Abstract
Mammalian transcriptome analysis has uncovered tens of thousands of novel transcripts of unknown function (TUFs). Classical and recent examples suggest that the majority of TUFs may underlie vital intracellular functions as non-coding RNAs because of their low coding potentials. However, only a portion of TUFs have been studied to date, and the functional significance of TUFs remains mostly uncharacterized. To increase the repertoire of functional TUFs, we screened for TUFs whose expression is controlled during differentiation of pluripotent human mesenchymal stem cells (hMSCs). The resulting six TUFs, named transcripts related to hMSC differentiation (TMDs), displayed distinct transcriptional kinetics during hMSC adipogenesis and/or osteogenesis. Structural and comparative genomic characterization suggested a wide variety of biologically active structures of these TMDs, including a long nuclear non-coding RNA, a microRNA host gene and a novel small protein gene. Moreover, the transcriptional response to established pathway activators indicated that most of these TMDs were transcriptionally regulated by each of the two key pathways for hMSC differentiation: the Wnt and protein kinase A (PKA) signaling pathways. The present study suggests that not only TMDs but also other human TUFs may in general participate in vital cellular functions with different molecular mechanisms.
INTRODUCTION
Deep sequencing and high-density tiling microarray studies of mammalian cDNAs recently revealed tens of thousands of novel transcripts (1–3). These transcripts differ in size, splicing mode, polyadenylation status and intracellular location. Because their functional roles are currently unclear, they are designated as transcripts of unknown function (TUFs) (4,5). In the post-genomic era, the functional significance of TUFs has received considerable attention. Do most TUFs play important roles in cellular mechanisms, or are they simply transcriptional noise and functionally irrelevant (6,7)? As the majority of human TUFs lack identifiable orthologs in other mammalian model organisms including mouse, answering these questions may improve our understanding of human-specific biology (8).
A common feature of TUFs is the low coding potential. In sharp contrast with mRNAs, which generally contain a single large open reading frame (ORF), TUFs have many start and stop codons scattered throughout their entire nucleotide sequences. Thus, TUFs always have several small ORFs, and yet they rarely resemble any protein families. Because of these features, it is widely believed that most TUFs are non-coding RNAs (ncRNAs). ncRNAs do have a variety of cellular roles (9,10). Certain ncRNAs elicit gene regulatory functions by producing small RNAs such as microRNAs and small nucleolar RNAs (snoRNAs) (11). For more than a decade, long and polyadenylated ncRNA genes such as XIST [X (inactive)-specific transcript] have been known to be functionally essential to gene regulation at a chromosomal and global scale (12). Recently, the number of functional ncRNAs has been expanded using different experimental approaches (13). For example, the identification of binding protein partners for TUFs is a straightforward strategy for understanding the molecular functions of TUFs, as was done for HSR1 (heat shock RNA-1) (14). Loss-of-function screening with short hairpin RNA libraries against conserved mammalian TUFs led to the discovery of NRON (non-coding repressor of transcription factor NFAT) (15). These studies indicate that there are far more TUFs for which the biological significance remains unknown.
In this study, we sought to identify human TUFs that are involved in intracellular signaling pathways. The intracellular signaling network sustains a variety of cell functions including homeostasis, tumorigenesis and differentiation, and to our current knowledge, is governed mostly by proteins. But one would expect that TUFs could play vital roles in the signaling network, potentially as ncRNAs, although no systematic effort has been made to seek such ‘signaling’ TUFs. Thus, this study attempted to identify human TUFs that are regulated by any of the major signaling pathways during human stem cell differentiation. In general, stem cells are transcriptionally hyperactive as compared with differentiated cells (16), and the induction of differentiation initiates global and stepwise activation and/or inactivation of many genes and signaling pathways (17,18). Human bone marrow-derived mesenchymal stem cells (hMSCs) are capable of ex vivo multi-lineage differentiation into adipocytes, osteocytes, chondrocytes and even cells of non-mesodermal origin, when treated with differentiation-inducing reagents (19,20). By taking advantage of this controllable cell differentiation system, we designed a microarray specific for human TUFs to screen for TUFs that exhibited changes of 5-fold or more in abundance in the early stage of hMSC adipogenic or osteogenic differentiation. These transcripts are referred to as transcripts related to hMSC differentiation (TMDs). Sequence analysis of the TMDs and their genomic loci allowed us to categorize and determine the biologically active structures. In order to identify the upstream-signaling pathways for TMDs, we compared TMD transcription levels in the presence and absence of established activators of the Wnt or protein kinase A (PKA) signaling cascades, which are known to govern hMSC differentiation (21–24). The present study demonstrates that TMDs may play vital roles in stem cell functions and moreover suggests that other human TUFs have the potential to contribute to many different cellular phenomena with versatile mechanisms.
MATERIALS AND METHODS
Cells, ex vivo differentiation and chemical induction of signaling pathways
hMSCs (CD105-, CD166-, CD29- and CD44-positive; <5% positive for CD14, CD34 and CD45) were purchased from Cambrex (PT-2501, East Rutherford, NJ, USA). In this study, hMSCs from two different donors were used for kinetic profiling of the TMDs and their neighboring protein genes, as shown in Figures 1A, 2A, 3A, 4A and S2: #4F1560 (donor 1; female, 23-year-old, African American) and #4F1301 (donor 2; male, 26-year-old, Caucasian). For the subsequent and detailed characterization of TMDs, only the hMSCs from donor 1 were used. hMSCs were maintained in MSC basal medium (MSCGM BulletKit, Lonza). hMSCs were expanded to passage 4 before differentiation induction. For adipogenic induction, sub-confluent or confluent cultures of cells were treated and maintained in adipogenic induction medium (hMSC Differentiation BulletKit, Adipogenic, Lonza). For osteogenic induction, hMSC cultures that were ∼10% confluent were treated and maintained in osteogenic induction medium (hMSC Differentiation BulletKit, Osteogenic, Lonza). Cell stimulation by dexamethasone (Dex), insulin, indomethacin, or isobutylmethylxanthine (IBMX) was performed at the concentration used in adipogenic induction medium. For the Wnt activation experiment, Wnt3A (150 ng/ml; R&D Systems) was added to adipogenic induction medium. For PKA activation experiments, forskolin (50 μM; Calbiochem) was added to MSC basal medium.
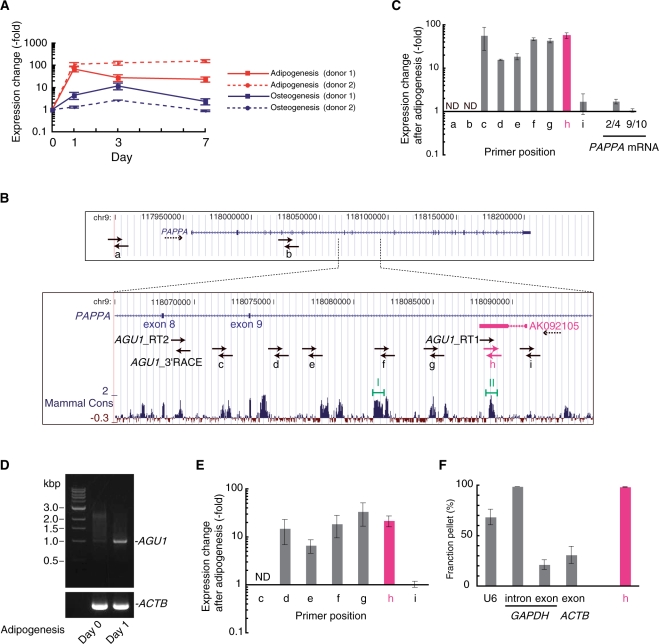
Figure 1.
