Abstract
The ardA gene, found in many prokaryotes including important pathogenic species, allows associated mobile genetic elements to evade the ubiquitous Type I DNA restriction systems and thereby assist the spread of resistance genes in bacterial populations. As such, ardA contributes to a major healthcare problem. We have solved the structure of the ArdA protein from the conjugative transposon Tn916 and find that it has a novel extremely elongated curved cylindrical structure with defined helical grooves. The high density of aspartate and glutamate residues on the surface follow a helical pattern and the whole protein mimics a 42-base pair stretch of B-form DNA making ArdA by far the largest DNA mimic known. Each monomer of this dimeric structure comprises three alpha–beta domains, each with a different fold. These domains have the same fold as previously determined proteins possessing entirely different functions. This DNA mimicry explains how ArdA can bind and inhibit the Type I restriction enzymes and we demonstrate that 6 different ardA from pathogenic bacteria can function in Escherichia coli hosting a range of different Type I restriction systems.
INTRODUCTION
Sequence analysis of bacterial genomes has demonstrated that horizontal gene transfer (HGT) is a fundamental mechanism for driving diversity and evolution. The transmission of DNA to bacterial cells that are not direct descendants of the donor is often achieved via mobile genetic elements such as bacteriophage, plasmids and conjugative transposons. Mobilization of these elements can lead to the spread of antimicrobial resistance in clinical environments and in the wider community (1–5). Human morbidity and mortality as a result of this trend is mirrored by the economic cost of, for example, the spread of herbicide resistance genes in crop pathogens (6).
Over 50% of eubacteria and archaea contain the genes for one or more of the four classes of known DNA restriction and restriction-modification (RM) systems (7–10). In laboratory experiments, it is clear that the function of these RM systems is to protect the host cell from invasion by foreign DNA. RM systems work by recognizing specific DNA sequences and triggering an endonuclease activity which rapidly cleaves the foreign DNA allowing facile destruction by exonucleases.
Despite the demonstrated efficacy of the RM systems, genome analysis of pathogenic bacteria from both clinical and environment settings make it abundantly clear that HGT by transduction, transformation and conjugation is extremely common. The process is not only widespread within species but also between species where RM systems are operative. It is established that HGT is directly responsible for the spread of resistance genes (11). It is therefore important in understanding and tackling antibiotic resistance to ascertain the mechanism by which HGT circumvents such an apparently effective defence. One most likely explanation has come from the identification of potential anti-RM genes within mobile genetic elements such as phage, plasmids and transposons (12,13). These anti-RM systems seem to have been acquired and maintained by the host organism and their occasional activation of such genes weakens or negates the RM defence system allowing further HGT (14,15).
Characterized anti-RM systems include the gene 0.3 protein, Ocr, of phage T7 (16–20) and the ArdA and ArdB proteins very commonly found on conjugative plasmids and transposons in a large range of prokaryotes (13,21–27). The Ocr protein is relatively restricted in its distribution (28,29) but is the most fully characterized of anti-RM proteins. Structural analysis suggests it to be a structural mimic of DNA and it, similar to most anti-RM systems characterized thus far, targets the Type I RM systems (17,18,30). Other mechanisms described for evading host RM systems include loss of DNA target sequences, modification of the DNA, proteolysis of RM systems and the hydrolysis of RM cofactors and have been extensively reviewed (10,12).
Type I RM systems are a very widespread and effective defence system against foreign DNA (31) perhaps explaining their selection as targets for anti-RM systems. Biochemically characterized Type I RM enzymes comprise two restriction (HsdR, R) subunits, two modification (HsdM, M) subunits and one DNA sequence specificity (HsdS, S) subunit in a single ∼440-kDa complex (32,33). The methyltransferase (MTase) core M2S1 modifies hemimethylated target ‘specificity’ sequences and triggers the R subunits to act when unmethylated targets are recognized. The restriction reaction involves extensive ATP hydrolysis to drive DNA translocation by the helicase motifs in each R subunit until an endonuclease activity is activated at a site on the DNA molecule distant from the initial specificity sequence. These RM systems can be grouped into different families, IA to IE so far, defined by subunit complementation in vivo, DNA hybridization, antibody cross reactivity and, to some extent, amino acid sequence comparison (34,35).
