Abstract
A major goal of molecular medicine is to target imaging agents or therapeutic compounds to a single organ. Targeting imaging agents to a single organ could facilitate the high-resolution, in vivo imaging of molecular events. In addition, genetic and acquired diseases primary to a single organ, such as cystic fibrosis, tuberculosis, lung cancer, pulmonary fibrosis, pulmonary hypertension, and acute respiratory distress syndrome, could be specifically targeted in the lung. By targeting and concentrating imaging agents or therapeutics to the lungs, deleterious side effects can be avoided with greater efficacy at much lower dosages. Pathologic changes can be identified earlier and followed over time. In addition, therapeutics that have been abandoned due to toxicities may find renewed utility when coupled with specific targeting agents such as antibodies. To achieve these goals, distinct molecular signatures must be found for each organ or disease-state.
Keywords: vascular endothelial cells, whole-body imaging, proteomics, intravital microscopy, transcytosis
MOLECULAR TARGETING
Most current targeted drugs are directed against specific proteins expressed by tissue parenchymal cells located deep within tissue—for example, function-altering antibodies or targeting antibodies linked to toxic agents. These treatments are very effective in vitro, where individual tissue cells are directly in contact with the media, and drugs, including antibodies, have the opportunity to bind their targets. However, the vast majority of these treatments have failed in vivo, where drug targeting is much more complex. Movement from the blood into tissue and even solid tumors is significantly limited in vivo by the vascular endothelium, a monolayer of attenuate endothelial cells that line all blood vessels (1). Poor delivery can prevent many carrier systems, including antibodies, nanoparticles, and gene vectors, from reaching their target tissue where they can be effective. The larger carriers are often rapidly scavenged from the circulating blood by the reticulo-endothelial system (i.e., liver and spleen), further preventing efficient binding and access in vivo.
Small drugs such as chemotherapeutics present an alternative to relatively large antibodies and nanoparticles because they can much more readily enter nearly all tissues. As a result, these agents are rapidly diluted, cleared from the blood, and excreted. Many of these drugs also lack specificity for any single organ or tissue. Their pervasive access can lead to high backgrounds and poor signal to noise during imaging and extensive side effects after systemic treatment. Chemotherapeutics, for example, act by killing cells that rapidly divide. They are effective against cancer cells, as well as other healthy cells that divide rapidly, such as those in bone marrow, the digestive tract, and hair follicles, which leads to many of the well-known side effects of these drugs.
For both large and small molecules, it appears that only a small proportion of the injected dose actually reaches the inside of diseased tissue and even most tumors where they can be effective. Thus, higher doses must be administered to reach effective levels within the diseased tissue, often leading to severe systemic side effects (2). Diseases of the lung could benefit greatly from improved specific delivery and penetration into the diseased tissue. For example, many patients with lung cancer or pneumonia suffer greatly because little chemotherapy gets into the diseased tissue. To bypass these problems, accessible new targets and ways to penetrate into a specific tissue such as a solid tumor are needed.
VASCULAR ENDOTHELIUM
The inner surface of blood vessels are lined by the endothelium, a thin layer of endothelial cells that help to control the permeability of blood vessels and also plays an important role in vasoregulation, coagulation, inflammation, and tissue metabolism, growth, survival, repair, and overall organ homeostasis and function. Disruption of the vascular endothelium and its normal barrier function can rapidly lead to tissue edema, hypoxia, pathology, and even organ death (3, 4).
The vascular endothelium offers a promising alternative target to tissue cells themselves. The vascular endothelium is in direct contact with the blood and is, therefore, inherently accessibly to probes or antibodies that are injected intravenously. In addition, endothelial cells are highly adapted to meet the needs of the underlying tissue and the microenvironment of the tissue surrounding the blood vessels significantly influences the phenotype of the endothelial cells (5–11). Defining the vascular proteome may uncover tissue- or disease-specific molecules that may be useful as targets for site-directed delivery of drugs, genes, or imaging agents.
PATHWAYS ACROSS THE ENDOTHELIUM
Sinusoidal endothelium (liver, spleen, and bone marrow) and fenestrated endothelium (kidneys, endocrine glands, and intestines) are relatively permissive barriers with large intracellular gaps or transcellular openings that allow the relatively rapid exchange of molecules from the blood into and out of tissue. Most tissues, including the lungs, have continuous endothelium. This is the most restrictive barrier with endothelial cells linked by intercellular junctions with various degrees of tightness (12).
Transport across the endothelial cell barrier is essential to support the underlying tissue and may provide pathways that can be exploited to target blood-borne molecules to tissue. Small molecules, water, and solutes can often be passively transported across the endothelial barrier by diffusion and convection through intracellular junctions, fenestrae, or transendothelial channels (13). Larger molecules may require active transport to move into or across endothelial cells.
Many endothelial cells exhibit two distinct forms of active transport: clathrin-coated vesicles and caveolae. Caveolae are 60-nm flask-shaped invaginations found at the plasma membrane in most continuous endothelia that may be involved in endocytosis and transcytosis (14–20). Caveolae are abundant; in vivo, they can occupy up to 50 to 70% of the endothelial cell plasma membrane (21–23). Caveolins are structural coat proteins that oligomerize around the bulb of caveolae and appear necessary for caveolae formation in vitro and in vivo (24–30).
CAVEOLAE AS TRANSPORT VESICLES
Caveolae were first identified in 1953 (20). Since then, researchers have debated over whether these membrane invaginations might play a role in transport (31). Evidence collected in the last two decades overwhelmingly favors a role as dynamic vesicular carriers. Caveolae contain proteins classically associated with vesicular transport, including vesicular SNAP receptors (v-SNAREs), N-ethylmaleimide–sensitive factor (NSF), soluble NSF attachment protein (SNAP), and GTPases. Electron microscopy of ultrathin section of rat lung confirmed that vesicle-associated membrane protein (VAMP) was localized within caveolae (32). These proteins are associated with caveolae under basal conditions; no further recruitment is needed, suggesting that rapid transport might be possible (33). Much like other vesicular pathways, caveolae-mediated endocytosis is sensitive to N-ethylmaleimide (NEM), a thioalkylating agent that inhibits the fusion of vesicles to target membranes (34, 35). Caveolae have also been functionally implicated in active transport of select ligands and even virus to specific locations within the cell (36–43).
More recently, isolated endothelial cell plasma membranes have been used to show that caveolae can bud and form free vesicles in the presence of GTP and ATP (44). GTP hydrolysis was needed for this budding. GTPγS, a nonhydrolyzable form of GTP, cannot induce budding and actually inhibits GTP-induced caveolar budding (45). This was confirmed on cultured cells, where electron microscopy showed that stimulation with GTP led to the loss of 78% of surface caveolae (45). GTP likely acts by stimulating the GTPase dynamin. Dynamin is necessary for caveolar fission; mutant forms of dynamin prevented caveolar fission (44, 46). Western analysis showed that dynamin is highly enriched in caveolae, and electron microscopy localized dynamin to the neck of caveolae, where it may act as a “pinchase” to mediate the final steps of vesicle budding (44, 46). Thus, key molecular machinery for budding, docking, and fusion are clearly present in caveolae. This caveolae-mediated internalization has been confirmed in hepatocytes (47), which have readily apparent caveolae in cell culture but few to no caveolae in vivo.
METHODS OF IDENTIFYING TISSUE-SPECIFIC MARKERS
To be effective, molecular targeting depends on the specific and unique expression of target proteins in different tissues and disease states. Multiple methods have been developed over the years to identify differential gene and protein expression, as summarized in Table 1. Monoclonal antibodies are one early method to investigate tissue-specific protein expression. Antibodies against unknown proteins can be generated by directly injecting target tissue, cells, or even tissue subfractions as an immunogen (48–52). Monoclonal antibodies can then be used to screen expression in cells and tissue as well as to immunoprecipitate the protein of interest for sequencing and identification. This is can be a slow and laborious process, but produces robust antibodies.
TABLE 1.