Structural characterization of the AGU1 locus and transcripts. (A) Expression change in AGU1 during the first 7 days of adipogenesis or osteogenesis from two different hMSC donors. The RNA levels were measured with real-time RT–PCR and normalized to 18S rRNA. Error bars show the standard deviations. (B) Schematic drawing showing two transcription units in the AGU1 locus, an AGU1 representative EST (AK092105) and PAPPA (pregnancy-associated plasma protein A) in the format of the UCSC Human Genome Browser. The transcriptional directions are denoted by dotted arrows. Solid arrows, which are out of scale, indicate the locations and orientations of primers used for reverse transcription, real-time PCR and 3′RACE. At the bottom, the level of sequence conservation among the mammalian species (Mammal Cons) is indicated (31). Two highly conserved regions (I and II) were predicted to fold into stable secondary structures (see Supplementary Figure S5). (C) Changes in transcriptional levels at nine different locations (a–i), shown in (B), and at two different locations on PAPPA mRNA (exons 2−4 and exons 9−10) were measured using real-time RT–PCR 24 h after hMSC adipogenic induction. cDNA was prepared by reverse transcription of total RNA with both random and oligo(dT) primers. Data were normalized to 18S rRNA abundance. Error bars show standard deviations. ND, not detected. (D) Agarose gel electrophoresis of 3′RACE products of AGU1 and ACTB (β-actin). The ∼1.0 kb band appeared only on Day 1 after adipogenic induction, and was confirmed to be derived from the AGU1 locus by sequencing (see Supplementary Figure S4). (E) AGU1-specific real-time RT–PCR. Transcriptional changes at seven different locations (c–i) 24 h after hMSC adipogenic induction were measured with cDNA prepared by reverse transcription of polyA+ RNA and AGU1_RT 2 primer. Data were normalized to GAPDH mRNA expression. Error bars show standard deviations. ND, not detected. (F) Cell fractionation analysis of AGU1 RNA. As controls, spliceosomal U6 snRNA and GAPDH intronic RNA were found in the pellet, whereas mRNAs of ACTB and GAPDH were in the supernatant. Error bars show standard deviations.
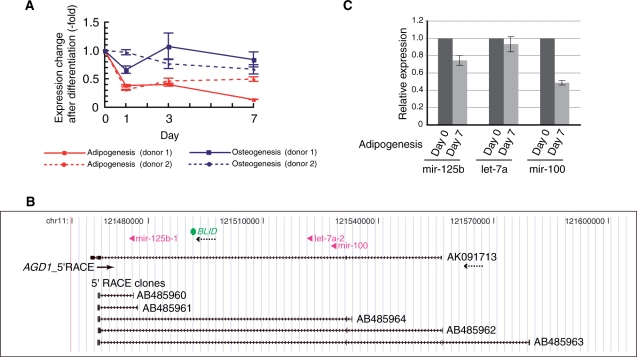
Figure 2.
Structural characterization of a microRNA host gene, AGD1. (A) Changes in expression of an AGD1 representative EST (AK091713) during the first 7 days of adipogenesis or osteogenesis in hMSCs from two different donors. RNA levels were measured by real-time RT–PCR and normalized to 18S rRNA. Error bars show standard deviations. (B) Genomic context of the AGD1 locus. A modified image from the UCSC Human Genome Browser showing an AGD1 representative EST (AK091713), BLID (BH3-like motif containing, cell death inducer), three intronic microRNAs (mir-125b, let-7a and mir-100) and the five different 5′RACE clones obtained in this study. The solid arrow and arrowheads, which are both out of scale, denote the position and orientation of the AGD1-specific PCR primer for 5′RACE (AGD1_5′RACE) and microRNAs, respectively. Dotted arrows indicate orientations of the transcripts. (C) TaqMan real-time RT–PCR measurement of the mature microRNAs before and after 7 days of adipogenic induction. Error bars show standard deviations.
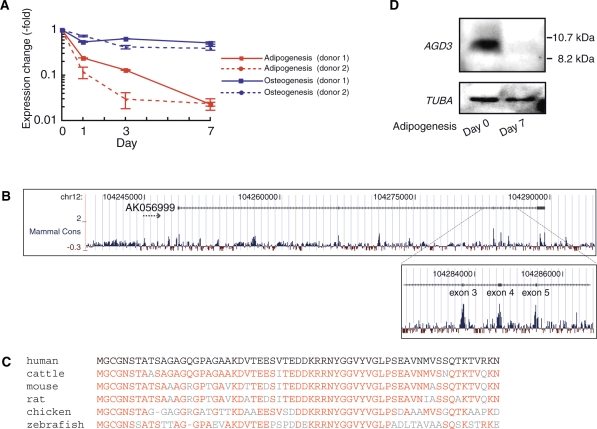
Figure 3.
Structural characterization of a novel small protein gene, AGD3. (A) Real-time RT–PCR measurement of AGD3 transcription level during the first 7 days of adipogenesis or osteogenesis in hMSCs from two different donors. RNA levels were normalized to 18S rRNA. Error bars show standard deviations. (B) Schematic drawing showing mammalian conservation of the nucleotide sequence in the AGD3 locus from the UCSC Human Genome Browser. (C) An alignment of amino-acid sequences of AGD3 vertebrate orthologs: human: NP_001138671, cattle: NP_001138674, mouse: NP_001138670, rat: EDM17086, chicken: NP_001138672, zebrafish: XP_001922116. Note that only the zebrafish ortholog contains an additional 30 amino-acid residues at the N terminus of the shown homologous sequences and that for all the others, the full-length sequences are shown. Residues identical to the human AGD3 amino acid are shown in red letters. (D) Western blotting analysis of the reduction in AGD3 protein after adipogenic induction. TUBA (tubulin, alpha) was chosen as an internal loading control.
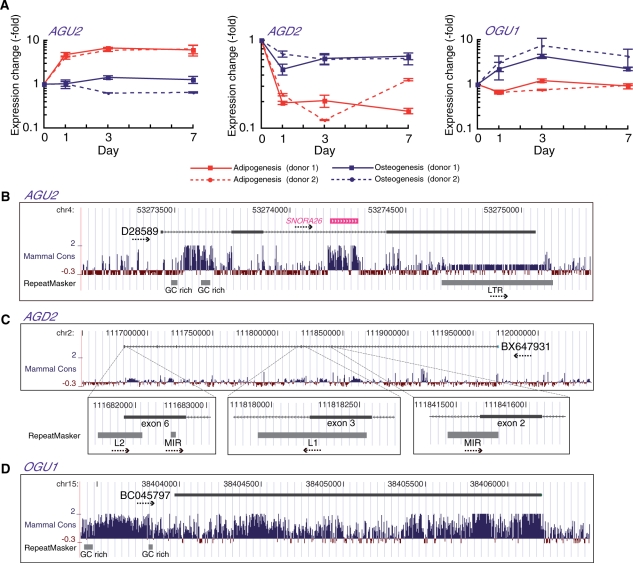
Figure 4.
Structural characterization of AGU2, AGD2 and OGU1. (A) Real-time RT–PCR measurement of AGU2, AGD2 and OGU1 during the first 7 days of adipogenesis or osteogenesis from two different hMSC donors. RNA levels were normalized to 18S rRNA. Error bars show standard deviations. (B–D) Modified views of the UCSC Human Genome Browser showing the genomic context of the AGU2, AGD2 and OGU1 loci. Exon–intron structures of representative ESTs of AGU2 (D28589), AGD2 (BX647931) and OGU1 (BC045797) are shown. Nucleotide sequence conservation and locations of genomic repeats are shown (31,43). The direction of transcripts and repeats are indicated by dotted arrows.
RNA extraction
Total RNA was extracted from hMSCs using the RNeasy kit (Qiagen) and was treated with RNase-free DNaseI (Qiagen) during extraction. For AGU1-specific cDNA preparation (Figure 1E), polyA+ RNA was isolated from total RNA using an Oligotex-dT30 <Super> mRNA Purification Kit (Takara). MicroRNAs as shown in Figure 2C were isolated from total RNA using a mirVana miRNA Isolation Kit (Ambion).
Microarray
A custom microarray for human TUFs was designed by OligoArray 2.1 and obtained from Agilent Technologies. The array included 14 915 and 15 092 probes for the sense and antisense of 5489 non-protein-coding transcripts in H-invitational Database (ver 2.0), respectively and 282 probes for 182 Erdmann ncRNAs. This array detects expression at two or three different locations on each strand of the targeted TUFs. Total RNA from hMSCs that were maintained in MSC basal medium or cultured in differentiation-inducing medium conditions for 1, 3, or 7 days before RNA extraction was labeled with Cy3. Samples were hybridized to the customized microarray according to the manufacturer's protocol. Arrays were scanned with a G2565BA Microarray Scanner System (Agilent Technologies). Data were analyzed using GeneSpring GX software (Agilent Technologies).
