measurements for the pure genotype classes CC, CT and TT. Analysis on the Sequenom platform used a primer/probe set selected to interrogate rs10065172; the primer and probe sequences are listed in Supplementary Table 1. PCR, mass extension and detection were done according to the manufacturer’s standard hME chemistry and protocol; the areas under the respective mass peaks corresponding to the two different alleles were used for analysis. Analysis on the TaqMan platform used a pre-designed SNP assay for rs10065172 (Applied Biosystems), in 5 μl reactions. To normalize for platform-and allele-specific components of the raw intensity measurements (for example, on Sequenom, the different masses of the two molecules used to detect the two alleles), we carried out multiplicative normalization using the measurements from control HapMap heterozygous DNAs by multiplying all intensity measurements by a batch-specific, allele-specific constant derived from the requirement that the median measurement for these control DNA samples be 100 for each allele. The two platforms yielded equivalent results for the same cell lines (Fig. 2c,f,g and Supplementary Fig. 2 online); Figure 2 includes data for both the mass-extension assay (b–d,f,g) and the quantitative PCR platform (e). On both platforms, we found that the precision of allelic-ratio measurements was improved by the use of larger quantities of cDNA as input to the reaction; 500 ng cDNA was used in the reactions for which results are shown in Figure 2.
Alternative methods of surveying IRGM expression
We considered alternative approaches (beyond allelic imbalance) for characterizing IRGM expression in cell lines, but found that the low abundance of the IRGM transcript made such results problematic to interpret. For example, analysis of published gene-expression microarray data for the HapMap lymphoblastoid cell lines (60 YRI and 60 CEU parents24) indicated a modest correlation (P < 0.05) between IRGM deletion genotype and IRGM expression measurements in the direction indicated by the allelic-imbalance data (Fig. 2e); however, the microarray measurements were in a low quantitative range for such measurements to be considered meaningful, and we considered the analysis inconclusive. Measurements of IRGM allelic imbalance (Fig. 2), which are highly internally controlled, were robust across all biological replicates tested (for both HeLa strains, HCT116 and the ten lymphoblastoid cell lines).
Cell lines and bacterial strain
HEK293T and HeLa (CCL2, from ATCC) cells were grown in DMEM (Gibco) with 10% iron-supplemented calf serum (CSFe) (Hyclone) at 37 °C and 5% CO2. The HeLa cell line stably expressing LC3-GFP (HeLa LC3-GFP) was generated by lentiviral transduction as previously described25; the LC3-GFP lentiviral vector was a gift from C. Münz (The Rockefeller University). The S. typhimurium strain SL1344 bearing a DsRed2 expression plasmid has been previously described5,26.
Plasmids and transfection
Complementary DNA encoding human IRGM isoform (a)7 was obtained from Open Biosystems and subcloned into pCMV-3xFlag vector using polymerase chain reaction (pCMV-3xFlag vector was generated by modifying ClonTech pCMV-HA vector with the appropriate epitope and MCS region modifications) (Supplementary Fig. 3 online). We confirmed the sequence of the entire cDNA (IRGM(a) with a C-terminal 3xFlag epitope tag) by DNA sequencing. Protein blotting was done using antibodies to Flag M2 and actin (Sigma), with appropriate HRP-conjugated secondary antibody (Covance).
IRGM-directed siRNA and validation
siRNA duplexes directed against IRGM were purchased from Invitrogen (Stealth siRNA, Invitrogen); sequences are listed in Supplementary Table 1. Control duplexes were purchased from the same supplier and were GC-matched, nontargeting sequences. We carried out siRNA validation against endogenous transcript by quantitative RT-PCR using IRGM-specific primers, normalized to GAPDH control reactions, 48 h after transfection of duplexes into HeLa cells. Duplexes si1 and si2 were able to effect robust knockdown of overexpressed IRGM when HEK293 cells were co-transfected with a 3xFlag-tagged IRGM expression construct; these two duplexes were used in all further experiments.