METHODS OF IDENTIFYING PROTEINS AT THE SURFACE OF ENDOTHELIAL CELLS
| Method | Advantages | Disadvantages |
|---|---|---|
| Methods of identifying proteins | ||
| Antibody-based | Creates topological map of protein expression | Slow and laborious, depends on quality antibodies, can be difficult to identify unknown antigens |
| Phage display | Unbiased, unknown targets can be identified, applicable in vivo and in vitro, large libraries can be screened | Phage are rapidly scavenged by the liver in vivo, short peptides lack specificity, not high-throughput |
| Genomics | Rapid, high throughput, nonbiased, many genomes are nearly complete for straightforward identification of genes | Large changes needed, protein expression does not always correlate with protein expression, large number of “hits” require laborious validation |
| Two-dimensional gels | Simple, rapid, separates complex samples. Can be directly used to show differences between tissues. | Large amounts of material are needed, some proteins do not migrate into gel |
| Mass spectrometry | Rapid, high throughput, nonbiased, requires small amounts of sample | Limited by complexity of sample, low abundance proteins can be lost, large number of hits can require extensive validation |
| Protein arrays | Rapid, sensitive, large-scale screen | Biased, requires specific probes for each protein |
| Methods for reducing complexity | ||
| Cultured cells | Large amounts of material, readily accessible | Cultured cells rapidly lose in vivo phenotype |
| Lectin analysis | Reveals differences in glycoproteins, can be applied in vitro or in vivo | Only applicable for specific glycoproteins |
| Radiolabeling | Verifies proteins at the surface of endothelial cells | Requires large amounts of radionuclides, no way to purify proteins, small radionuclides readily permeate into tissue |
| Biotinylation | Labels proteins at the cell surface, strong interaction between avidin and biotin can be used to purify proteins for identification | Difficult to control the degree of biotinylation, small molecules can permeate into endothelial cells and tissue |
| Silica nanoparticles | Selectively label surface proteins, do not enter into tissue, entire luminal surface and its caveolae can be isolated | Technically complicated, small amounts of tissue are obtained |
Phage display libraries are another way to discover novel targets. George Smith first suggested that bacteriophages could be used to display proteins or antibody fragments in 1985 (53). Because the approach can use a large, random library, it is unbiased, and unknown targets can be identified. In a process called panning, phage that bind to a target (antibody, protein, or even tissue) are repeatedly isolated and amplified. Phage can be purified and used as probes themselves to isolate the binding partner for identification. Phage can also be injected intravenously, to circulate and presumably to bind a single protein at the endothelial surface (54, 55). They can then be isolated from each organ or tissue of interest. However, the liver and spleen rapidly scavenge phage from the blood, mostly before they have a chance to circulate through each organ and bind sufficiently (56). In addition, short peptides can lack specificity and may bind a large range of proteins in a multitude of organs, requiring additional ex vivo validation. These problems can be partially overcome by avoiding direct in vivo panning but rather screening antibody phage libraries on key membranes and then creating antibody-like fusion proteins, which unlike the phage, are not rapidly removed from the circulating blood and can indeed immunotarget successfully in vivo (57). Phage display libraries have revealed some promising targets, but currently this method may not yet be optimally suited for the high-throughput needed to comprehensively map protein expression and identify tissue-specific proteins.
Genomics approaches can be used to comprehensively define expression patterns and identify differences among samples in a relatively rapid manner. These approaches have identified unique gene expression on vascular endothelial cells derived from different tissues, from angiogenic tissue, and from tumor tissue (58, 59). Large changes are needed to detect differences between tissues. Unfortunately, changes in gene expression do not always correlate with changes in protein expression, nor does genomic data offer information about the location of a protein. Obviously, the location of a protein affects its accessibility and can also affect its function. Post-translational modifications make distinct proteins that cannot be assessed through genomic analysis and can also alter protein location and function. To truly identify accessible, tissue-specific targets, protein expression itself must be characterized at a very large-scale level that comprehensively identifies tens of thousands of proteins.
Two-dimensional (2D) gels are one simple and rapid way to visualize differences between tissues. In these gels, mixtures of proteins are separated by two distinct properties, providing better separation between proteins (60). Spots that are unique to one tissue can be excised and identified by mass spectrometry (61, 62). For these approaches, successful identification of proteins requires that the proteins migrate onto the gel. Many proteins simply do not separate well on such gels and can be underrepresented or lost altogether. Mass spectrometry–based techniques can rapidly identify large numbers of proteins based on the presence of digested peptides. Highly complex samples are difficult to separate, and low abundance proteins can be lost (63). Protein arrays offer a third alternative to identify differences between tissues. These use antibodies or peptides to identify the proteins present in the sample but are limited by affinity of the probes and the complexity of the sample (64).
REDUCING COMPLEXITY BY FOCUSING ON ENDOTHELIAL CELLS
Each of the above methods is limited by the complexity of the starting sample. Though it is clearly important to define the proteins at the endothelial cell surface, these cells are only a tiny part of any whole tissue homogenate. Even highly enriched endothelial cell proteins can be missed when the total organ homogenate is analyzed (65). Studying endothelial cells requires a reliable method to isolate these cells from the entire tissue. Several methods are described in Table 1.
Endothelial cells were first isolated and successfully cultured in the 1970s (66, 67). This allowed tremendous insights into the molecular components and functions of these cells and showed that they were rapidly responsive to vasoactive cytokines, shear stress, and inflammatory compounds (4, 68–70).
Numerous chemical techniques were developed in the late 1980s and early 1990s to label surface proteins of endothelial cells. Because proteins at the surface of endothelial cells in vitro and in vivo are directly exposed to the media or to blood, they also can be labeled by reagents added to the media, perfused through the vasculature, or injected intravenously (71–74). Different lectins can bind to different sets of surface glycoproteins, which has been used to compare different segments of vasculature (75), different organs (72), and in vitro versus in situ protein expression (74). Radio-iodination of surface compounds in vitro identified albumin-binding proteins (37, 76). In situ radiolabeling is inefficient and requires high amounts of Iodine-125, often exceeding 10 mCi, making this process difficult (74). Moreover, the small radionuclides can readily cross the vascular wall to enter the tissue and radiolabel other nonendothelial cells. Radiolabeling is most often used to verify the presence of known proteins, as there is no simple way to separate and identify radiolabeled proteins, significantly limiting the utility of this approach.
Biotinylation can chemically label proteins for rapid purification. It can label proteins at the surface of vascular endothelial cells, especially cells grown in cultures in which conditions can be well controlled (71). The strong interaction between biotin and avidin can be used to purify the biotinylated proteins, which can then be identified with mass spectrometry (77, 78) or antibodies (79–83). This method has been used to identify proteins at the endothelial cell surface (77, 84) and cell junctions (79), to determine differences between luminal and abluminal surfaces of cells (82, 85), to identify signaling components present at the cell surface (83), and to determine the location of proteins at different membrane subdomains (80). In vivo biotinylation identifies a unique pattern of proteins from the whole tissue homogenate, showing that this method isolates a subset of proteins (77, 78, 84, 86). These studies provide a significant advancement toward vascular mapping in vivo; however, it is difficult to control the degree and the location of biotinylation, especially in vivo. Biotin may not have equal access to all parts of the cell surface. All proteins may not be biotinylated similarly. Biotinylation reagents are quite small. They can readily enter almost all tissues and even cross lipid membranes to get inside cells. They can readily permeate throughout a tissue to label and ultimately help identify perivascular proteins inside the surrounding tissue and even possibly within endothelial cells (77). Using polar or charged biotinylation reduces entry inside cells, but access into tissue beyond the endothelial cell surface remains. This somewhat pervasive access and labeling may be appropriate for identifying candidate targets accessible to small targeting agents, but can greatly complicate identification of tissue-specific endothelial cell surface proteins useful for direct targeting of antibodies, proteins, nanoparticles, and gene vectors.
Colloidal silica particles offer a promising alternative to more permeable reagents. When perfused through the vasculature in vivo, these nanoparticles are too large to penetrate into the tissue and thus can quite selectively coat the endothelial cell surface. After cross-linking and tissue homogenization, the dense silica-coated luminal endothelial cell plasma membranes are easy to separate from other components of the tissue, and even other components of the endothelial cells (87). The endothelial plasma membrane can be further subfractionated to study functional microdomains such as caveolae (87). The silica nanoparticles are too large to readily enter into the caveolae. Caveolae can be separated mechanically from the luminal membrane by shear stress or the addition of GTP and then isolated by buoyant density centrifugation (19, 44, 87). Electron microscopy of the isolated caveolae shows a homogenous population of appropriately sized, 60- to 80-nm vesicles. Western analysis shows that this isolate is enriched for caveolae markers such as caveolin but depleted of markers for other subcellular organelles. In addition, magnetic immunobeads labeled with caveolin antibodies showed that the population was highly pure (> 95%) (88).
Studying endothelial cells in vivo is essential to define relevant molecular targets. Though distinct in vivo, once in culture, endothelial cells isolated from different organs de-differentiate into a more common phenotype. They lose expression of tissue-specific proteins and the number of surface caveolae decreases 30- to 100-fold (5, 89–91). Very recent mass spectrometry analysis shows profound differences in proteins expression in vitro versus in vivo. Approximately 40% of the proteins expressed in vivo are not found in vitro (61). In light of this, tissue-specific vascular targets must, at the very least, be validated in vivo.
MAPPING THE VASCULAR ENDOTHELIAL CELL PROTEOME
Early attempts in the 1980s and 90s to define the endothelial cell proteome focused on using antibodies and other specific probes to identify individual proteins (as detailed above). These methods provided valuable insight into the functional components of endothelial cells, but such approaches are difficult and necessarily piecemeal. Mass spectrometry–based approaches can provide a much more comprehensive profile of endothelial cell proteins. Initial studies using mass spectrometry–based approaches were done on endothelial cells in vitro using 2D gel electrophoresis or protein arrays followed by mass spectrometry analysis to identify proteins (64, 92). Though most cell types, including endothelial cells, are predicted to contain very large numbers of distinct proteins, these studies only identified a handful of proteins (< 60). Clearly, more comprehensive approaches are needed to fully map the vascular endothelial cells, and especially the proteins present at the plasma membrane.