RT–PCR
Total RNAs were reverse-transcribed using a QuantiTect reverse transcription kit (Qiagen). The obtained cDNA was amplified using ExTaq HS DNA polymerase (Takara) and PCR primers listed in Supplementary Table S1, and the resultant PCR products were analyzed by 1% agarose gel electrophoresis as shown in Supplementary Figure S1.
Real-time RT–PCR
cDNA was prepared with the QuantiTect reverse transcription kit (Qiagen) or the PrimeScript kit (Takara). In both cases, random and oligo(dT) primers were used except as follows. For AGU1-specific cDNA preparation (Figures 1A, 1F, 5, 6 and S2A), total RNAs were reverse-transcribed with AGU1-specific primers (AGU1_RT 1) as well as 18S ribosomal RNA-specific (Figures 1A, 5, 6 and S2A) or Luc RNA-specific (Figure 1F) primers as a normalization control, using the PrimeScript kit (Takara). For AGU1-specific cDNA preparation (Figure 1E), polyA+ RNAs were reverse-transcribed with an AGU1-specific RT primer (AGU1_RT 2) as well as a GAPDH-specific primer (GAPDH exon reverse) as a normalization control. Real-time PCR analysis was performed with a LightCycler 480 (Roche) or a Thermal Cycler Dice Real Time System (Takara), using SYBR Premix EX Taq (Takara) as the reaction reagent. Results were analyzed based on the second derivative method, and the resultant cycle threshold (Ct) values for transcripts of interest were normalized to those for 18S ribosomal RNA, GAPDH mRNA, or Luc RNA only as shown in Figure 1F (see below for details). Two different sets of primers were used for the individual TMDs to confirm reproducibility of the transcription kinetics (Figures S2 and S3). The primer sequences are listed in Table S1.
Figure 5.
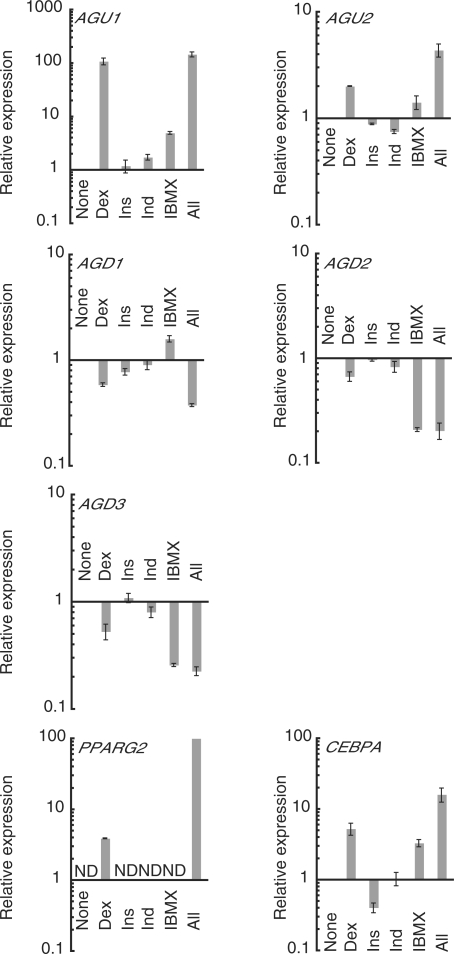
Effects of the individual adipogenesis-inducing reagents upon TMD expression changes during adipogenesis. Relative expression changes of adipogenesis-related TMDs and two master transcription factors (PPARG2 and CEBPA) after 24 h treatment with none, each, or all of the four adipogenesis-inducing reagents (dexamethasone, Dex; insulin, Ins; indomethacin, Ind; isobutylmethylxanthine, IBMX) were quantified by real-time RT–PCR. For the TMDs and CEBPA, expression levels induced by any of the reagents are indicated relative to those in hMSCs that were treated with none of the adipogenesis-inducing reagents (None). Because PPARG2 showed a detectable expression level only with Dex and all four reagents, the former expression level (Dex) is indicated relative to the latter one (All), which is normalized to 100. ND, not detected. See Table S1 for primer sequences. Error bars show standard deviations.
Figure 6.
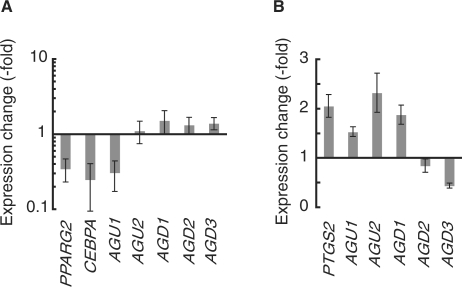
Dissection of upstream-signaling pathways regulating TMD transcription. (A) TMD expression changes by Wnt activation. Expression levels of all the adipogenesis-related TMDs and Wnt target genes (PPARG2 and CEBPA) were measured using real-time RT–PCR 24 h after adipogenic induction in the absence or presence of Wnt3A (150 ng/ml). The log ratio of the individual TMD expression with versus without Wnt3A is shown on the y-axis. (B) TMD expression changes by PKA activation. Expression levels of all the adipogenesis-related TMDs and a PKA target gene (PTGS2) were measured using real-time RT–PCR after 7-day treatment of hMSCs with forskolin (50 μM) or vehicle control (dimethyl sulfoxide). The ratio of the individual TMD expression with versus without forskolin is shown on the y-axis. See Table S1 for primer sequences. Error bars show standard deviations.
To confirm target specificities of the primer sets, real-time PCR products after the 40 cycle reactions were gel purified, cloned into pCRII-TOPO (Invitrogen), and sequenced with the ABI PRISM BigDye Terminator Cycle Sequencing kit (Applied Biosystems) using an ABI 3100 DNA sequencer (data not shown).
The intracellular abundance of three differently mature microRNAs, hsa-mir-125b, hsa-let-7a and hsa-mir-100, was measured using a TaqMan MicroRNA Assay (Applied Biosystems) and normalized to RNU48.
5′ and 3′RACE
To determine the sequence of the 3′-end of the AGU1 transcript, total RNAs from undifferentiated hMSCs or hMSCs cultured in adipogenic induction medium for 24 h were reverse-transcribed with the 3′RACE_RT primer using a Superscript III first-strand cDNA synthesis kit (Invitrogen). The cDNAs obtained were amplified with the AGU1_3′RACE and 3′RACE PCR primers. To determine the sequence of the 5′-end of the AGD1 transcript, total RNAs from undifferentiated hMSCs were reverse-transcribed using the FirstChoice RLM-RACE kit (Ambion). The cDNAs obtained were amplified with the AGD1_5′RACE and 5′RACE outer primers (Ambion).
The resulting PCR products were analyzed on a 1% agarose gel, purified using a gel extraction kit (Qiagen) and cloned into pCRII-TOPO (Invitrogen). The inserts were sequenced with the ABI PRISM BigDye Terminator Cycle Sequencing kit (Applied Biosystems) using an ABI 3100 DNA sequencer. These sequences were registered in GenBank (AB485960-AB485964 for AGD1 5′RACE sequences, AB485715 for AGU1 3′RACE sequence).
Fractionation of total RNA from adipogenic hMSCs
hMSCs in a T25 flask (∼106 cells) at 24 h after adipogenic induction as described above were trypsinized, washed twice with ice-cold PBS, resuspended in lysis buffer [10 mM Tris–HCl (pH 8.0), 150 mM NaCl, 0.5% NP-40, 1 mM DTT, 1× Complete Protease Inhibitor Cocktail (Roche), 100 U/ml RNAsin (Promega)] and centrifuged at 1000 × g for 3 min at 4°C. The supernatant was saved. For preparation of the total cell fraction, hMSCs in a T25 flask (∼106 cells) at 24 h after adipogenic induction were trypsinized, washed twice with ice-cold PBS and resuspended in lysis buffer.
For normalization of RNA loss during RNA extraction, both cytoplasmic and total cell fractions were spiked with an equal amount (∼1 ng) of Renilla reniformis luciferase RNA (Luc RNA). Luc RNA was synthesized by in vitro transcription from pRL-CMV (Promega) with T7 RNA Polymerase (TOYOBO) and purified with the RNeasy kit (Qiagen) as specified by the manufacturers. After total RNA extraction from both supernatant and total cell fractions using an RNeasy kit (Qiagen) and RNase-free DNaseI (Qiagen), real-time RT–PCR was performed as described above to obtain Ct values for AGU1, four different control RNAs (mRNAs for ACTB, GAPDH, GAPDH intron and U6 snRNA) and Luc RNA in the cytoplasmic and total cell fractions. Ct values for AGU1 and the control RNAs were normalized to those for Luc RNA. The resultant Ct values (Ct′) were used to calculate the fractions in the pellet using the formula 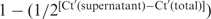
Prediction of RNA secondary structure
Secondary structures of the highly conserved regions within the AGU1 transcripts were predicted using the RNA Mfold program (version 3.2) (25), using default parameters. In all cases, the folding structures with the lowest free energy are shown.
