Abstract
DNA nanotechnology exploits the programmable specificity afforded by base-pairing to produce self-assembling macromolecular objects of custom shape. For building megadalton-scale DNA nanostructures, a long ‘scaffold’ strand can be employed to template the assembly of hundreds of oligonucleotide ‘staple’ strands into a planar antiparallel array of cross-linked helices. We recently adapted this ‘scaffolded DNA origami’ method to producing 3D shapes formed as pleated layers of double helices constrained to a honeycomb lattice. However, completing the required design steps can be cumbersome and time-consuming. Here we present caDNAno, an open-source software package with a graphical user interface that aids in the design of DNA sequences for folding 3D honeycomb-pleated shapes A series of rectangular-block motifs were designed, assembled, and analyzed to identify a well-behaved motif that could serve as a building block for future studies. The use of caDNAno significantly reduces the effort required to design 3D DNA-origami structures. The software is available at http://cadnano.org/, along with example designs and video tutorials demonstrating their construction. The source code is released under the MIT license.
INTRODUCTION
In 1982, Nadrian Seeman laid the theoretical framework for the use of DNA as a nanoscale building material (1,2). Subsequently, DNA has been used in the construction of increasingly complex shapes and lattices (3–8). In 2006, Paul Rothemund introduced DNA origami, a versatile method for constructing arbitrary 2D shapes and patterns with dimensions of 100 nm in diameter and 6 nm spatial resolution (9). The method uses hundreds of short oligonucleotide ‘staple’ strands to direct the folding of a long, single ‘scaffold’ strand of DNA into a programmed arrangement.
Since its introduction, DNA origami has been used for applications such as label-free RNA-hybridization probes (10), seeds for algorithmic assembly (11,12), and liquid-crystalline alignment media for NMR structure determination of membrane proteins (13). Toward increasing the size of DNA-origami design space, we recently extended DNA origami to construction of 3D shapes (14). While implementing DNA-origami shapes, we found it useful to develop computer-aided design (CAD) software to minimize tedious and error-prone tasks; similar efforts have been reported previously for oligonucleotide-based DNA nanostructures (15) or for planar DNA origami similar to Rothemund's original designs (16).
Here we describe our open-source software package, caDNAno, for use in the design of 3D DNA-origami shapes constrained to a honeycomb framework (14). We have used caDNAno to generate seven DNA-origami designs of 3D rectangular blocks of varying cross-section dimensions. Analysis of the folded blocks by agarose-gel electrophoresis and negative-stain transmission electron microscopy revealed that a block design specifying six-helices-per-x-raster row yields the greatest fraction of defect-free objects.
METHODS
Folding and purification of DNA-origami shapes
Each sample was prepared by combining 20 nM scaffold (p7560 or p8064, derived from M13mp18), 100 nM of each staple oligonucleotide, buffer and salts including 5 mM Tris, 1 mM EDTA (pH 7.9 at 20°C), and 22 mM MgCl2, except for the 30-helix-per-x-raster block, which was folded with 15 mM MgCl2. Folding was carried out by rapid heat denaturation followed by slow cooling from 80 to 61°C over 80 min, then 60 to 24°C over 173 h. Samples were electrophoresed on 2% agarose gels (0.5× TBE, 11 mM MgCl2, 0.5 μg/ml ethidium bromide) at 70 V for 4 h in an ice-water bath. Leading monomer bands were visualized with ultraviolet light, physically excised, crushed with a pestle (17) and filtered through a cellulose-acetate spin column for 3 min at 15 000 × g, 4°C.
Negative stain electron microscopy
Purified samples were adsorbed for 5 min onto glow-discharged formvar- and carbon-coated copper grids, stained for 1 min with 2% uranyl formate, 25 mM NaOH, and visualized at 68 000× magnification with an FEI Tecnai T12 BioTWIN operating at 120 kV.