As described above, the luminal surface of endothelial cells can be isolated in vivo using a silica-coating technique. Though far simpler than total tissue homogenate, the membrane isolate is still a highly complex mixture of proteins. Recent advances in proteomics techniques have made it possible to analyze complex samples, in part by pairing one- or two-dimensional liquid chromatography (1D- or 2D-LC) with mass spectrometry. 2D-LC separation can greatly improve sensitivity by diminishing peptide co-elution and thus enhancing identification of less abundant peptides not previously detected. Even so, each mass spectrometry measurement only identifies a portion of the proteins present in a sample. Therefore, replicate measurements are absolutely necessary to maximize the ability of a single method to identify proteins. When a sample of endothelial cell plasma membrane was repeatedly analyzed, 7 to 10 replicates were needed to reach 95% analytical completeness (61). After this point, additional replicates failed to identify significant amounts of new proteins. Comprehensive measurement is clearly necessary to define the endothelial cell proteins within a given tissue and to identify differences between tissues.
Using a statistically defined endpoint not only helps eliminate the inherent variance between single mass spectrometry measurements, but also increases the replicability of findings between different laboratories, as well as improves the utility of future mass spectrometry–based analyses (93). This requires heavy instrument time, but more clearly identifies proteins in a sample. The extra mass spectrometry measurement days may well be time well spent to avoid a long list of candidates that may be temporarily satisfying, yet ultimately overwhelming with few actual true positives.
LUNG VASCULAR ENDOTHELIUM
Our initial large-scale attempt to map the luminal surface of lung vascular endothelial cells using shotgun proteomics with multiple replicate measurements identified 450 distinct proteins (61). As expected, many of these proteins were associated with the plasma membrane. Proteins that peripherally associate with the inner leaflet of the plasma membrane made up 35% of the proteins, while 31% were integral membrane proteins or proteins with lipid anchors, 25% were cytoskeletal or junctional proteins, and 8% were externally bound. Structural and signaling proteins made up more than half of the proteins found associated with the luminal plasma membrane. Trafficking and adhesion proteins, extracellular enzymes, and transporter enzymes were all found in abundance as well (61). It is unlikely that this is the full complement of membrane proteins. This study failed to identify several known endothelial cell markers, including specific enzymes, adhesion molecules, and growth factor receptors, many of which are integral membrane proteins. Indeed, only 15% of the proteins we identified were integral membrane proteins. Many more are expected.
Integral membrane proteins are especially challenging to identify with current large-scale shotgun proteomic methods. They are intrinsically hydrophobic, resistant to extraction and solubilization, and different proteins occur across a wide concentration range. In the most comprehensive profiling of the luminal plasma membrane of lung vascular endothelial cells to date, we have combined multiple mass spectrometry methods, each with multiple replicates to identify 1,833 proteins, including more than 500 lipid-embedded proteins (63). To help preserve lipid-embedded proteins, we used harsh detergents to solubilize proteins and separated these proteins on SDS-PAGE gels. After gel prefractionation, proteins were digested in gel and then analyzed via 1D-LC or 2D-LC paired with mass spectrometry. All methods were compared with the more commonly used shotgun approach, namely gel-free 2D-LC paired with mass spectrometry, which we had used previously. Several benefits of gel prefractionation were seen. First, less sample was needed. For traditional gel-free approaches, 150 μg was used. For each of the gel-based approaches, 40 μg of sample was sufficient. Larger sample volumes did not lead to the identification of greater numbers of proteins. Second, fewer replicates were needed to reach 95% analytical completeness. For gel-free approaches, 7 to 10 replicate measurements were needed, while the gel-based approaches only required 4 to 5 replicates (63).
Most importantly, gel-based approaches provided superior protein identification by identifying greater numbers of proteins, providing better protein coverage, and identifying proteins across a broad dynamic range. Together, the three gel-based methods more than quintupled the protein identifications relative to the gel-free method, accounting for over 98% of the total protein identifications. Gel-based methods identified more peptides per protein, leading to greater protein coverage. The lower limit of meaningful protein identification for gel-free methods was estimated at greater than 2 ng, but each gel-based method has much better sensitivity, down to 0.1 ng. The three gel-based methods could detect proteins over about five orders of magnitude. Though the gel-based methods identified more proteins than the gel-free method, each method identified unique proteins (63).
Integral membrane proteins are thought to comprise at least 20% of all open reading frames in the genome (94). Protein prefractionation by SDS-PAGE greatly enhanced detection by mass spectrometry analysis. Each gel-based method detected two- to sixfold more integral membrane proteins than the gel-free approach. Even when normalized to the number of soluble proteins identified, gel-based methods identified a greater amount of integral membrane proteins (63). This may be predictable because soluble proteins are far more amenable to in-solution digestion than integral membrane proteins and therefore ideal for analysis with gel-free approaches.
Gel-free mass spectrometry methods usually require that samples be precipitated and resolubilized, often in urea. Many proteins may be lost during this process. To determine where proteins were lost, some samples were simply solubilized in cell lysis buffer. Others were processed further and precipitated, then resolubilized in cell lysis buffer or in urea to determine the relative effects of precipitation and resolubilization. Protein abundance was compared by Western analysis of 64 known proteins. Most soluble proteins were equivalently present, regardless of preparation. Integral membrane proteins and lipid-anchored proteins were often lost during either the precipitation step or the resolubilization step. When samples were prepared as they are for gel-free mass spectrometry, some lipid-embedded proteins decreased by as much as 90%, which can prevent downstream identification by mass spectrometry (63). Thus, increased protein identification by gel-based methods arises in part from the overall reduced sample complexity for each mass spectrometry measurement as afforded by the gel prefractionation and in part from the improved retention and separation of integral membrane and lipid-embedded proteins in the gels for digestion.
QUANTIFICATION AND NORMALIZATION OF MASS SPECTROMETRY DATA
Multiple replicate measurements and even multiple mass spectrometry–based approaches are likely necessary to comprehensively define the proteins present in a sample. Mass spectrometry measurements contain inherent biases and variations. Replicate samples, regardless of the abundance feature used, will usually show variation in protein abundance, which is likely not a reflection of biological change. Proper quantification and normalization is needed to minimize inherent experimental bias and variability so that real changes between distinct samples can be reliably detected.
Currently used quantitative mass spectrometry experiments often rely on chemically (95) or biosynthetically (96) tagged systems. Relative differences in protein expression can then be quantified (97), but comparisons between multiple datasets and with previously existing data are impossible. Newer label-free methods are being developed that eliminate the need for expensive labeling reagents and extra analytical capabilities, and permit comparison between datasets. These rely on mass spectrometry output future of abundance, such as spectral or peptide counts (98–101) or chromatographic peak area or height values (102–105) to determine relative protein abundance. These methods correlate with protein abundance (101–103, 106), but detailed comparative analysis and extensive validation is needed to determine which is more accurate.
We tested various methods to quantify, normalize, and compare complex label-free proteomic data using features intrinsic to typical mass spectrometry measurements such as spectral and peptides counts, fragment ion (ms/ms) intensities (unpublished data). These features are much easier to extract from the mass spectrometry data than peak area measurements and are much more suitable for quantification when using a low-resolution mass spectrometer. Alone, no single feature successfully normalized replicate mass spectrometry measurements. Replicates of a single liver sample were significantly different. Two published normalization methods that use spectral counts, NSAF (107) and Rsc (108), were also unable to sufficiently normalize the liver data. We incorporated these features and protein length into a single index called SIN. SIN was able to successfully normalize mass spectrometry measurements from replicate measurements, from different sample loads, and from different instruments.
The novel SIN method was validated in several ways. SIN does not over-normalize the mass spectrometry data, because real differences were still readily detectable. Significant differences could be detected between heart and kidney samples. In addition, densitometry of Coomassie blue–stained gels correlated with mass spectrometry estimates of protein abundance. When the relative enrichment of a given protein found at the lung plasma membrane was compared with the protein abundance in the general tissue homogenate, both densitometry of Western blots and SIN values from mass spectrometry analysis showed excellent positive correlation. SIN clearly compensated for experimental and random bias and noise across a wide array of techniques and samples (unpublished data). The ability to use SIN to accurately determine the relative abundance of thousands of proteins in complex samples is impressive and will hopefully facilitate comparison of data between labs. In addition, by providing a simple and amenable method to quantify and normalize mass spectrometry data, we hope that replicate mass spectrometry measurements and the more comprehensive data they generate will become the norm.