Preparation of polyclonal antibody against AGD3 protein
Three different synthetic peptides derived from the N and C terminus and the middle region of the AGD3 protein (CGNSTATSAGAGQGP, CVNMVSSQTKTVRKN and CTEDDKRRNYGGVYV, respectively) were conjugated with Inject Maleimide Activated mcKLH (Pierce) and mixed with Freund's Complete Adjuvant (Thermo Scientific). Rabbits were immunized with a cocktail of these three peptides and maintained for 8 weeks. Antisera generated were purified by affinity chromatography using the C-terminal peptide coupled to Activated Thiol−Sepharose 4B (Pharmacia).
Immunoblotting
Undifferentiated hMSCs and hMSCs cultured in adipogenic induction medium for 7 days were washed with 1× PBS, lysed in lysis buffer [50 mM Tris–HCl, (pH 8.0), 120 mM NaCl, 1 mM EDTA, 0.5% NP40, 1 mM PMSF, 1× complete protease inhibitor cocktail (Roche)] and centrifuged at 14 000 rpm for 15 min. Protein concentration was determined by DC protein assay (Bio-Rad). Each protein sample (90 µg) was analyzed by 15% SDS–PAGE and electroblotted onto Immobilon-P membrane (Millipore). The filters were blocked with 0.3% (w/v) skim milk in 1× PBS/0.1% Tween 20 for 1 h at room temperature and then incubated with the anti-hAGD3 rabbit polyclonal antibody (1:100) or a mouse anti-TUBA monoclonal antibody (1:500; sc-8035, SantaCruz). Finally, the blots were incubated with peroxidase-labeled donkey anti-rabbit IgG (1:5000; NA934V, Amersham) or sheep anti-mouse IgG (1:2000; NA931V, Amersham). Immunoreactive proteins were detected with Immobilon Western Chemiluminescence HRP substrate (Millipore) and the LAS-3000 system (Fuji Film).
RESULTS
Stem cell-based screening for functional human TUF candidates
hMSCs from two different donors were expanded in growth medium until the fourth passage and then were differentiated into one of the two lineages using established protocols. Adipogenesis was induced by supplementation with four different reagents (Dex, insulin, indomethacin and IBMX), whereas osteogenesis was initiated by a different set of reagents (Dex, ascorbic acid 2-phosphate and β-glycerophosphate). As hMSCs were maintained in each of the differentiation media, the differentiation marker genes for the individual lineages were transcriptionally induced: ADIPOQ (adiponectin, C1Q and collagen domain−containing) for adipogenesis and ALPL (alkaline phosphatase liver/bone/kidney) for osteogenesis (Supplementary Figure S1).
To monitor TUF expression, we developed a DNA microarray in which the probes target 182 Erdmann ncRNAs (26) and 5489 transcripts annotated as non-protein-coding transcripts in the full-length human cDNA H-invitational (H-inv) Database (ver. 2.0) (27). This microarray included two or three different probes for each strand of the targeted loci. Because the H-inv transcripts possess only small ORFs (<240 bp, corresponding to <80 amino acids), and the longest ORF for each transcript encodes no functional protein sequence or motif, this microarray was customized specifically for TUFs that could be novel genes for ncRNAs or small proteins. In addition, these transcripts varied in length, splicing mode, polyadenylation status and retroelement content so that our screen was unbiased within the context of TUF primary sequences.
The initial screening by the customized microarray allowed us to obtain TUFs for which the expression levels increased or decreased by at least 5-fold on Day 1, 3 or 7 after hMSC adipogenic or osteogenic induction. To add to the stringency of this screen, we selected only TUFs for which all the probes on either strand indicated similar changes in expression. We next performed manual inspection of mRNAs and ESTs assigned within and around the transcribed regions of the obtained TUFs in the UCSC Human Genome Browser (28) so as to exclude non-full-length transcripts from the resultant TUFs. Although the majority of the cDNA collections in the H-inv Database are full-length transcripts cloned mostly by oligo-capping (29), the collection probably includes truncated transcripts from neighboring genes. Indeed, many genomic regions producing TUFs overlapped annotated genes, and some of these TUFs could represent truncated forms of these overlapping gene transcripts that were incorrectly annotated as independent full-length transcripts. To exclude such experimental artifacts and to acquire only the genuine independent full-length TUFs for further characterization, the TUFs obtained by the microarray screening were examined using the following procedures. First, the relative transcriptional orientations of the individual TUFs and their adjacent or overlapping NCBI Reference Sequence (RefSeq) genes were evaluated by confirming the position of the canonical polyadenylation (polyA) signal (AATAAA or ATTAAA) and/or polyA tails at the 3′-termini of the mRNAs and ESTs assigned to the corresponding loci. Anti-parallel orientation was regarded as evidence that the TUF transcription units were independent from the neighboring genes. When the transcriptional orientation was parallel to adjacent RefSeq genes, nucleotide distances were also considered. In this study, TUFs that were ≥5 kb away from the nearest annotated gene were labeled as independent transcripts. When TUFs overlapped with any RefSeq genes in the same orientation, exon–intron structures were assessed to certify TUF transcriptional independence: when a TUF was wholly included within an intron of a RefSeq gene or vice versa, both were considered independent transcriptional units, even if they were transcribed on the same strand. After this intensive manual evaluation of the genomic and transcript structures, the TUFs that were likely to have originated from independent transcription units were finally subjected to quantitative RT–PCR to confirm the expression changes observed on the microarray.
This manual evaluation process identified six functional TUF candidates, which are referred to as TMDs. TMDs exhibited distinct expression kinetics within the two different lineages (see below). Five TMD loci were either up- or downregulated more drastically during adipogenesis than during osteogenesis, and thus these were named adipogenesis upregulating transcripts 1 and 2 (AGU1 and AGU2) and adipogenesis downregulating transcripts 1, 2 and 3 (AGD1, AGD2 and AGD3), respectively. The last TMD locus was upregulated only during osteogenesis and was thus named osteogenesis upregulating transcript 1 (OGU1).
AGU1: a 20 kb ncRNA that is upregulated during adipogenesis
Because the representative EST for AGU1 (AK092105) is located on the antisense strand of intron 9 of PAPPA (pregnancy-associated plasma protein A), the expression profiles during adipogenesis and osteogenesis were measured using cDNAs prepared with strand-specific AGU1_RT 1 primer (Figure 1B). Transcription of the EST locus was dramatically increased during adipogenesis, more than during osteogenesis (Figure 1A). Antisense transcripts are often involved in cis-repression of transcription of their sense counterparts, which usually results in a negative correlation between the two transcriptional states (30). Unexpectedly, however, this was not the case for AGU1. PAPPA mRNA levels changed by <5-fold throughout the first 7 days in each of the two lineages (Supplementary Figure S2A). The discordant transcriptional responses of AGU1 and PAPPA imply that, unlike many examples of sense-antisense gene pairs, AGU1 may not be a gene silencer of PAPPA, at least not during hMSC adipogenesis. More importantly, the AGU1 locus responded to adipogenesis induction by increasing transcription independently of PAPPA, indicating that AGU1 has distinct cis-regulatory elements controlled by upstream signaling pathway(s) during adipogenesis.
The boundary of the AGU1 transcription unit was not clear. Unlike the other TMDs, there was no EST cluster in the AK092105 region in the UCSC Human Genome Browser (31). To approximate both boundaries of the AGU1 transcribed region, we took advantage of the dramatic induction of AGU1 transcription during hMSC adipogenesis. Expression changes at eight different locations upstream or downstream of the AGU1 EST (AK092105) were measured 24 h after adipogenic induction (Figure 1B and C). As noted above, the AK092105 region [location (h)] displayed ∼50-fold induction; furthermore, the transcriptionally activated region spanned location (c) shown in Figure 1B. On the contrary, the upstream position [Figure 1B, location (i)] exhibited only a minor transcriptional response. Two different sets of PCR primers for different exonic portions of PAPPA mRNA (exons 2−4 and exons 9−10) showed only modest expression changes, confirming that transcription of the PAPPA strand was largely unchanged. These results indicate that AGU1 transcription starts around the original EST region and ends between location (b) and (c) shown in Figure 1B.