Chemicals and supplies
Sigma: EDTA, 2xYT Microbial Medium. Fisher Scientific: magnesium chloride, polyethylene glycol 8000 (PEG8000), sodium chloride (NaCl), Tris base, sodium hydroxide, potassium acetate, lauryl sulfate, glacial acetic acid. BD: LB broth, Bacto agar. Molecular BioProducts: 8-well PCR strip tubes. Invitrogen: agarose. Bio-Rad: Freeze ‘N Squeeze DNA gel-extraction spin columns. Kimble-Chase: pellet pestles. SPI: carbon/formvar copper grids, uranyl formate. Bioneer: RPC-purified deoxyribonucleotides.
Recombinant M13 filamentous bacteriophage construction
Recombinant phages were prepared by replacement of the BamHI-XbaI segment of M13mp18 by a PCR amplification fragment from a previously generated random sequence (18). Double-stranded (replicative form) bacteriophage M13 DNA bearing inserts were prepared as described (13). The inserts were verified by a double-restriction digest with BamHI and XbaI, followed by sequencing. The design: scaffold pairings are as follows: i: p8064, ii: p7560, iii: p8064, iv: p7560, v: p8064, vi: p7560, vii: p7560.
Gel-based yield estimation
ImageJ (http://rsb.info.nih.gov/ij/) was used for gel-image analysis. The percentage of scaffold that partitioned as a monomeric species was estimated as the background-subtracted integrated intensity value of a selection box enclosing the leading band of each lane divided by the background-subtracted integrated intensity value of a selection box enclosing the material from the well, down to the bottom of the leading band.
RESULTS AND DISCUSSION
In a fully occupied honeycomb lattice, each staple helix has three nearest neighbors (e.g. helices 1, 3, 7, 8, 9, 10, 14, 16 in Figure 1). Our default rules allow antiparallel crossovers between adjacent staple helices only where the strand backbones arrive at points of closest proximity, which repeat every 21 base pairs if the helical twist is fixed at 10.5 base pairs per turn. Thus for a given staple helix, potential staple-crossover positions occur every seven base pairs, or two-thirds of a turn. Our default rules allow antiparallel crossovers between adjacent scaffold helices to occur five base pairs, or half a turn, upstream or downstream of allowed crossover positions for the associated staple helices. However, caDNAno permits the user to force crossovers between any two staple bases or between any two scaffold bases. Users should take care when forcing crossovers, as departure from the default rules may lead to folding failure if too much deviation from canonical DNA geometry is implied.
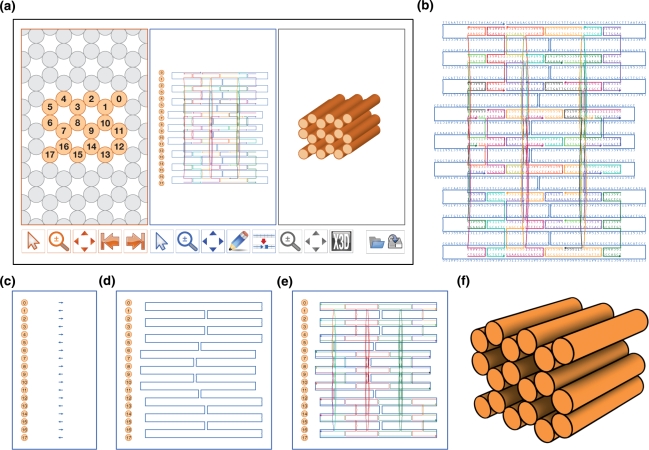
Figure 1.
caDNAno Interface and design pipeline. (a) Screenshot of caDNAno interface. Left, Slice panel displays a cross-sectional view of the honeycomb lattice where helices can be added to the design. Middle, Path panel provides an interface to edit an unrolled 2D schematic of the scaffold and staple paths. Right, Render panel provides a real-time 3D model of the design. (b) Exported SVG schematic of example design from a, with scaffold (blue) and staple (multi-color) sequences. (c) Path panel snapshot during first step of the design process. Short stretches of scaffold are inserted into the Path panel as helices are added via the Slice panel. (d) The Path panel editing tools are used to stitch together a continuous scaffold path. (e) The auto-staple button is used to generate a default set of continuous staple paths, including crossovers. The breakpoint tool is subsequently used to split the staple paths into lengths between 18 and 49 bases. Finally, the scaffold sequence is applied to generate the list of staple sequences. (f) Exported X3D model from the Render panel.