NOVEL CAVEOLAR TARGETS IN LUNG TISSUE
Our lab has used several approaches to map the vascular proteome of the lung and other organs to identify tissue-specific proteins. Initial monoclonal antibodies and phage display antibody libraries were constructed using the isolated luminal surface of endothelial cells as an immunogen (56). These were then screened against Western blots or directly on the isolated endothelial cell membranes. Specific probes were used to identify proteins by immunoprecipitation and mass spectrometry. Endothelia cell membranes isolated from major organs were analyzed on 2D gels to produce high-resolution “maps” of the proteins present in the sample (61, 62). In many cases, antigens that were difficult to detect in the whole tissue homogenate were readily apparent in the isolated plasma membranes, showing that silica-based tissue fractionation revealed an enriched population of proteins. Furthermore, each organ showed a specific and characteristic array of proteins, strongly suggesting that tissue-specific markers could be identified. We used a systems biology approach to identify lung-specific endothelial cell surface proteins. Cells rapidly de-differentiate when removed from their natural environment and cultured. Therefore, we hypothesized that proteins found in cultured endothelial cells might be more universally expressed, while those expressed only in vivo might rely on the specific tissue microenvironment for expression and may represent possible tissue-specific targets. We isolated the luminal plasma membrane from lung vascular endothelial cells in vivo or in culture and analyzed each sample using replicate mass spectrometry measurements. Of the 450 proteins identified at the luminal surface of lung vascular endothelial cells in vivo, only 263 were found at the surface of cultured lung endothelial cells. The other 187 were found only in vivo. To focus on proteins that were only expressed in vivo, we applied strict identification criteria to rat lung endothelial plasma membranes isolated in vivo, while low-stringency criteria were applied to membranes from cultured endothelial cells. Only endothelial cell surface proteins that extend into the blood are readily accessible to circulating antibodies. By analyzing the sequences of identified proteins, we identified 11 differentially expressed proteins likely to have domains outside the cell. To confirm organ specificity, we used specific antibodies to determine the presence of proteins in various organs and in culture. Consistent with the mass spectrometry findings, all 11 proteins were enriched in lung endothelial plasma membranes but were not found at the surface of cultured lung endothelial cells. In vivo, many of these proteins were expressed in other organs, including heart, kidney, and liver. Two proteins, aminopeptidase P (APP) and OX-45, were detected in endothelial cells only from lung. Unfortunately, OX-45 is also expressed on various leukocytes (109). Thus, radiolabeled antibodies against OX-45 showed little targeting to the lung but instead remained in the blood (61).
MEANS TO VALIDATION
Successful molecular targeting likely depends on several factors. Tissue-specific probes must be specific for a single organ and their target proteins must be accessible. The first step in validation of possible targeting systems is to verify the specificity of the probe. Analyzing Western blots of the endothelial cell surface membranes should show one band of the appropriate weight.
Western analysis can be used initially to test tissue specificity and localization of the target protein. To verify restricted tissue expression, the probe can be tested against proteins isolated from many different organs (62). In addition, the localization of the target protein can be assessed by analyzing whole tissue homogenates as well as different subfractions (110). In this way, the presence of the protein at the endothelial cell surface or within caveolae can be determined. Localization of the probe to a type of cell or even sublocation within the cell can be further verified in tissue slices by using immunohistochemistry, immunofluorescence, and electron microscopy (110).
One of the most vital questions to answer is whether the protein target is accessible to intravenously injected probes. The classical way to determine this is to inject radiolabeled probes. At various times after intravenous injection, major tissue types can be dissected and analyzed for radioactivity. This allows a quantitative assessment of the tissue distribution of the probe and also an approximate time course (111). Immunospecificity and localization of the probe can be compared using different indices (51, 56, 62, 110). This is especially relevant for antibody-based targeting system, because specific antibodies can be compared with control IgG. Comparing antibody accumulation in a target organ for a specific antibody versus a control antibody can give an immunospecificity index. Comparing organ accumulation to antibody remaining in blood can give a localization index. Specific localization can be calculated by comparing localization of the specific antibody to the localization of the control antibody. Although robust and quantitative, such analyses are highly invasive and static, requiring the killing of many animals at each time point.
Noninvasive in vivo imaging may be the new gold standard for in vivo targeting of probes. New methodologies such as γ-scintigraphic (112), positron emission tomography (PET) (113), and luminescence imaging (114) provide visual as well as quantitative analysis of probe targeting. These methods are noninvasive and can be used over time in the same animal. This makes these methods the most objective, dynamic, and comprehensive tools to date. Rapid dynamic imaging can create a true time course with far fewer animals than those needed for standard biodistribution studies. In addition, minor tissues that are difficult to isolate can also be studied with in vivo imaging. The above methods can provide important quantitative data for assessing targeting grossly at the whole body and organ level but not so much at the cell or subcellular level.
Traditionally, the final validation of successful endothelial cell targeting involves intravenous injection of the probe. At set times after injection, tissue is excised and the probe is localized with immunohistochemistry, immunofluorescence, and/or electron microscopy. This can provide valuable confirmation of the ability of antibody to target the endothelial cell surface, to be transcytosed across the endothelial cell barrier, and to penetrate deep into the tissue; however, these are static methods and can only provide “snapshots” over time. Using this approach for initial validation can be problematic. For example, simply showing tissue histology with probe signal in one tissue but not another does not constitute validation of tissue-specific targeting. A proper titration curve determining signal range and sensitivity of the tissue staining assay is mandatory, but not usually performed, to ascertain the dynamic range of the assay and the degree of specificity in targeting. Immunohistochemistry usually does not have a large dynamic range. Thus, it is not surprising that even only two- to threefold higher expression in a tissue can render immunostaining that appears specific but rather is false-negative for many other tissues. To minimize this problem, the signal should be maximally developed (even to saturation) in the highest expressing tissue before stopping the reaction in other tissues. Then, other tissue can be declared negative within the dynamic range of the tissue. It is fairly clear that using more than one method to validate seems most wise.
Continuous, dynamic, noninvasive imaging in live animals is clearly the desired standard. Intravital microscopy is a valuable tool for visualizing, at the level of light microscopy, specific organ endothelial cell targeting and processing of candidate antibody probes (i.e., transendothelial transport by caveolae with interstitial accumulation). Especially when paired with tissue growing in dorsal skinfold chambers, intravital microscopy can present a high-resolution, dynamic view of endothelial cell targeting as well as the movement of probes into tissue, ultimately to better assess tissue/organ targeting, uptake, and accumulation in live animals (110).
LUNG-SPECIFIC TARGETING IN VIVO
Several proteins, including angiotensin-converting enzyme (ACE) (115), platelet/endothelial cell adhesion molecule (PECAM) (116, 117), and intercellular adhesion molecule (ICAM) (117), have been used to specifically target the lung and to deliver therapeutic compounds in vivo. Many of these proteins are not lung specific and instead rely on the extensive lung vasculature and the lung receiving the vast majority of cardiac output for specific targeting. A partial list of pulmonary markers can be found in Table 2. In addition, without a means to bypass the endothelial barrier, many of these antibodies and their cargo will remain bound at the endothelial cell surface and unable to reach cells within the underlying tissue, which is often the therapeutic target. Though these proteins can effectively retarget intravenously injected probes, proteins with restricted lung expression and that provide a portal into underlying tissue might expand the utility of vascular targeting.
TABLE 2.
MARKERS OF NORMAL AND PATHOLOGICAL PULMONARY ENDOTHELIUM
| Markers | Lung | Tumor | Other Tissue |
|---|---|---|---|
| Lung endothelial markers | |||
| ACE | Highly enriched | Present | Heart, kidney, liver |
| APP | Highly enriched | Not expressed | None |
| Aquaporin 1 | Present | Present | Heart, kidney |
| Carbonic anhydrase | Highly enriched | Not expressed | Heart, kidney, liver |
| DDPIV | Enriched | Enriched | Liver |
| ECE | Present | Present | Heart, kidney, liver |
| OX-45 | Highly enriched | Not expressed | None |
| PV-1 | Highly enriched | Highly enriched | Heart, kidney, liver |
| RAGE | Present | Present | Heart, liver |
| STR | Present | Present | Heart, kidney, liver |
| TM | Present | Present | Heart, kidney, liver |
| Tumor endothelial markers | |||
| AnnA1 | None | Highly enriched | None |
| AnnA8 | Low | Highly enriched | None |
| APN | Present | Enriched | Heart |
| C-CAM | Enriched | Present | Liver |
| Endoglin | Present | Present | Heart, liver |
| EphrinA5 | Present | Present | Heart, kidney, liver |
| EphrinA7 | Low | Present | Heark, kidney, liver |
| MPO | Low | Enriched | Heart |
| Neuropilin | Low | Present | Heart, liver |
| nucleolin | Present | Enriched | Heart |
| TfnR | Present | Enriched | Heart, liver |
| Tie2 | Low | Enriched | Kidney, liver |
| VEGFR1 | Low | Highly enriched | Kidney |
| VEGFR2 | Low | Present | Heart, kidney, liver |
| VitDBP | None | Enriched | Liver |
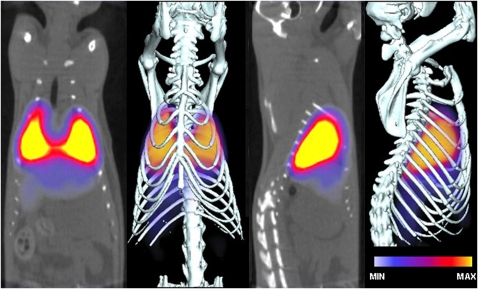
Aminopeptidase P (APP) may fulfill both these needs. Electron microscopy showed that APP antibodies specifically immunolabel caveolae in vascular lung endothelium and can target colloidal gold nanoparticles to caveolae after pulmonary artery perfusion in situ (51, 110). Little to no labeling of caveolae was seen with control antibodies. To assess the specificity and rapidity of lung targeting of APP antibodies in live animals, we injected rats with radiolabeled APP or control antibodies via the tail vein (62, 110). Whole-body γ-scintigraphic imaging revealed that APP antibodies could rapidly target the lungs. The lung silhouette was first discernible 10 seconds after injection, and the heart could be seen as a dark, signal-free shadow. In contrast, control IgG was detected throughout the entire animal and exhibited little to no specific organ targeting. SPECT-CT imaging revealed that antibodies against APP produced the highest-intensity lung image, which remained essentially unchanged from 30 minutes (Figure 1) through 48 hours. We also tested several other antibodies against proteins that show relative lung enrichment, but are not found in caveolae. Consistent with reports in the literature, CD34, ACE, podocalyxin, and PECAM antibodies also specifically targeted the lung, though at lower levels than APP, and the lung-specific signal quickly dissipated after 1 hour. Signal from radiolabeled ACE antibodies was distributed through the thoracic cavity, consistent with ACE expression in both lung and heart luminal endothelial cell membranes. Podocalyxin and PECAM antibodies also targeted other organs, including kidney and liver (62, 110).