To identify the exact 3′ terminus of the AGU1 transcription unit, 3′RACE was performed using an AGU1_3′RACE primer, which was targeted just downstream of location (c) (Figure 1B). Only the cDNA prepared from hMSCs after adipogenic induction produced a ∼1.0 kb PCR product as a discrete band in an agarose gel (Figure 1D). Cloning and sequencing confirmed that this PCR product had indeed originated from the 3′ terminus of the AGU1 transcript because it included the polyA signal hexamer AATAAA close to the 3′-terminal polyA tail (Supplementary Figure S4). We then examined whether this entire AGU1 region produced one consecutive transcript. Northern blotting is commonly used to estimate transcript length. We were, however, unable to detect AGU1 transcripts, which was probably due to the low cognate expression of AGU1, similar to many other ncRNAs (data not shown). Therefore, we applied real-time RT–PCR for this purpose. The AGU1-specific reverse transcription primer AGU1_RT 2, which was designed to hybridize to the upstream region of the AGU1 3′ terminus that we determined, was used to synthesize AGU1 cDNA with polyA+ RNAs from undifferentiated and adipogenic hMSCs. Both cDNAs were then used for real-time PCR with seven primer sets for locations (c) to (i). All the expected locations except (c) and (i) showed significant increases in transcription levels after adipogenic induction (Figure 1E). Similar to what is shown in Figure 1C, location (i) showed no change, confirming that the AGU1 transcription start site is located between locations (h) and (i). The absence of a transcriptional effect at (c) might be explained by the existence of any spliced intron including location (c). In sum, the AGU1 locus spans the antisense strand of introns 8 and 9 of PAPPA and quickly responds to hMSC adipogenesis by producing at least one ∼20 kb polyA+ transcript.
Many kinds of regulatory RNAs are compartmentalized in a particular intracellular location, and many known ncRNAs reside in the nucleus and function as gene regulators (10). To learn more about the molecular characteristics of the AGU1 transcript, we fractionated total RNAs from hMSCs cultured in adipogenesis induction medium for 24 h into detergent-soluble supernatant and detergent-insoluble pellet. We then examined the distribution of AGU1 RNA using real-time RT–PCR (Figure 1F). Whereas nuclear RNAs, U6 spliceosomal snRNA and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) intronic RNA, were predominantly found in the pellet fraction, cytoplasmic control RNAs, ACTB (β-actin) mRNA and GAPDH mRNA were located in the supernatant fraction. With this nuclear/cytoplasmic fractionation method, AGU1 RNA was found almost exclusively in the pellet fraction, which suggests that the AGU1 transcript may be localized to the nucleus. Although the overall sequence conservation of the AGU1 locus is fairly low, this locus includes partial regions that are highly conserved among mammals. Indeed, two of the conserved regions [i.e. regions (I) and (II) shown in Figure 1B] could code for RNA transcripts with relatively stable secondary structures (Supplementary Figure S5). AGU1 RNA may interact with other nuclear molecules, through these conserved structures, to fulfill its important role as a ncRNA in hMSC adipogenesis.
AGD1: a host gene of clustered microRNAs
Despite slightly different fold changes in the transcription levels between hMSCs from different donors, AGD1 transcription was reduced more strongly during adipogenesis than osteogenesis (Figure 2A). As shown in Figure 2B, the AGD1 locus includes a known protein-encoding gene BLID (BH3-like motif containing, cell death inducer), as well as three microRNA loci (mir-125b, let-7a and mir-100) within the AGD1 introns. All five transcribed regions are on the same strand. BLID possesses its own transcriptional promoter and terminator regions, but these mature microRNAs could be generated by processing of spliced AGD1 intron RNAs. This idea is supported by the 5′RACE experiments, which showed that AGD1 cDNAs started at different sites in undifferentiated hMSCs and that longer cDNAs covered all three microRNA loci in their introns (Figure 2B). We next performed TaqMan real-time RT–PCR and quantified expression changes in the three individual mature microRNAs during hMSC adipogenesis (Figure 2C). Of the three microRNAs, only mir-100 showed a significant decrease in intracellular abundance, which was ∼2-fold at 7 days after adipogenesis induction. While AGD1 is the unique locus for mir-100 in the human genome, there are two paralogous loci for each of the other two microRNAs. Therefore, assuming that the paralogous loci of let-7a and mir-125b are expressed at higher levels than AGD1, we could explain the uneven fold changes in expression levels of clustered microRNAs. Accordingly, expression reduction at the AGD1 locus may contribute to hMSC adipogenesis by balancing intracellular levels of its intronic microRNAs with other microRNA loci.
AGD3: a novel small protein-encoding gene
AGD3 displayed a drastic reduction in expression by ∼50-fold at Day 7 after adipogenic induction (Figure 3A). The AGD3 transcript is the only TMD that was previously investigated: it was reported to be an ncRNA that is overexpressed in cancer tissues (overexpressed in colon carcinoma-1; OCC-1), although its molecular function is still unknown (32). To assess the functions of the AGD3 transcript, we investigated the degree of sequence conservation across the entire AGD3 locus. The consecutive exons 3, 4 and 5 of an AGD3-representative EST were highly conserved among mammals (Figure 3B). Sequence analysis revealed that this region of the EST does not possess any stop codons and thus that the conserved region spanning from exons 1 to 5 constitutes a small 63-amino-acid ORF. Surprisingly, the amino-acid sequences were highly conserved that were deduced from orthologous genomic loci of other vertebrates (Figure 3C). Human AGD3 protein shows 86% and 68% sequence identity with the mouse and zebrafish orthologs, respectively. To examine whether this protein was indeed expressed, we prepared a polyclonal antibody against a peptide epitope derived from the putative C-terminal region of human AGD3 protein. Western blotting confirmed that AGD3 expression is reduced during hMSC adipogenesis at the protein level as well (Figure 3D). This ∼10-kDa band was not detected in cell lysates from hMSCs that were treated with anti-AGD3 siRNAs (data not shown), demonstrating the specificity of our antibody. Therefore, it is highly possible that AGD3 RNA plays a vital role, not as a ncRNA but as an mRNA for stem cell function and/or adipogenesis, and that this role is maintained across vertebrates.
AGU2, AGD2 and OGU1: ncRNA candidates with different structural aspects
The other three TMDs also exhibited >5-fold changes in expression during hMSC adipogenesis or osteogenesis (Figure 4A). Individual TMD expression was controlled separately from the expression of neighboring genes, suggesting a function for the observed changes in TMD transcription during hMSC differentiation (Supplementary Figure S2). AGU2 includes one H/ACA box snoRNA, SNORA26, in one of the two introns (Figure 4B). This small RNA, however, showed little change in its intracellular abundance during hMSC adipogenesis by northern blotting (data not shown). AGU2 contains another conserved GC-rich region in the first intron, which may be processed as a not-yet-annotated small functional RNA. This notion does, however, require more intensive analysis of the function of AGU2.
From an evolutionary perspective, it is of interest that transcription units of AGU2 as well as AGD2 are shaped by retrotransposons. AGU2 contains a long terminal repeat (LTR)-like element, which provides its polyA signal. Similarly, for AGD2, an ancient class of long interspersed nuclear element, LINE2 (L2), serves as a transcription terminator and two other repeats, LINE1 (L1) and mammalian-wide interspersed repeat (MIR), were integrated as splice donor sites (Figure 4C). These examples demonstrate the involvement of retrotransposon-derived sequences in TUF evolution and imply functional linkages of the genomic repeats with ncRNAs. On the contrary, the entire OGU1 locus including the promoter region is highly conserved among mammals and does not contain any retrotransposon-derived elements (Figure 4D). Unlike AGD3, no conserved ORFs were seen in the transcribed region of OGU1, which may function as a ncRNA.