The design process has four main steps. First, a target shape is approximated by selecting a raster-style scaffold path that passes between neighboring helices along antiparallel crossovers at allowed positions. Second, staple paths complementary to scaffold are assigned. By default, all permitted staple crossovers are included, except for those that would be five base pairs away from a scaffold crossover between the same two helices. Third, the staple paths are broken into shorter segments 18 to 49 bases long, usually with a mean length of 30 to 35 bases. Finally, the scaffold path is populated with the DNA sequence of the desired template (e.g. 7–8 kb M13-genome-based vector), and the complementary staple sequences are determined.
This design pipeline is integrated from start to finish in the caDNAno three-panel interface (Figure 1a). The z-axis is defined as parallel to the helical axes. The Slice panel (orange border) provides an x–y cross-section view of the honeycomb helix lattice for any z-depth, with helices represented as circles. When the user clicks on an empty circle, that helix position is made available for routing of scaffold and staple strands by adding a schematic side view of the same helix to the Path panel (blue border). The Path panel is used for nucleotide-level editing of scaffold- and staple-path connectivity, assigning DNA sequences to scaffold paths, and reading out of staple DNA sequences. The Render panel (grey border) provides a real-time, 3D cylinder model for visualizing the shape as it is constructed. In each panel, pan and zoom tool buttons allow the user to view or edit the shape at different positions and magnifications. The Slice and Path panels have specialized tools for making additions, edits, rearrangements or deletions to a design (detailed descriptions of the tool buttons are found in the Supplementary Note 1). Completion of the design pipeline results in a list of staple DNA sequences corresponding to the schematics shown in each panel; the result also can be represented as a detailed SVG schematic (Figure 1b).
The process of approximating a 3D shape with a scaffold path begins with selection of helices in the Slice panel to approximate a 2D projection of that shape. When a helix is added to the design in the Slice panel, the same helix also is made active in the Path panel and is populated with a three-base-long scaffold path by default. Thus, once the desired helices are added to the design via the Slice panel (Figure 1a, orange panel), several short, disconnected scaffold paths are visible in the Path panel (Figure 1c). The Path-panel editing tools are used to extend the scaffold paths in the z-direction and to connect neighboring helices with Holliday-junction crossovers. The goal is to complete a continuous raster-style traversal of the target shape using a scaffold path (Figure 1d).
Once the scaffold path is complete, complementary staple paths are assigned by clicking the ‘Auto-staple’ tool button beneath the Path panel. Staple paths are created wherever scaffold is present, according to an algorithm that follows the aforementioned rules for crossover spacing (Figure 1e). Staple paths that fall outside the preferred length range (18–49 bases) are highlighted, and the user is responsible for using the editing tools to break the staple paths into shorter segments. After all staples are edited into a satisfactory arrangement, the scaffold path is populated with a DNA sequence using the ‘Add Sequence’ tool. Several default sequences are provided, or the user can input his or her own. Additionally, a 3D model can be exported in X3D format, with double helices represented as cylinders of 2 nm diameter and 0.34 nm per base-pair length (Figure 1f).