Figure 1.
SPECT-CT imaging of aminopeptidase P antibodies shows antibody accumulation in the lungs in vivo. Radiolabeled antibodies were injected intravenously, and SPECT-CT images were acquired 30 minutes later.
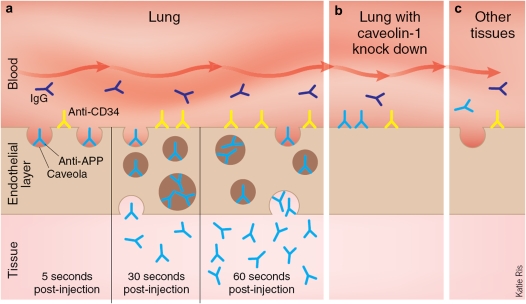
To dynamically follow antibody targeting in real time at high resolution, we engrafted rat lung tissue into dorsal skin fold window chambers on nude mice (110). The lung tissue revascularized within 1 to 2 weeks. After tail vein injection of fluorophore-conjugated antibody, the rat lung tissue was monitored continuously with intravital microscopy. Within 10 seconds, APP antibodies bound to the lung endothelium, but did not bind to vasculature outside of the engrafted rat lung tissue or to engrafted rat heart or liver tissue. This shows that engrafted tissue retained both its species- and tissue-specific markers, even when growing in a mouse host. APP antibodies bound to the vascular endothelium within the lung tissue in approximately 10 seconds. Lower-resolution images showed that the entire engrafted lung tissue was fluorescent within 10 minutes. APP antibody appeared to be actively pumped into the lung tissue, even acting against a concentration gradient. Control IgG did not block binding and transcytosis of APP antibodies. IgG also did not bind the endothelium or extravasate in the tissue, even when much higher concentrations were used. Targets that were accessible to the blood and enriched in caveolae were clearly necessary to see rapid transcytosis. The caveolin antibody did not bind to the lung endothelium and was not transcytosed. This is not surprising, because caveolin is inaccessible to the blood. Antibodies against CD34 rapidly bound the lung microvessel surface but did not cross the endothelial cell barrier, even when higher amounts were used and imaging was extended to 30 minutes. Caveolae themselves appear necessary to transport antibodies across the endothelial cell barrier and into tissue. We used lentivirally expressed short hairpin RNAs against the caveolin-1 gene to lower the amount of caveolae. APP antibodies could still bind to the lung microvessel surface in this tissue, but the antibody was not transcytosed or pumped into the lung tissue (110). Figure 2 provides a schematic summary of events at endothelial cell surface. This cartoon illustrates caveolae-mediated trafficking of antibodies and comes from a News and Views articles written about our 2007 Nature Biotechnology paper documenting the pumping ability of caveolae to enable antibodies to target and penetrate specifically into the lung after intravenous injection.
Figure 2.
Overview of caveolae-dependent trafficking of antibodies. (a) Antibodies targeted against caveolae (anti-APP, light blue) are rapidly transported into the lung tissue, antibodies that bind outside of caveolae (anti-CD34, yellow) accumulate at the cell surface, and nonspecific IgG molecules (black) stay in the blood. (b) When caveolin-1 protein is decreased, aminopeptidase P antibodies remain at the cell surface, implicating caveolae in their transendothelial transport. (c) According to this model, aminopeptidase P antibodies are specific for lung endothelial cells and do not accumulate in blood vessels from other tissues. Reprinted by permission from Reference 119.
TARGETING LUNG TUMORS
Clinical utility depends on finding specific endothelial markers for disease states. Endothelial cells are extremely sensitive to the tissue environment and can rapidly change when removed from their normal environment (61, 62). Less drastic changes, such as inflammation and disease, may also alter tissue enough for changes to be reflected in the surrounding vasculature. Caveolae are found at the luminal surface of most endothelium and may facilitate transport to many different organs and even solid tumors, thus providing a universal pathway that can be exploited to deliver drugs or imaging agents to diseased, as well as healthy tissue.
When injected into rats bearing multiple tumors in the lungs, APP antibodies did not traffic to the tumors but instead targeted the surrounding normal lung tissue (62). When the luminal endothelial surface of vasculature from lung tumors was analyzed by Western analysis and with mass spectrometry, APP expression was lost. Tumor vascular endothelium expressed a distinct array of proteins, clearly different from healthy lungs, suggesting that the solid tumors might form a distinct type of tissue that influences protein expression in its neovasculature. Several possible markers for tumor endothelium are listed in Table 2. We again applied a subtractive proteomic approach and mass spectrometric analysis of these plasma membranes and identified 15 proteins that were differentially expressed. As expected, several known tumor angiogenesis markers were up-regulated, including vascular endothelial growth factor (VEGF) receptors-1 and -2, Tie2, aminopeptidase-N, endoglin, carcino-embryonic antigen-related cell adhesion molecule 1, and neuropilin-1. Previously unknown tumor-induced vascular proteins (annexin A1, annexin A8, ephrin A5, ephrin A7, myeloperoxidase, nucleolin, transferrin receptor, and vitamin D–binding protein) were also detected. Most of the proteins (12 of 15) were much more enriched in endothelial cell plasma membranes isolated from the tumors in the lung versus normal lung tissue. Unfortunately, almost all of these proteins exist in the healthy tissue of one or more major organs, though at lower levels. One surprising result was the induction of a novel protein at the endothelial cell surface. AnnexinA1 (AnnA1) is normally found intracellularly; however, AnnA1 could be detected at the luminal surface of endothelial cells taken from tumor tissue. This protein was specific for tumor endothelium and did not appear in the endothelium of normal organs. Immunohistochemistry showed that AnnA1 was also found in vascular endothelium of human solid tumors of the prostate, liver, breast, and lung, but not matched normal tissue (62).
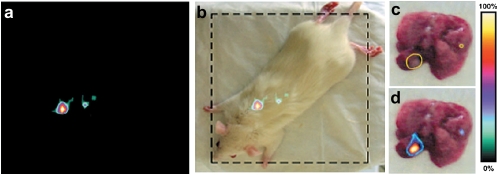
To test whether AnnA1 was expressed on the luminal surface of endothelial cells and thus accessible to circulating antibodies, we performed whole-body imaging using radiolabeled monoclonal antibodies. Gamma-scintigraphic planar imaging of rats with tumors captured 4 hours after injection showed a distinct focus of radioactivity in the lung and little signal elsewhere (Figure 3). No targeting of normal organs was seen. Even lung targeting was absent in tumor-free rats. Nontargeting control IgG did not show detectable selective tumor uptake. When the lungs were excised and imaged, the radioactive hot spots labeled by AnnA1 antibodies corresponded to visible tumors (Figure 3). Surprisingly, 80% of the animals treated with radiolabeled AnnA1 antibodies survived 8 days or longer, while all IgG-treated and untreated rats died within 7 days. This increased survival is striking because in this model, many animals die within 2 to 4 days of treatment and thus may lack sufficient time to benefit from the treatment. A single injection of antibody caused significant remission even in advanced disease (62).
Figure 3.
(a and b) Tumor-bearing rats were intravenously injected with 125I-AnnA1 antibodies and imaged 4 hours later. Tumor-bearing lungs were excised for imaging. (c) Tumors, circled in yellow, could be seen to overlap with (d) hot spots from planar imaging. Reprinted by permission from Reference 62.
FUTURE DIRECTIONS
The rapid targeting and transcytosis shown above goes well beyond traditional vascular targeting and exploits a natural pathway to deliver imaging agents or therapeutic compounds across the endothelial barrier, actually concentrating them in tissue where they can be effective. By delivering these agents to specific sites, harmful side effects can be decreased (110, 118). In addition, active pumping of antibodies and conjugated molecules across the endothelial cell layer and into tissue may prevent degradation (110), further increasing therapeutic potential.
Lung-specific antibodies can be used to target many drugs to the lungs and may also be effective against cancer and other infectious lung diseases such as pneumonia and influenza. Targeting caveolar proteins could also direct imaging agents, nanoparticles, or even genetic material to a specific organ or disease state. Targeting imaging agents to the lung is clearly beneficial for early disease detection, but is also likely to facilitate molecular imaging of lung function in vivo. Caveolae are rich in signaling molecules, and targeted small molecules, siRNAs, peptides, or nanoparticles could be used to activate or inhibit specific pathways. Thus, pumping agents into a specific tissue may finally allow the goals of molecular targeting to be realized as well as offer new windows into the function of organs in vivo.