Chemical stimuli for TMD expression alteration
During the process of in vitro adipogenesis used in this study, the four different reagents (Dex, insulin, indomethacin and IBMX) individually and cooperatively induce activation and/or inactivation of multiple signaling pathways. We speculated that TMD expression could be functionally controlled by any of these signaling pathways during adipogenesis. We thus determined which adipogenesis-inducing reagent(s) were responsible for the observed expression changes of the individual AGUs and AGDs. hMSCs were incubated for 24 h in the presence of each or all of the adipogenesis-inducing reagents, and the TMD transcriptional levels under these five different conditions were quantified relative to those in the undifferentiated state.
Interestingly, the TMDs showed various responses to the individual reagents (Figure 5). AGU1 was mostly Dex responsive, similar to the adipogenesis marker gene PPARG2 (peroxisome proliferator-activated receptor gamma 2) (19,33,34), which also showed residual but detectable transcriptional induction with Dex alone. Like other adipogenesis master genes such as CEBPA (CCAAT/enhancer binding protein, alpha) (35,36), AGU2 responded weakly to Dex and IBMX and showed higher and significant expression with all four reagents. This implies that the AGU2 induction seen in adipogenesis might be due to cooperative stimulation by these two chemicals. For all three adipogenesis downregulating transcripts, Dex induced ∼2-fold reductions, insulin and indomethacin produced little change and the combination of all four reagents induced a significant reduction in transcription. In contrast, IBMX induced the opposite changes: stimulation of AGD1 and repression of AGD2 and AGD3. Overall, these TMDs exhibited different patterns of transcriptional responses to the individual reagents, suggesting that the transcription of each TMD is controlled by a distinct set of upstream signaling cascades.
Upstream signaling pathways of TMDs
Based on the chemical requirements for the individual TMD responses, we attempted to determine the specific pathways that regulate each of the TMDs. Of the adipogenesis-inducing reagents, Dex is a glucocorticoid hormone and is involved in various signaling pathways including the Wnt pathway (37,38). We tested whether activation of the Wnt pathway moderated expression of TMDs, especially Dex-responsive AGU1, during hMSC adipogenesis (Figure 6A). Wnt activation by Wnt3A was verified by suppressed expression of PPARG2 and CEBPA, which are downstream transcription factors in the Wnt pathway (39–41). As expected, Wnt3A significantly reduced Dex-responsive AGU1 levels, suggesting that AGU1 is a Wnt target gene. Transcriptional levels of the other TMDs were less affected by Wnt3A or Dex alone (Figures 5 and 6A), suggesting the involvement of other pathways in their expression regulation.
IBMX inhibits phosphodiesterase, prevents cAMP degradation and leads to activation of the PKA pathway (24). We thus next investigated the effect of the PKA pathway on IBMX-responsive TMD expression by examining the effects of another PKA-activating chemical, forskolin. Forskolin mostly mediated TMD expression responses that were observed with IBMX (Figure 6B). In particular, AGU2 and AGD1 were induced in a similar manner to a known PKA downstream gene, PTGS2 (prostaglandin-endoperoxide synthase 2) (42), whereas AGD3 was significantly suppressed. On the contrary, AGD2 demonstrated a striking reduction in abundance only with IBMX, which suggests that this TMD might be regulated by IBMX-sensitive and forskolin-insensitive signaling pathways. Overall, it appears that the PKA pathway is one of the upstream regulators of these three TMDs.
DISCUSSION
Six different transcripts that responded to hMSC differentiation by undergoing dramatic changes in intracellular abundance were newly identified from our unbiased search for functional TUFs. Our homemade microarray contained probes that target TUF collections that have a wide variety of sequence characteristics such as the degree of sequence conservation and repeat content. Genomic screens for functional elements typically involve removing repeated elements from the initial collections using RepeatMasker (43). However, the intergenic regions of the human genome, where TUFs are found, contain many retrotransposon-derived repeats, with close to 50% of the genome containing such repeats (44). Also, there is accumulating evidence showing that many retrotransposon-derived elements are used as promoters, enhancers, splice sites and polyA signals for known protein genes, as was seen for AGU2 and AGD2 (Figure 4) (45,46). Knowing these impacts of repeats on gene evolution and regulation, the observed transcriptional responses of these two TUFs during hMSC differentiation indicate their functional importance. It is also possible that repeats embedded chimerically in TUFs could be involved in TUF function.
Although the extent of sequence conservation across species of the entire TUF nucleotide sequence seems low, TUFs possess partially conserved regions, enabling us to identify structures of potentially functional entities in TMDs. One of the most striking and unexpected findings in this study is that AGD3 encodes a novel protein that is conserved from humans to zebrafish (Figure 3C). It was previously reported that this transcript was incapable of translating protein from any of its possible ORFs in vitro (32). Our results clearly show, however, that a protein product is derived from the AGD3 transcript in hMSCs. Previously, some ncRNAs have turned out to be mRNAs for small regulatory proteins during fruit fly development (47,48). In mammals, the bioinformatic survey of the FANTOM collection of mouse cDNAs estimated the presence of at least 1200 new genes coding for small proteins with <100 amino acids, most of which seemed to be translated from putative ncRNAs (49). However, to our knowledge, no studies have confirmed the expression of endogenous protein products from mammalian TUFs. Thus, AGD3 is the first human TUF that has been demonstrated to encode a novel protein.
The high degree of conservation of the AGD3 amino acid sequence across species strongly supports an evolutionarily conserved role for this protein (Figure 3). In addition, AGD3 was suppressed by the PKA pathway during adipogenesis (Figure 6B). PKA signaling is one of the key pathways in determining the commitment of hMSCs to differentiate into either an adipogenic or osteogenic lineage. The activation of PKA facilitates adipogenesis (23,24). Moreover, the PKA pathway is critical to cell growth and is likely involved in tumorigenesis (50). In agreement with this idea, the AGD3 transcript was initially observed to be highly expressed in colon tumors, indicating its involvement in cell proliferation and/or the cell cycle (32). Future studies will be needed to reveal mechanistic features of the AGD3 protein in hMSC biology via the PKA pathway.
AGD1 is a polycistronic microRNA host gene, which encodes mir-125b-1 (fly lin-4), let-7a-2 and mir-100 in its introns. Only mir-100 was downregulated concomitantly with AGD1 reduction after hMSC adipogenesis (Figure 2C). Any mechanistic links between mir-100 and hMSC adipogenesis, however, remain speculative because of a lack of functional information about mir-100 itself, although the entire microRNA cluster seems linked with hMSC-related biology. The intracellular abundance of these three mature microRNAs was positively correlated with neural differentiation; these microRNAs are expressed predominantly in brain and are upregulated during murine and human neural differentiation by retinoic acid (51,52). Although the results were not definitive, hMSCs seem capable of neural differentiation (53,54). Furthermore, the master transcriptional factor for adipogenesis, PPARG, also controls neural stem cell proliferation and differentiation (55). Taken together, one can hypothesize that AGD1 reduction may be involved in the fate of hMSCs (adipogenesis, self-proliferation, or perhaps neurogenesis) through mir-100 and/or the other spliced exonic transcripts.
AGU1 is a genuine non-coding RNA gene suppressed by Wnt signaling (Figure 6A). Wnt signaling is important for embryonic development and stem cell fate decision into various lineages (56–59), and the Wnt pathway stimulates osteogenesis and inhibits adipogenesis (60–62). Our observation that adipogenic induction robustly upregulated AGU1 by more than 50-fold only during adipogenesis is consistent with the hypothesis that AGU1 is one of the genes driving hMSC adipogenesis (Figure 1). AGU1 RNA was likely to be restricted to the nucleus and thus may be untranslated and functional as an ncRNA (Figure 1F) (10). Walking-primer RT–PCR with AGU1-specific cDNA revealed that AGU1 spanned ∼20 kb (Figure 1E). The ∼20 kb transcribed region was included in the antisense portion of intron 9 of PAPPA, the transcriptional level of which remained unchanged during hMSC adipogenesis. Therefore, we do not consider AGU1 to be an antisense regulator of PAPPA expression; rather, the AGU1 transcript may be a signaling ncRNA that receives signals from the Wnt pathway for inactivation from the cytoplasm, potentially through its conserved secondary structure(s), and contributes to hMSC adipogenesis progression by increasing its own nuclear abundance.