We used caDNAno to design seven different honeycomb-pleated-origami rectangular blocks (Figure 2a, top row), creating a simple scaffold-path trajectory that followed the same approximate path through each structure: as viewed down the helical axes, close-packing rows of helices were arrayed within the honeycomb framework in an x-raster pattern (i.e. left to right, then down, then right to left, then down, etc.); the connectivity of neighboring scaffold helices is more apparent in partially folded cylinder models (Figure 2b, top row). The x-raster rows within the honeycomb framework are corrugated; they stagger up and down and encompass helices that are actually at two different y-positions. Similarly, virtual y-oriented layers can be defined that stagger left and right and encompass helices that are at two different x-positions. The shapes were folded either from a 7560-base scaffold into 60 parallel helices or from an 8064-base scaffold into 64 parallel helices to create number-of-rows versus number-of-helices-per-x-raster-row combinations of 15 × 4, 10 × 6 (analyzed independently in ref. 14), 8 × 8, 6 × 10, 4 × 16, 3 × 20, 2 × 30. Each helix was allotted 126 bases of scaffold. Of those 126 bases, 98 were paired with complementary staples, and the remaining 28 bases were divided into front and rear unpaired loop fragments at the ends of each helix (detailed schematics and staple lists are included in Supplementary Notes 2 and 3, respectively).
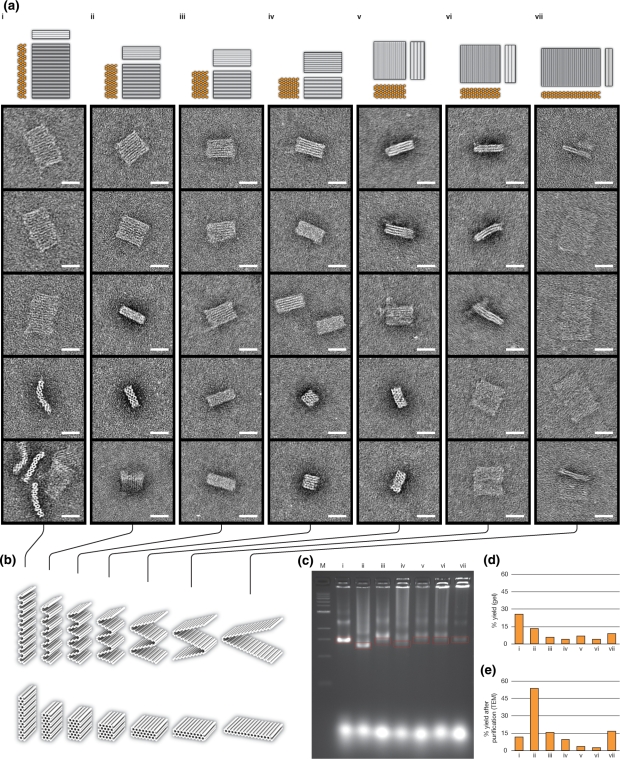
Figure 2.
Transmission electron microscopy (TEM) and agarose-gel analysis of DNA-origami blocks. The nomenclature of the designs is m × n, where m is the number of x-raster rows, and n is the number of helices per x-raster row. (i), 15 × 4 motif; (ii) 10 × 6 motif; (iii) 8 × 8 motif; (iv) 6 × 10 motif; (v) 4 × 16 motif; (vi) 3 × 20 motif; (vii) 2 × 30 motif. (a) Cylinder-model projections and transmission-electron micrographs for rectangular-block designs. (b) Partially folded models, which do not represent the actual folding pathway, are displayed above fully folded models. Scaffold crossovers only occur between helices that are neighbors in the partially folded models. Thus, these models capture an important feature of the design: the path of the scaffold stays within a 2D surface. (c) Agarose-gel analysis of folding of blocks. Marker is a 1 kb ladder. Red boxes indicate the region of each lane that was counted as the fastest-migrating monomeric species for yield estimates in option d and that was physically extracted from the gel during purification before TEM imaging. The 6 × 10 design displays the fastest gel mobility. (d) Fraction of scaffold incorporated into fastest-migrating monomeric species, as estimated by ethidium-bromide-fluorescence intensity. (e) Fraction of well-folded species after gel purification, as estimated by image analysis of 100 randomly selected particles for each shape. Scale bars: 25 nm.