This system could also be used to target drugs to any single organ with a specific, blood-accessible biomarker. To fulfill this goal, the vascular endothelial cell proteome of all major organs must be comprehensively mapped and targets must be validated. The benefits of targeting caveolae are just becoming clear. To truly exploit this pathway, we must understand its limitations. Are there size constraints? How are molecules processed and where do they go? Ultimately, targeted therapies may have a tremendous impact by increasing the efficacy of drug therapies while decreasing dose, treatment length, and side effects as well as allowing powerful in vivo imaging of both healthy and diseased tissue.
Supported by the following NIH, NCI, and NHLBI grants: PO1CA104898, R01CA083989, R01CA115215, R01CA119378, R01HL058216, R01HL074063, R24CA095893, R33CA118602, and R01HL052766.
Conflict of Interest Statement: K.A.M. received grant support from the NIH $10,001 to $50,000 (Kirschstein Pre-doctoral Fellowship, 2006-2008). J.E.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Dvorak HF, Nagy JA, Dvorak AM. Structure of solid tumors and their vasculature: implications for therapy with monoclonal antibodies. Cancer Cells 1991;3:77–85. [PubMed] [Google Scholar]
- 2.Vitetta ES. Immunotoxins and vascular leak syndrome. Cancer J 2000;6:S218–S224. [PubMed] [Google Scholar]
- 3.Fajardo LF. The complexity of endothelial cells. Am J Clin Pathol 1989;92:241–250. [DOI] [PubMed] [Google Scholar]
- 4.Jaffe EA. Cell biology of endothelial cells. Hum Pathol 1987;18:234–239. [DOI] [PubMed] [Google Scholar]
- 5.Madri JA, Williams SK. Capillary endothelial cell culture: phenotype modulation by matrix components. J Cell Biol 1983;97:153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goerdt S, Steckel F, Schulze-Osthoff K, Hagemeier HH, Macher E, Sorg C. Characterization and differential expression of an endothelial cell- specific surface antigen in continuous and sinusoidal endothelial, in skin vascular lesions and in vitro. Exp Cell Biol 1989;57:185–192. [DOI] [PubMed] [Google Scholar]
- 7.Gumkowski F, Kaminska G, Kaminski M, Morrissey LW, Auerbach R. Heterogeneity of mouse vascular endothelium. Blood Vessels 1987;24:11–23. [PubMed] [Google Scholar]
- 8.Hagemeier HH, Vollmer E, Goerdt S, Schulze-Osthoff K, Sorg C. A monoclonal antibody reacting with endothelial cells of budding vessels in tumors and inflammatory tissues, and non-reactive with normal adult tissues. Int J Cancer 1986;38:481–488. [DOI] [PubMed] [Google Scholar]
- 9.Aird WC, Edelberg JM, Weiler-Guettler H, Simmons WW, Smith TW, Rosenberg RD. Vascular bed-specific expression of an endothelial cell gene is programmed by the tissue microenvironment. J Cell Biol 1997;138:1117–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janzer RC, Raff MC. Astrocytes induce blood-brain barrier properties in endothelial cells. Nature 1987;325:253–257. [DOI] [PubMed] [Google Scholar]
- 11.Stewart PA, Wiley MJ. Developing nervous tissue induces formation of blood-brain barrier characteristics in invading endothelial cells: a study using quail-chick transplantation chimeras. Dev Biol 1981;84:183–192. [DOI] [PubMed] [Google Scholar]
- 12.Jennings MA, Florey L. An investigation of some properties of endothelium related to capillary permeability. Proc R Soc Lond B Biol Sci 1967;167:39–63. [DOI] [PubMed] [Google Scholar]
- 13.Wagner RC, Chen S-C. Transcapillary transport of solute by the endothelial vesicular system: evidence from thin serial section analysis. Microvasc Res 1991;42:139–150. [DOI] [PubMed] [Google Scholar]
- 14.Milici AJ, Watrous NE, Stukenbrok H, Palade GE. Transcytosis of albumin in capillary endothelium. J Cell Biol 1987;105:2603–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghitescu L, Bendayan M. Transendothelial transport of serum albumin: a quantitative immunocytochemical study. J Cell Biol 1992;117:745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghitescu L, Fixman A, Simionescu M, Simionescu N. Specific binding sites for albumin restricted to plasmalemmal vesicles of continuous capillary endothelium: receptor-mediated transcytosis. J Cell Biol 1986;102:1304–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghinea N, Mai TV, Groyer-Picard MT, Milgrom E. How protein hormones reach their target cells. receptor-mediated transcytosis of hcg through endothelial cells. J Cell Biol 1994;125:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnitzer JE, Oh P, Jacobson BS, Dvorak AM. Caveolin-enriched caveolae purified from endothelium in situ are transport vesicles for albondin-mediateed transcytosis of albumin. Mol Biol Cell 1994;5:A75. [Google Scholar]
- 19.Jacobson BS, Stolz DB, Schnitzer JE. Identification of endothelial cell-surface proteins as targets for diagnosis and treatment of disease. Nat Med 1996;2:482–484. [DOI] [PubMed] [Google Scholar]
- 20.Palade GE. Fine structure of blood capillaries. J Appl Phys 1953;24:1424. [Google Scholar]
- 21.Bruns RR, Palade GE. Studies on blood capillaries: I. General organization of blood capillaries in muscle. J Cell Biol 1968;37:244–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansson BR. Size and distribution of endothelial plasmalemmal vesicles in consecutive segments of the microvasculature in cat skeletal muscle. Microvasc Res 1979;17:107–117. [DOI] [PubMed] [Google Scholar]
- 23.Simionescu M, Simionescu N, Palade GE. Morphometric data on the endothelium of blood capillaries. J Cell Biol 1974;60:128–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fra AM, Williamson E, Simons K, Parton RG. Detergent-insoluble glycolipid microdomains in lymphocytes in the absence of caveolae. J Biol Chem 1994;269:30745–30748. [PubMed] [Google Scholar]
- 25.Parolini I, Topa S, Sorice M, Pace A, Ceddia P, Montesoro E, Pavan A, Lisanti MP, Peschle C, Sargiacomo M. Phorbol ester-induced disruption of the cd4-lck complex occurs within a detergent-resistant microdomain of the plasma membrane: involvement of the translocation of activated protein kinase c isoforms. J Biol Chem 1999;274:14176–14187. [DOI] [PubMed] [Google Scholar]
- 26.Drab M, Verkade P, Elger M, Kasper M, Lohn M, Lauterbach B, Menne J, Lindschau C, Mende F, Luft FC, et al. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science 2001;293:2449–2452. [DOI] [PubMed] [Google Scholar]
- 27.Razani B, Woodman SE, Lisanti MP. Caveolae: from cell biology to animal physiology. Pharmacol Rev 2002;54:431–467. [DOI] [PubMed] [Google Scholar]
- 28.Peters KRCW, Palade GE. Endothelial plasmalemmal vesicles have a characteristic striped bipolar surface structure. J Cell Biol 1985;101:2233–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothberg KG, Heuser JE, Donzell WC, Ying YS, Glenney JR, Anderson RG. Caveolin, a protein component of caveolae membrane coats. Cell 1992;68:673–682. [DOI] [PubMed] [Google Scholar]
- 30.Griffoni C, Spisni E, Santi S, Riccio M, Guarnieri T, Tomasi V. Knockdown of caveolin-1 by antisense oligonucleotides impairs angiogenesis in vitro and in vivo. Biochem Biophys Res Commun 2000;276:756–761. [DOI] [PubMed] [Google Scholar]
- 31.Severs NJ. Caveolae: static inpocketings of the plasma membrane, dynamic vesicles or plain artifact? J Cell Sci 1988;90:341–348. [DOI] [PubMed] [Google Scholar]
- 32.McIntosh DP, Schnitzer JE. Caveolae require intact VAMP for targeted transport in vascular endothelium. Am J Physiol 1999;277:H2222–H2232. [DOI] [PubMed] [Google Scholar]
- 33.Schnitzer JE, Liu J, Oh P. Endothelial caveolae have the molecular transport machinery for vesicle budding, docking, and fusion including VAMP, NSF, SNAP, annexins, and GTPases. J Biol Chem 1995;270:14399–14404. [DOI] [PubMed] [Google Scholar]
- 34.Predescu D, Horvat R, Predescu S, Palade GE. Transcytosis in the continuous endothelium of the myocardial microvasculatrue is inhibited by N-ethylmaleimide. Proc Natl Acad Sci USA 1994;91:3014–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schnitzer JE, Allard J, Oh P. NEM inhibits transcytosis, endocytosis, and capillary permeability: implication of caveolae fusion in endothelia. Am J Physiol 1995;268:H48–H55. [DOI] [PubMed] [Google Scholar]
- 36.Parton RG, Joggerst B, Simons K. Regulated internalization of caveolae. J Cell Biol 1994;127:1199–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schnitzer JE, Carley WW, Palade GE. Specific albumin binding to microvascular endothelium in culture. Am J Physiol 1988;254:H425–H437. [DOI] [PubMed] [Google Scholar]
- 38.Tran D, Carpentier JL, Sawano F, Gorden P, Orci L. Ligands internalized through coated or noncoated invaginations follow a common intracellular pathway. Proc Natl Acad Sci USA 1987;84:7957–7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelkmans L, Kartenback J, Helenius A. Caveolar endocytosis of simian virus 40 reveals a novel two-step vesicular transport pathway to the er. Nat Cell Biol 2001;3:473–483. [DOI] [PubMed] [Google Scholar]
- 40.Norkin LC. Simian virus 40 infection via mhc class i molecules and caveolae. Immunol Rev 1999;168:13–22. [DOI] [PubMed] [Google Scholar]
- 41.Empig CJ, Goldsmith MA. Association of the caveola vesicular system with cellular entry by filoviruses. J Virol 2002;76:5266–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mackay RL, Consigli RA. Early events in polyoma virus infection: attachment, penetration, and nuclear entry. J Virol 1976;19:620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richterova Z, Liebl D, Horak M, Palkova Z, Stokrova J, Hozak P, Korb J, Forstova J. Caveolae are involved in the trafficking of mouse polyomavirus virions and artificial vp1 pseudocapsids toward cell nuclei. J Virol 2001;75:10880–10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oh P, McIntosh DP, Schnitzer JE. Dynamin at the neck of caveolae mediates their budding to form transport vesicles by GTP-driven fission from the plasma membrane of endothelium. J Cell Biol 1998;141:101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnitzer JE, Oh P, McIntosh DP. Role of gtp hydrolysis in fission of caveolae directly from plasma membranes. Science 1996;274:239–242. [published erratum appears in Science 1996;274:1069.] [DOI] [PubMed] [Google Scholar]