Before concluding that these TMDs were under the control of these pathways, we confirmed the independence of the TMDs from their neighboring genes in terms of both genomic structures and transcriptional responses (Supplementary Figure S2). The exon–intron structures and strand orientations of both the TMDs and their neighboring genes were examined to ensure that the TMD loci were transcription units that are independent from adjacent and/or overlapping genes. Transcriptional independence tends to be ignored, but it is very important in screening for functional TUFs. Transcription of more than one gene located in the same chromosomal locus is often concomitantly controlled, for example, by bidirectional promoters or global chromatin modifications (63), and yet all of these genes do not always contribute to downstream signaling (64). In other words, transcripts expressed simultaneously with neighboring genes could be transcriptional noise and thus functionally irrelevant. Therefore, we validated non-correlated profiles of transcriptional kinetics between the individual TMDs and the corresponding adjacent gene(s) to confirm TMD independence in transcriptional regulation. Screening for TUFs for which the transcriptional levels are changed by physiologically active reagents is a useful strategy to identify functional TUF candidates and subsequently validate transcriptional independence of TUFs (65,66).
John Mattick's group used mouse embryonic stem cells and identified 174 polyadenylated TUFs that are differentially expressed by at least 2-fold during differentiation to embryoid bodies (66). Although there is a difference in the number of identified TUFs between the previous report and ours (possibly because our threshold of a 5-fold expression difference was more stringent), both studies demonstrate the utility of stem cells for screening and subsequent characterization of functional TUF candidates. Stem-cell differentiation involves not only fundamental but also lineage-specific cellular mechanisms. hMSCs and other human stem cells, including embryonic or induced pluripotent stem cells, are useful in screening for human-specific TUFs (67–69).
In conclusion, we have identified novel transcripts under the control of two different pathways in the intracellular-signaling network. In general, functional redundancy, which often interferes with the elucidation of gene functions, is due to the presence of parallel pathways producing the observed phenotypes. Therefore, our identification of signaling pathways involving these transcripts will assist in the experimental design for their functional characterization. Understanding the signaling networks in which TUFs and TMDs participate will help uncover their molecular function and may help integrate TUFs into a new paradigm of gene function.
ACCESSION NUMBERS
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
New Energy and Industrial Technology Development Organization (NEDO) of Japan for Functional RNA Project; Tokyo Institute of Technology for Tokyo Tech Award for Challenging Research (to Y.A.). Funding for open access charge: Tokyo Institute of Technology.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Kimitsuna Watanabe and Norihiro Okada for their scientific support and continued encouragement; Ryoko Watanabe for her technical assistance; Toshiyuki Sato, Goro Terai and Aya Kojima for microarray probe design; Jef Boeke, Jeffery Han, Sarah Wheelan and Masaki Kajikawa for their critical reading of this manuscript, Hiroshi Ichinose for his technical support, and all the other Aizawa Lab members for their valuable discussions. The authors thank the Japan Biological Informatics Consortium (JBIC) for its strong support through its management of the ‘Functional RNA Project’.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 2.Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 3.Bertone P., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 4.Kapranov P., Willingham A.T., Gingeras T.R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 5.Willingham A.T., Gingeras T.R. TUF love for ‘junk’ DNA. Cell. 2006;125:1215–1220. doi: 10.1016/j.cell.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Brosius J. Waste not, want not—transcript excess in multicellular eukaryotes. Trends. Genet. 2005;21:287–288. doi: 10.1016/j.tig.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 7.Huttenhofer A., Schattner P., Polacek N. Non-coding RNAs: hope or hype? Trends. Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Pang K.C., Frith M.C., Mattick J.S. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends. Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Mattick J.S., Makunin I.V. Non-coding RNA. Hum. Mol. Genet. 2006;15(Spec No 1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 10.Prasanth K.V., Spector D.L. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- 11.Kawaji H., Hayashizaki Y. Exploration of small RNAs. PLoS Genet. 2008;4:e22. doi: 10.1371/journal.pgen.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wutz A. Xist function: bridging chromatin and stem cells. Trends. Genet. 2007;23:457–464. doi: 10.1016/j.tig.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Goodrich J.A., Kugel J.F. Non-coding-RNA regulators of RNA polymerase II transcription. Nat. Rev. Mol. Cell Biol. 2006;7:612–616. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- 14.Shamovsky I., Ivannikov M., Kandel E.S., Gershon D., Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 15.Willingham A.T., Orth A.P., Batalov S., Peters E.C., Wen B.G., Aza-Blanc P., Hogenesch J.B., Schultz P.G. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 16.Efroni S., Duttagupta R., Cheng J., Dehghani H., Hoeppner D.J., Dash C., Bazett-Jones D.P., Le Grice S., McKay R.D., Buetow K.H., et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindsley R.C., Gill J.G., Kyba M., Murphy T.L., Murphy K.M. Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development. 2006;133:3787–3796. doi: 10.1242/dev.02551. [DOI] [PubMed] [Google Scholar]
- 18.Prudhomme W., Daley G.Q., Zandstra P., Lauffenburger D.A. Multivariate proteomic analysis of murine embryonic stem cell self-renewal versus differentiation signaling. Proc. Natl Acad. Sci. USA. 2004;101:2900–2905. doi: 10.1073/pnas.0308768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pittenger M.F., Mackay A.M., Beck S.C., Jaiswal R.K., Douglas R., Mosca J.D., Moorman M.A., Simonetti D.W., Craig S., Marshak D.R. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 20.Chamberlain G., Fox J., Ashton B., Middleton J. Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells. 2007;25:2739–2749. doi: 10.1634/stemcells.2007-0197. [DOI] [PubMed] [Google Scholar]
- 21.Boland G.M., Perkins G., Hall D.J., Tuan R.S. Wnt 3a promotes proliferation and suppresses osteogenic differentiation of adult human mesenchymal stem cells. J. Cell Biochem. 2004;93:1210–1230. doi: 10.1002/jcb.20284. [DOI] [PubMed] [Google Scholar]
- 22.De Boer J., Wang H.J., Van Blitterswijk C. Effects of Wnt signaling on proliferation and differentiation of human mesenchymal stem cells. Tissue Eng. 2004;10:393–401. doi: 10.1089/107632704323061753. [DOI] [PubMed] [Google Scholar]
- 23.Siddappa R., Martens A., Doorn J., Leusink A., Olivo C., Licht R., van Rijn L., Gaspar C., Fodde R., Janssen F., et al. cAMP/PKA pathway activation in human mesenchymal stem cells in vitro results in robust bone formation in vivo. Proc. Natl Acad. Sci. USA. 2008;105:7281–7286. doi: 10.1073/pnas.0711190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang D.C., Tsay H.J., Lin S.Y., Chiou S.H., Li M.J., Chang T.J., Hung S.C. cAMP/PKA regulates osteogenesis, adipogenesis and ratio of RANKL/OPG mRNA expression in mesenchymal stem cells by suppressing leptin. PLoS ONE. 2008;3:e1540. doi: 10.1371/journal.pone.0001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szymanski M., Erdmann V.A., Barciszewski J. Noncoding regulatory RNAs database. Nucleic Acids Res. 2003;31:429–431. doi: 10.1093/nar/gkg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imanishi T., Itoh T., Suzuki Y., O'Donovan C., Fukuchi S., Koyanagi K.O., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:e162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karolchik D., Hinrichs A.S., Kent W.J. The UCSC Genome Browser. Curr. Protoc. Bioinformatics. 2007 doi: 10.1002/0471250953.bi0104s17. Chapter 1, Unit 1.4. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki Y., Yoshitomo-Nakagawa K., Maruyama K., Suyama A., Sugano S. Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene. 1997;200:149–156. doi: 10.1016/s0378-1119(97)00411-3. [DOI] [PubMed] [Google Scholar]