Each of the shapes was folded in separate chambers by heat denaturation followed by cooling for renaturation, and analyzed by agarose-gel electrophoresis (Figure 2c). The seven shapes varied significantly in leading band yield, mobility, and sharpness, as well as amount of undesired formation of higher-order aggregates. We estimated folding yields as integrated intensity of material that migrated as a leading band divided by total intensity of material in the lane up to and including the well (Figure 2d). Material in each of the leading bands was isolated by physical extraction and analyzed by negative-stain transmission electron microscopy (Figure 2a). For each shape, 100 randomly selected individual-particle images were collected, and folding yields were estimated (Figure 2e). A particle was judged to be well-folded if its outline could be aligned with a semi-transparent projection model of the corresponding design and it exhibited no obvious defects such as missing, broken, disrupted, or smeared out sections more than 3 nm away from the unpaired scaffold loops at the front and rear interfaces. For example, of the five particle images shown for the 4 × 16 design in Figure 2a(v), only the topmost particle was counted as well-folded.
Only folding with three of the seven designs—four-helix-per-x-raster or 15 × 4 (two y-layers), six-helix-per-x-raster or 10 × 6 (three y-layers), thirty-helix-per-x-raster or 2 × 30 (two x-layers)—produced sharp leading monomer bands by agarose-gel electrophoresis (Figure 2c). Thus designs with a smaller number of x-layers or y-layers may have a folding advantage due to fewer numbers of highly embedded helices, which may be more difficult to assemble, and perhaps also due to the lower crossover densities. Consistent with this trend, single-layer shapes fold much faster and to higher yield (9). Folding with the six-helix-per-x-raster (10 × 6) design produced the leading band with the greatest mobility, while folding with the four-helix-per-x-raster (15 × 4) design produced the leading band with the greatest intensity, indicating the best yield. Our previous results suggested that faster gel mobility of the same design under different folding conditions correlates with fewer defects (14), although it is more difficult to interpret mobility differences across designs with inherently different shapes.
The six-helix-per-x-raster (10 × 6) shape appeared the most robust of the seven designs in terms of yielding particles that are intact after folding, staining, and drying (Figure 2e). We also have found that this six-helix-x-raster design performs well when used to construct shapes with as few as three x-raster rows (i.e. 18 helices total) and longer lengths of helices (data not shown). Interestingly, the 15 × 4 and 2 × 30 designs produced particles that appeared bent when adhering to the grid surface with a perpendicular orientation of the helical axes; it is possible that the positively charged stain is deforming these particles, but that the other designs produce particles that are sufficiently thick to resist such deformation. Thinner objects such as the 15 × 4 and 2 × 30 designs might be suitable for some applications if a staining artifact is the cause of the observed deformations. Further studies will be necessary for optimizing design parameters that might affect folding yield, such as staple-break-point distribution, scaffold routing, and scaffold- versus staple-crossover densities (19).
The construction of complex, 3D DNA nanostructures will increase the range of applications that can be addressed, but also will add complexity to the design process. By restricting design space to the honeycomb-lattice framework, we reduce the number of choices that need to be made when implementing a 3D DNA-origami shape while retaining a significant amount of flexibility. Our caDNAno software package relieves the user from completing the tedious conversion of a creative design to oligonucleotide sequences. We have found that caDNAno performs favorably when compared to ad hoc methods for generating staple sequences for a new shape design, typically reducing the time required for monotonous sequence assignment from days or weeks down to a few hours.