- 46.Oh P, Schnitzer JE. Dynamin-mediated fission of caveolae from plasma membranes. Mol Biol Cell 1996;7:83a. [Google Scholar]
- 47.Henley JR, Krueger EW, Oswald BJ, McNiven MA. Dynamin-mediated internalization of caveolae. J Cell Biol 1998;141:85–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaplan KL, Weber D, Cook P, Dalecki M, Rogozinski L, Sepe O, Knowles D, Butler VP. Monoclonal antibodies to e92, an endothelial cell surface antigen. Arteriosclerosis 1983;3:403–412. [DOI] [PubMed] [Google Scholar]
- 49.Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975;256:495–497. [DOI] [PubMed] [Google Scholar]
- 50.Parks WM, Gingrich RD, Dahle CE, Hoak JC. Identification and characterization of an endothelial, cell-specific antigen with a monoclonal antibody. Blood 1985;66:816–823. [PubMed] [Google Scholar]
- 51.McIntosh DP, Tan X-Y, Oh P, Schnitzer JE. Targeting endothelium and its dynamic caveolae for tissue-specific transcytosis in vivo: a pathway to overcome cell barriers to drug and gene delivery. Proc Natl Acad Sci USA 2002;99:1996–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Testa JE, Chrastina A, Li Y, Oh P, Schnitzer JE. Ubiquitous yet distinct expression of podocalyxin on vascular surfaces in normal and tumor tissues in the rat. J Vasc Res 2009;46:311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 1985;228:1315–1317. [DOI] [PubMed] [Google Scholar]
- 54.Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature 1996;380:364–366. [DOI] [PubMed] [Google Scholar]
- 55.Rajotte D, Arap W, Hagedorn M, Koivunen E, Pasqualini R, Ruoslahti E. Molecular heterogeneity of the vascular endothelium revealed by in vivo phage display. J Clin Invest 1998;102:430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valadon P, Garnett JD, Testa JE, Bauerle M, Oh P, Schnitzer JE. Screening phage display libraries for organ-specific vascular immunotargeting in vivo. Proc Natl Acad Sci USA 2006;103:407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valadon P, Nussbaum G, Oh J, Scharff MD. Aspects of antigen mimicry revealed by immunization with a peptide mimetic of cryptococcus neoformans polysaccharide. J Immunol 1998;161:1829–1836. [PubMed] [Google Scholar]
- 58.Seaman S, Stevens J, Yang MY, Logsdon D, Graff-Cherry C, St Croix B. Genes that distinguish physiological and pathological angiogenesis. Cancer Cell 2007;11:539–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, Lal A, Riggins GJ, Lengauer C, Vogelstein B, et al. Genes expressed in human tumor endothelium. Science 2000;289:1197–1202. [DOI] [PubMed] [Google Scholar]
- 60.Righetti PG. Electrophoresis: the march of pennies, the march of dimes. J Chromatogr A 2005;1079:24–40. [DOI] [PubMed] [Google Scholar]
- 61.Durr E, Yu J, Krasinska KM, Carver LA, Yates JRI, Testa JE, Oh P, Schnitzer JE. Direct proteomic mapping of the lung microvascular endothelial cell surface in vivo and in cell culture. Nat Biotechnol 2004;22:985–992. [DOI] [PubMed] [Google Scholar]
- 62.Oh P, Li Y, Yu J, Durr E, Krasinska KM, Carver LA, Testa JE, Schnitzer JE. Subtractive proteomic mapping of the endothelial surface in lung and solid tumours for tissue-specific therapy. Nature 2004;429:629–635. [DOI] [PubMed] [Google Scholar]
- 63.Li Y, Yu J, Wang YZ, Griffin NM, Long F, Shore S, Oh P, Schnitzer JE. Enhancing identifications of lipid-embedded proteins by mass spectrometry for improved mapping of endothelial plasma membranes in vivo. Mol Cell Proteomics 2009;8:1219–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Templin MF, Stoll D, Schrenk M, Traub PC, Vohringer CF, Joos TO. Protein microarray technology. Drug Discov Today 2002;7:815–822. [DOI] [PubMed] [Google Scholar]
- 65.Schnitzer JE. Caveolae: from basic trafficking mechanisms to targeting transcytosis for tissue-specific drug and gene delivery in vivo. Adv Drug Deliv Rev 2001;49:265–280. [DOI] [PubMed] [Google Scholar]
- 66.Gimbrone MA Jr, Cotran RS, Folkman J. Endothelial regeneration: studies with human endothelial cells in culture. Ser Haematol 1973;6:453–455. [PubMed] [Google Scholar]
- 67.Jaffe EA, Nachman RL, Becker CG, Minick CR. Culture of human endothelial cells derived from umbilical veins: identification by morphologic and immunologic criteria. J Clin Invest 1973;52:2745–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nagel T, Resnick N, Atkinson WJ, Dewey CF Jr, Gimbrone MA Jr. Shear stress selectively upregulates intercellular adhesion molecule-1 expression in cultured human vascular endothelial cells. J Clin Invest 1994;94:885–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Resnick N, Gimbrone MA Jr. Hemodynamic forces are complex regulators of endothelial gene expression. FASEB J 1995;9:874–882. [DOI] [PubMed] [Google Scholar]
- 70.Topper JN, Gimbrone MA Jr. Blood flow and vascular gene expression: fluid shear stress as a modulator of endothelial phenotype. Mol Med Today 1999;5:40–46. [DOI] [PubMed] [Google Scholar]
- 71.Cole SR, Ashman LK, Ey PL. Biotinylation: an alternative to radioiodination for the identification of cell surface antigens in immunoprecipitates. Mol Immunol 1987;24:699–705. [DOI] [PubMed] [Google Scholar]
- 72.Belloni PN, Nicolson GL. Differential expression of cell surface glycoproteins on various organ-derived microvascular endothelia and endothelial cell cultures. J Cell Physiol 1988;136:398–410. [DOI] [PubMed] [Google Scholar]
- 73.Merker MP, Carley WW, Gillis CN. Molecular mapping of pulmonary endothelial membrane glycoproteins of the intact rabbit lung. FASEB J 1990;4:3040–3048. [DOI] [PubMed] [Google Scholar]
- 74.Schnitzer JE, Shen CP, Palade GE. Lectin analysis of common glycoproteins detected on the surface of continuous microvascular endothelium in situ and in culture: identification of sialoglycoproteins. Eur J Cell Biol 1990;52:241–251. [PubMed] [Google Scholar]
- 75.Schnitzer JE, Siflinger-Birnboim A, Del Vecchio PJ, Malik AB. Segmental differentiation of permeability, protein glycosylation, and morphology of cultured bovine lung vascular endothelium. Biochem Biophys Res Commun 1994;199:11–19. [DOI] [PubMed] [Google Scholar]
- 76.Ghinea N, Eskenasy M, Simionescu M, Simionescu N. Endothelial albumin binding proteins are membrane-associated components exposed on the cell surface. J Biol Chem 1989;264:4755–4758. [PubMed] [Google Scholar]
- 77.Rybak JN, Ettorre A, Kaissling B, Giavazzi R, Neri D, Elia G. In vivo protein biotinylation for identification of organ-specific antigens accessible from the vasculature. Nat Methods 2005;2:291–298. [DOI] [PubMed] [Google Scholar]
- 78.Scheurer SB, Roesli C, Neri D, Elia G. A comparison of different biotinylation reagents, tryptic digestion procedures, and mass spectrometric techniques for 2-d peptide mapping of membrane proteins. Proteomics 2005;5:3035–3039. [DOI] [PubMed] [Google Scholar]
- 79.Alexander JS, Blaschuk OW, Haselton FR. An N-cadherin-like protein contributes to solute barrier maintenance in cultured endothelium. J Cell Physiol 1993;156:610–618. [DOI] [PubMed] [Google Scholar]
- 80.Fridlich R, David A, Aviram I. Membrane proteinase 3 and its interactions within microdomains of neutrophil membranes. J Cell Biochem 2006;99:117–125. [DOI] [PubMed] [Google Scholar]
- 81.Fujimoto T, Nakade S, Miyawaki A, Mikoshiba K, Ogawa K. Localization of inositol 1,4,5-trisphosphate receptor-like protein in plasmalemmal caveolae. J Cell Biol 1992;119:1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sargiacomo M, Lisanti M, Graeve L, Le Bivic A, Rodriguez-Boulan E. Integral and peripheral protein composition of the apical and basolateral membrane domains in MDCK cells. J Membr Biol 1989;107:277–286. [DOI] [PubMed] [Google Scholar]
- 83.Walker J, Bossman P, Lackey BR, Zimmerman JK, Dimmick MA, Hilderman RH. The adenosine 5′,5′′,p1,p4-tetraphosphate receptor is at the cell surface of heart cells. Biochemistry 1993;32:14009–14014. [DOI] [PubMed] [Google Scholar]
- 84.Scheurer SB, Rybak JN, Roesli C, Brunisholz RA, Potthast F, Schlapbach R, Neri D, Elia G. Identification and relative quantification of membrane proteins by surface biotinylation and two-dimensional peptide mapping. Proteomics 2005;5:2718–2728. [DOI] [PubMed] [Google Scholar]
- 85.Roberts LM, Black DS, Raman C, Woodford K, Zhou M, Haggerty JE, Yan AT, Cwirla SE, Grindstaff KK. Subcellular localization of transporters along the rat blood-brain barrier and blood-cerebral-spinal fluid barrier by in vivo biotinylation. Neuroscience 2008;155:423–438. [DOI] [PubMed] [Google Scholar]
- 86.De La Fuente EK, Dawson CA, Nelin LD, Bongard RD, McAuliffe TL, Merker MP. Biotinylation of membrane proteins accessible via the pulmonary circulation in normal and hyperoxic rats. Am J Physiol 1997;272:L461–L470. [DOI] [PubMed] [Google Scholar]
- 87.Schnitzer JE, McIntosh DP, Dvorak AM, Liu J, Oh P. Separation of caveolae from associated microdomains of gpi-anchored proteins. Science 1995;269:1435–1439. (see comments). [DOI] [PubMed] [Google Scholar]
- 88.Oh P, Schnitzer JE. Immunoisolation of caveolae with high affinity antibody binding to the oligomeric caveolin cage: toward understanding the basis of purification. J Biol Chem 1999;274:23144–23154. [DOI] [PubMed] [Google Scholar]
- 89.Schnitzer JE. The endothelial cell surface and caveolae in health and disease. In: Born GVR, Schwartz CJ, editors. Vascular endothelium: physiology, pathology and therapeutic opportunities. Stuttgart: Schattauer; 1997. pp. 77–95.