- 30.Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 31.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pibouin L., Villaudy J., Ferbus D., Muleris M., Prosperi M.T., Remvikos Y., Goubin G. Cloning of the mRNA of overexpression in colon carcinoma-1: a sequence overexpressed in a subset of colon carcinomas. Cancer Genet. Cytogenet. 2002;133:55–60. doi: 10.1016/s0165-4608(01)00634-3. [DOI] [PubMed] [Google Scholar]
- 33.Tontonoz P., Hu E., Graves R.A., Budavari A.I., Spiegelman B.M. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 34.Mbalaviele G., Abu-Amer Y., Meng A., Jaiswal R., Beck S., Pittenger M.F., Thiede M.A., Marshak D.R. Activation of peroxisome proliferator-activated receptor-gamma pathway inhibits osteoclast differentiation. J. Biol. Chem. 2000;275:14388–14393. doi: 10.1074/jbc.275.19.14388. [DOI] [PubMed] [Google Scholar]
- 35.Freytag S.O., Paielli D.L., Gilbert J.D. Ectopic expression of the CCAAT/enhancer-binding protein alpha promotes the adipogenic program in a variety of mouse fibroblastic cells. Genes Dev. 1994;8:1654–1663. doi: 10.1101/gad.8.14.1654. [DOI] [PubMed] [Google Scholar]
- 36.Jaiswal R.K., Jaiswal N., Bruder S.P., Mbalaviele G., Marshak D.R., Pittenger M.F. Adult human mesenchymal stem cell differentiation to the osteogenic or adipogenic lineage is regulated by mitogen-activated protein kinase. J. Biol. Chem. 2000;275:9645–9652. doi: 10.1074/jbc.275.13.9645. [DOI] [PubMed] [Google Scholar]
- 37.Ohnaka K., Tanabe M., Kawate H., Nawata H., Takayanagi R. Glucocorticoid suppresses the canonical Wnt signal in cultured human osteoblasts. Biochem. Biophys. Res. Commun. 2005;329:177–181. doi: 10.1016/j.bbrc.2005.01.117. [DOI] [PubMed] [Google Scholar]
- 38.Smith E., Frenkel B. Glucocorticoids inhibit the transcriptional activity of LEF/TCF in differentiating osteoblasts in a glycogen synthase kinase-3beta-dependent and -independent manner. J. Biol. Chem. 2005;280:2388–2394. doi: 10.1074/jbc.M406294200. [DOI] [PubMed] [Google Scholar]
- 39.Kennell J.A., MacDougald O.A. Wnt signaling inhibits adipogenesis through beta-catenin-dependent and -independent mechanisms. J. Biol. Chem. 2005;280:24004–24010. doi: 10.1074/jbc.M501080200. [DOI] [PubMed] [Google Scholar]
- 40.Kawai M., Mushiake S., Bessho K., Murakami M., Namba N., Kokubu C., Michigami T., Ozono K. Wnt/Lrp/beta-catenin signaling suppresses adipogenesis by inhibiting mutual activation of PPARgamma and C/EBPalpha. Biochem. Biophys. Res. Commun. 2007;363:276–282. doi: 10.1016/j.bbrc.2007.08.088. [DOI] [PubMed] [Google Scholar]
- 41.Kang S., Bennett C.N., Gerin I., Rapp L.A., Hankenson K.D., Macdougald O.A. Wnt signaling stimulates osteoblastogenesis of mesenchymal precursors by suppressing CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma. J. Biol. Chem. 2007;282:14515–14524. doi: 10.1074/jbc.M700030200. [DOI] [PubMed] [Google Scholar]
- 42.Sayasith K., Lussier J.G., Sirois J. Role of upstream stimulatory factor phosphorylation in the regulation of the prostaglandin G/H synthase-2 promoter in granulosa cells. J. Biol. Chem. 2005;280:28885–28893. doi: 10.1074/jbc.M413434200. [DOI] [PubMed] [Google Scholar]
- 43.Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996–2004. Available at http://www.repeatmasker.org.
- 44.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 45.van de Lagemaat L.N., Landry J.R., Mager D.L., Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends. Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 46.Makalowski W. Genomics. Not junk after all. Science. 2003;300:1246–1247. doi: 10.1126/science.1085690. [DOI] [PubMed] [Google Scholar]
- 47.Kondo T., Hashimoto Y., Kato K., Inagaki S., Hayashi S., Kageyama Y. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat. Cell Biol. 2007;9:660–665. doi: 10.1038/ncb1595. [DOI] [PubMed] [Google Scholar]
- 48.Hanyu-Nakamura K., Sonobe-Nojima H., Tanigawa A., Lasko P., Nakamura A. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature. 2008;451:730–733. doi: 10.1038/nature06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frith M.C., Forrest A.R., Nourbakhsh E., Pang K.C., Kai C., Kawai J., Carninci P., Hayashizaki Y., Bailey T.L., Grimmond S.M. The abundance of short proteins in the mammalian proteome. PLoS Genet. 2006;2:e52. doi: 10.1371/journal.pgen.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho-Chung Y.S., Pepe S., Clair T., Budillon A., Nesterova M. cAMP-dependent protein kinase: role in normal and malignant growth. Crit. Rev. Oncol. Hematol. 1995;21:33–61. doi: 10.1016/1040-8428(94)00166-9. [DOI] [PubMed] [Google Scholar]
- 51.Sempere L.F., Freemantle S., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laneve P., Di Marcotullio L., Gioia U., Fiori M.E., Ferretti E., Gulino A., Bozzoni I., Caffarelli E. The interplay between microRNAs and the neurotrophin receptor tropomyosin-related kinase C controls proliferation of human neuroblastoma cells. Proc. Natl Acad. Sci. USA. 2007;104:7957–7962. doi: 10.1073/pnas.0700071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grove J.E., Bruscia E., Krause D.S. Plasticity of bone marrow-derived stem cells. Stem Cells. 2004;22:487–500. doi: 10.1634/stemcells.22-4-487. [DOI] [PubMed] [Google Scholar]
- 54.Tondreau T., Dejeneffe M., Meuleman N., Stamatopoulos B., Delforge A., Martiat P., Bron D., Lagneaux L. Gene expression pattern of functional neuronal cells derived from human bone marrow mesenchymal stromal cells. BMC Genomics. 2008;9:166. doi: 10.1186/1471-2164-9-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wada K., Nakajima A., Katayama K., Kudo C., Shibuya A., Kubota N., Terauchi Y., Tachibana M., Miyoshi H., Kamisaki Y., et al. Peroxisome proliferator-activated receptor gamma-mediated regulation of neural stem cell proliferation and differentiation. J. Biol. Chem. 2006;281:12673–12681. doi: 10.1074/jbc.M513786200. [DOI] [PubMed] [Google Scholar]
- 56.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 57.Logan C.Y., Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 58.Nusse R. Wnt signaling and stem cell control. Cell Res. 2008;18:523–527. doi: 10.1038/cr.2008.47. [DOI] [PubMed] [Google Scholar]
- 59.Davis L.A., Zur Nieden N.I. Mesodermal fate decisions of a stem cell: the Wnt switch. Cell Mol. Life Sci. 2008;65:2658–2674. doi: 10.1007/s00018-008-8042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farmer S.R. Regulation of PPARgamma activity during adipogenesis. Int. J. Obes. (Lond.) 2005;29(Suppl. 1):S13–S16. doi: 10.1038/sj.ijo.0802907. [DOI] [PubMed] [Google Scholar]
- 61.Prestwich T.C., Macdougald O.A. Wnt/beta-catenin signaling in adipogenesis and metabolism. Curr. Opin. Cell Biol. 2007;19:612–617. doi: 10.1016/j.ceb.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou H., Mak W., Zheng Y., Dunstan C.R., Seibel M.J. Osteoblasts directly control lineage commitment of mesenchymal progenitor cells through Wnt signaling. J. Biol. Chem. 2008;283:1936–1945. doi: 10.1074/jbc.M702687200. [DOI] [PubMed] [Google Scholar]
- 63.Sproul D., Gilbert N., Bickmore W.A. The role of chromatin structure in regulating the expression of clustered genes. Nat. Rev. Genet. 2005;6:775–781. doi: 10.1038/nrg1688. [DOI] [PubMed] [Google Scholar]
- 64.Ebisuya M., Yamamoto T., Nakajima M., Nishida E. Ripples from neighbouring transcription. Nat. Cell Biol. 2008;10:1106–1113. doi: 10.1038/ncb1771. [DOI] [PubMed] [Google Scholar]
- 65.Louro R., Nakaya H.I., Amaral P.P., Festa F., Sogayar M.C., da Silva A.M., Verjovski-Almeida S., Reis E.M. Androgen responsive intronic non-coding RNAs. BMC Biol. 2007;5:4. doi: 10.1186/1741-7007-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dinger M.E., Amaral P.P., Mercer T.R., Pang K.C., Bruce S.J., Gardiner B.B., Askarian-Amiri M.E., Ru K., Solda G., Simons C., et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 68.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 69.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.