Infection and autophagy assays
HeLa cells (either parental or stably transduced to express LC3-GFP) were plated in 12-well plates containing 18 mm glass coverslips at a density of 1 × 106 cells per well. After 24 h, 20 pmol of modified RNA oligo duplexes (Stealth RNAi, Invitrogen) were transfected into each well using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. We carried out co-transfections using a similar protocol, with the addition of 500 ng of plasmid DNA to the RNA duplexes before transfection. IRGM titrations were done in the absence of siRNA duplexes, with 0, 100, 200 or 500 ng of 3xFlag-IRGM and an appropriate quantity of empty vector to equalize DNA amounts at 500 ng. After 24–48 h, cells were infected with SL1344 DsRed2 at a multiplicity of infection (m.o.i.) of 100 as previously described5,27. After one hour of infection, cells were washed in PBS and fixed with 4% formalin. Following permeablization with 0.1% Triton X-100 in PBS, actin was stained with Alexa6333-conjugated phalloidin (Invitrogen) and coverslips were mounted in aqueous mountant (Polymount, PolySciences). Counting was done at ×100 magnification on a wide-field fluorescence microscope (Zeiss Axioplan, Carl Zeiss MicroImaging). We counted the number of bacteria per cell, along with the number of bacteria enclosed within LC3-GFP membranes. At least 50 cells were counted for each experimental condition. The numbers of LC3-GFP positive bacteria were then calculated as a percentage of total bacteria. We assessed significance using the two-tailed, unequal variance Student’s t-test. Images were obtained using laser scanning confocal microscopy (BioRad Radiance 2000) as high-resolution z-stacks, which were subsequently projected onto single images using LSM Image software (Carl Zeiss MicroImaging).
Supplementary Material
Acknowledgments
The current work was funded by a US National Institute of Allergy and Infection Diseases grant, AI062773, HL088297, DK43351, and CCIB developmental funds to R.J.X. and by a Lilly Life Sciences Research Fellowship to S.A.M. The National Institute of Diabetes and Digestive and Kidney Disease (NIDDK) IBD Genetics Consortium is funded by the following grants: DK62431 (S.R.B.), DK62422 (J.H.C.), DK62420 (R.H.D.), DK62432 and DK064869 (J.D.R.), DK62423 (M.S.S.), DK62413 (K.D.T.), and DK62429 (J.H.C.). Additional support was provided by the Burroughs Wellcome Foundation (J.H.C.), the Crohn’s and Colitis Foundation of America (S.R.B., J.H.C., J.D.R.), and the NIDDK, DK064869 (J.D.R.). M. Garber and C. Bekpen provided helpful discussion; C. Patil and J. Korn provided thoughtful comments on the manuscript. LC3-GFP lentiviral vector was a gift from C. Münz (The Rockefeller University).
Footnotes
URLs. MACH software, http://www.sph.umich.edu/csg/abecasis/MaCH
Accession codes. GenBank: human IRGM, A1A4Y4.
Note: Supplementary information is available on the Nature Genetics website.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkes M, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 4.Hampe J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 5.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarroll SA, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 7.Bekpen C, et al. The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol. 2005;6:R92. doi: 10.1186/gb-2005-6-11-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duerr RH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowles CR, Hirschhorn JN, Altshuler D, Lander ES. Detection of regulatory variation in mouse genes. Nat Genet. 2002;32:432–437. doi: 10.1038/ng992. [DOI] [PubMed] [Google Scholar]
- 10.Campbell CD, Kirby A, Nemesh J, Daly MJ, Hirschhorn JN. A survey of allelic imbalance in F1 mice. Genome Res. 2008;18:555–563. doi: 10.1101/gr.068692.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosokawa N, Hara Y, Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2007;581:2623–2629. doi: 10.1016/j.febslet.2007.05.061. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bejerano G, et al. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 14.Kamal M, Xie X, Lander ES. A large family of ancient repeat elements in the human genome is under strong selection. Proc Natl Acad Sci USA. 2006;103:2740–2745. doi: 10.1073/pnas.0511238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishihara H, Smit AF, Okada N. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 2006;16:864–874. doi: 10.1101/gr.5255506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowe CB, Bejerano G, Haussler D. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc Natl Acad Sci USA. 2007;104:8005–8010. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santangelo AM, et al. Ancient exaptation of a CORE-SINE retroposon into a highly conserved mammalian neuronal enhancer of the proopiomelanocortin gene. PLoS Genet. 2007;3:1813–1826. doi: 10.1371/journal.pgen.0030166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruda VM, et al. Tissue specificity of enhancer and promoter activities of a HERV-K(HML-2) LTR. Virus Res. 2004;104:11–16. doi: 10.1016/j.virusres.2004.02.036. [DOI] [PubMed] [Google Scholar]
- 19.Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat Genet. 2006;38:82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- 20.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 21.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conrad DF, Hurles ME. The population genetics of structural variation. Nat Genet. 2007;39:S30–S36. doi: 10.1038/ng2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet. 2007;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 24.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmid D, Pypaert M, Münz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from auto-phagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niess JH, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 27.Beuzón CR, et al. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 2000;19:3235–3249. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macville M, et al. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res. 1999;59:141–150. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.