- 90.Schnitzer JE. Transport functions of the glycocalyx, specific proteins, and caveolae in endothelium. In: Bassingthwaite J, Goresky CA, Linehan JH, editors. Capillary permeation, cellular transport and reaction kinetics. London: Oxford Press; 1997. pp. 31–69.
- 91.Thum T, Haverich A, Borlak J. Cellular dedifferentiation of endothelium is linked to activation and silencing of certain nuclear transcription factors: implications for endothelial dysfunction and vascular biology. FASEB J 2000;14:740–751. [DOI] [PubMed] [Google Scholar]
- 92.Obermeyer N, Janson N, Bergmann J, Buck F, Ito WD. Proteome analysis of migrating versus nonmigrating rat heart endothelial cells reveals distinct expression patterns. Endothelium 2003;10:167–178. [DOI] [PubMed] [Google Scholar]
- 93.Service RF. Proteomics: proteomics ponders prime time. Science 2008;321:1758–1761. [DOI] [PubMed] [Google Scholar]
- 94.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci 1998;7:1029–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shiio Y, Donohoe S, Yi EC, Goodlett DR, Aebersold R, Eisenman RN. Quantitative proteomic analysis of myc oncoprotein function. EMBO J 2002;21:5088–5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, silac, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 2002;1:376–386. [DOI] [PubMed] [Google Scholar]
- 97.Chen X, Sun L, Yu Y, Xue Y, Yang P. Amino acid-coded tagging approaches in quantitative proteomics. Expert Rev Proteomics 2007;4:25–37. [DOI] [PubMed] [Google Scholar]
- 98.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 2003;426:570–574. [DOI] [PubMed] [Google Scholar]
- 99.Gilchrist A, Au CE, Hiding J, Bell AW, Fernandez-Rodriguez J, Lesimple S, Nagaya H, Roy L, Gosline SJ, Hallett M, et al. Quantitative proteomics analysis of the secretory pathway. Cell 2006;127:1265–1281. [DOI] [PubMed] [Google Scholar]
- 100.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (EMPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 2005;4:1265–1272. [DOI] [PubMed] [Google Scholar]
- 101.Liu H, Sadygov RG, Yates JR III. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 2004;76:4193–4201. [DOI] [PubMed] [Google Scholar]
- 102.Bondarenko PV, Chelius D, Shaler TA. Identification and relative quantitation of protein mixtures by enzymatic digestion followed by capillary reversed-phase liquid chromatography-tandem mass spectrometry. Anal Chem 2002;74:4741–4749. [DOI] [PubMed] [Google Scholar]
- 103.Chelius D, Bondarenko PV. Quantitative profiling of proteins in complex mixtures using liquid chromatography and mass spectrometry. J Proteome Res 2002;1:317–323. [DOI] [PubMed] [Google Scholar]
- 104.Cutillas PR, Vanhaesebroeck B. Quantitative profile of five murine core proteomes using label-free functional proteomics. Mol Cell Proteomics 2007;6:1560–1573. [DOI] [PubMed] [Google Scholar]
- 105.Silva JC, Denny R, Dorschel C, Gorenstein MV, Li GZ, Richardson K, Wall D, Geromanos SJ. Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome: a sweet tale. Mol Cell Proteomics 2006;5:589–607. [DOI] [PubMed] [Google Scholar]
- 106.Gao BB, Stuart L, Feener EP. Label-free quantitative analysis of one-dimensional page lc/ms/ms proteome: application on angiotensin II-stimulated smooth muscle cells secretome. Mol Cell Proteomics 2008;7:2399–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res 2006;5:2339–2347. [DOI] [PubMed] [Google Scholar]
- 108.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics 2005;4:1487–1502. [DOI] [PubMed] [Google Scholar]
- 109.Henniker AJ, Bradstock KF, Grimsley P, Atkinson MK. A novel non-lineage antigen on human leucocytes: characterization with two cd-48 monoclonal antibodies. Dis Markers 1990;8:179–190. [PubMed] [Google Scholar]
- 110.Oh P, Borgstrom P, Witkiewicz H, Li Y, Borgstrom BJ, Chrastina A, Iwata K, Zinn KR, Baldwin R, Testa JE, et al. Live dynamic imaging of caveolae pumping targeted antibody rapidly and specifically across endothelium in the lung. Nat Biotechnol 2007;25:327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Muzykantov VR, Puchnina EA, Atochina EN, Hiemish H, Slinkin MA, Meertsuk FE, Danilov SM. Endotoxin reduces specific pulmonary uptake of radiolabeled monoclonal antibody to angiotensin-converting enzyme. J Nucl Med 1991;32:453–460. [PubMed] [Google Scholar]
- 112.Wirrwar A, Schramm N, Vosberg H, Muller-Gartner HW. High resolution spect in small animal research. Rev Neurosci 2001;12:187–193. [DOI] [PubMed] [Google Scholar]
- 113.Rossin R, Muro S, Welch MJ, Muzykantov VR, Schuster DP. In vivo imaging of 64cu-labeled polymer nanoparticles targeted to the lung endothelium. J Nucl Med 2008;49:103–111. [DOI] [PubMed] [Google Scholar]
- 114.Weissleder R, Ntziachristos V. Shedding light onto live molecular targets. Nat Med 2003;9:123–128. [DOI] [PubMed] [Google Scholar]
- 115.Danilov SM, Martynov AV, Klibanov AL, Slinkin MA, Sakharov I, Malov AG, Sergienko VB, Vedernikov A, Muzykantov VR, Torchilin VP. Radioimmunoimaging of lung vessels: an approach using indium-111-labeled monoclonal antibody to angiotensin-converting enzyme. J Nucl Med 1989;30:1686–1692. [PubMed] [Google Scholar]
- 116.Garnacho C, Shuvaev V, Thomas A, McKenna L, Sun J, Koval M, Albelda S, Muzykantov V, Muro S. RhoA activation and actin reorganization involved in endothelial cam-mediated endocytosis of anti-PECAM carriers: critical role for tyrosine 686 in the cytoplasmic tail of PECAM-1. Blood 2008;111:3024–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kozower BD, Christofidou-Solomidou M, Sweitzer TD, Muro S, Buerk DG, Solomides CC, Albelda SM, Patterson GA, Muzykantov VR. Immunotargeting of catalase to the pulmonary endothelium alleviates oxidative stress and reduces acute lung transplantation injury. Nat Biotechnol 2003;21:392–398. [DOI] [PubMed] [Google Scholar]
- 118.Red-Horse K, Ferrara N. Vascular targeting via caveolae. Nat Biotechnol 2007;25:431–432. [DOI] [PubMed] [Google Scholar]