In addition to supporting the design of basic shapes such as rectangular blocks, caDNAno provides tools to introduce deviations from the basic honeycomb architecture, such as forced crossovers, to create very complicated designs. Additional software development will be required to make designs of these non-standard motifs more natural, for example for caDNAno to predict the structural consequences of these changes. More work is also needed to see what design rules lead to stable structures; for examples of designs that folded successfully, although with varying yields, see the gallery section at http://cadnano.org/.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [grant number 1DP2OD004641-01]; the Claudia Adams Barr Program in Innovative Cancer Research. Funding for open access charge: National Institutes of Health Grant 1DP2OD004641-01.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Hendrik Dietz, Tim Liedl, Björn Högberg, Katrina Galkina, Andrew Leifer and Nathan Derr for feature suggestions and beta-testing. We thank Xingping Su for cloning the p7560 and p8064 scaffold vectors. We thank Alexander Wait for providing server support. This work was supported by Claudia Adams Barr Program Investigator and NIH New Investigator awards to W.M.S.
REFERENCES
- 1.Seeman N.C. Nucleic acid junctions and lattices. J. Theor. Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 2.Seeman N.C. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 3.Chen J.H., Seeman N.C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 4.Fu T.J., Seeman N.C. DNA double-crossover molecules. Biochemistry. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 5.Li X.J., Yang X.P., Qi J., Seeman N.C. Antiparallel DNA double crossover molecules as components for nanoconstruction. J. Am. Chem. Soc. 1996;118:6131–6140. [Google Scholar]
- 6.Winfree E., Liu F., Wenzler L.A., Seeman N.C. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 7.Shih W. M., Quispe J. D., Joyce G. F. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 8.He Y., Ye T., Su M., Zhang C., Ribbe A.E., Jiang W., Mao C. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 9.Rothemund P.W. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 10.Ke Y., Lindsay S., Chang Y., Liu Y., Yan H. Self-assembled water-soluble nucleic acid probe tiles for label-free RNA hybridization assays. Science. 2008;319:180–183. doi: 10.1126/science.1150082. [DOI] [PubMed] [Google Scholar]
- 11.Fujibayashi K., Hariadi R., Park S.H., Winfree E., Murata S. Toward reliable algorithmic self-assembly of DNA tiles: a fixed-width cellular automaton pattern. Nano Lett. 2008;8:1791–1797. doi: 10.1021/nl0722830. [DOI] [PubMed] [Google Scholar]
- 12.Barish R.D., Schulman R., Rothemund P.W.K., Winfree E. An information-bearing seed for nucleating algorithmic self-assembly. Proc. Natl Acad. Sci. USA. 2009;106:6054–6059. doi: 10.1073/pnas.0808736106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Douglas S.M., Chou J.J., Shih W.M. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Natl Acad. Sci. USA. 2007;104:6644–6648. doi: 10.1073/pnas.0700930104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douglas S.M., Dietz H., Liedl T., Högberg B., Graf F., Shih W.M. Self-assembly of DNA into nanoscale three dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birac J.J., Sherman W.B., Kopatsch J., Constantinou P.E., Seeman N.C. Architecture with GIDEON, a program for design in structural DNA nanotechnology. J. Mol. Graph. Model. 2006;25:470–480. doi: 10.1016/j.jmgm.2006.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen E.S., Dong M., Nielsen M.M., Jahn K., Lind-Thomsen A., Mamdouh W., Gothelf K.V., Besenbacher F., Kjems J. DNA origami design of dolphin-shaped structures with flexible tails. ACS Nano. 2008;2:1213–1218. doi: 10.1021/nn800215j. [DOI] [PubMed] [Google Scholar]
- 17.Kurien B.T., Kaufman K.M., Harley J.B., Scofield R.H. Pellet pestle homogenization of agarose gel slices at 45°C for deoxyribonucleic acid extraction. Anal. Biochem. 2001;296:162–166. doi: 10.1006/abio.2001.5299. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J., Russell D. Molecular Cloning: A Laboratory Manual, 3rd. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 19.Jungmann R., Liedl T., Sobey T.L., Shih W., Simmel F.C. Isothermal assembly of DNA origami structures using denaturing agents. J. Am. Chem. Soc. 2008;130:10062–10063. doi: 10.1021/ja8030196. [DOI] [PubMed] [Google Scholar]