Despite the wide distribution of ardA genes in many important pathogens (24), there is very little biochemical data on their mode of action and behaviour in solution (26,27) and no atomic structure. We now demonstrate that ArdA from a number of pathogens is very effective against the archetypal Type I RM systems of Escherichia coli. We also report the first structure of an ArdA protein and its implications for the structure of Type I RM enzymes. ArdA forms an extremely elongated molecule with a highly charged surface. Its structure is reminiscent of ∼42 bp of B-form DNA making it the largest DNA mimic yet characterized. It has already been demonstrated that ArdA binds to the core MTase of a Type I RM enzyme (26,27). The ArdA structure allows a rationalization of this behaviour.
MATERIALS AND METHODS
Escherichia coli strains
JM109 was used as a general cloning strain (Promega Madison, WI, USA). The expression strain BL21(DE3) was purchased from Invitrogen (Groningen, The Netherlands). NM1261 (restriction and modification negative; r–m–), NM1049 (EcoKI RM system, Type IA), NK354 (EcoAI RM system, Type IB), NK402 (EcoR124I RM system, Type IC) and NM1009 (StySBLI RM system, Type ID) were a kind gift of Professor Noreen E. Murray (School of Biology, University of Edinburgh, UK).
The NK and NM strains were converted to DE3 lysogens using the λDE3 lysogenization kit (Invitrogen) with some modifications. Where necessary, the Selection phage provided in the kit was replaced by NM848 (h82 imm21 cI, a kind gift from Noreen Murray) which possesses the appropriate host range. Integration of λDE3 was verified by assessing their ability to support growth of the T7 Tester phage (supplied in the kit).
Genes for production of ArdA protein
Four ardA genes were identified in the public sequence database held by the National Center for Biotechnology Information (Bethesda, MD, USA) (Table 1). The gene sequences were then synthesized by GeneArt (Regensburg, Germany) using an optimized codon usage pattern for expression in E. coli. To facilitate cloning, an NdeI restriction site was engineered to overlap the ATG start codon of each synthetic gene and a HindIII site was also included 6 bp downstream of the stop codon. The synthesized genes were ligated into pET24a at the NdeI and HindIII sites of the expression vector. The anticipated sequence was subsequently verified by DNA sequencing on both strands. Two further ardA genes from transposon Tn916 (open reading frame 18 in Tn916) and Bacteroides fragilis NCTC9343 (open reading frame BF1222) (25,36) were amplified and ligated into pTrc99a (37) as previously described (27). Amino acid substitutions were constructed by site-directed mutagenesis using a QuikChange kit (Stratagene, La Jolla, CA, USA) as per the manufacturer's instructions. Substitutions were confirmed by sequencing.
Table 1.
The source of the ardA genes investigated
| Reference | Organism | Gene ID | Amino acid length | Predicted pI | Optimal protein expression temperature,°C |
|---|---|---|---|---|---|
| 25 | Enterococcus faecalis | Tn916 (ORF18) | 165 | 3.91 | 37 |
| 36 | Enterococcus faecalis V583 | EF2335 | 166 | 3.91 | 30 |
| 37 | Staphylococcus aureus Mu50 | SAV0405 | 166 | 3.92 | 30 |
| 38 | Clostridium difficile 630 | CD0376 | 167 | 3.78 | 37 |
| 39 | Streptococcus agalactiae 2603V/R | SAG2011 | 160 | 4.05 | 30 |
| 40 | Bacteroides fragilis NCTC 9343 | BF1222 | 177 | 3.98 | 37 |
Escherichia coli BL21(DE3) were transformed with the pET24a or pTrc99a-based expression constructs and plated onto LB agar containing 100 μg/ml kanamycin or 100 μg/ml carbenicillin, respectively. Single colonies were picked and grown in LB medium supplemented with 100 μg/ml of the appropriate antibiotic to an OD600 of 0.5. Heterologous gene expression was induced by addition of IPTG to a final concentration of 1 mM. Growth was continued for up to 4 h before harvesting. The amount of recombinant protein was initially assessed by SDS–PAGE (4–15% gradient gel) analysis. In each case, a prominent band of the anticipated molecular mass was clearly visible in the induced cell extract, which was absent from the control experiments conducted using the corresponding plasmid vector without insert (data not shown). In order to purify the recombinant proteins, cells were grown in 10-l cultures and harvested 4 h post IPTG induction. Typically we obtained ∼40 g wet cell paste which was stored at –20°C until required.
Assessment of in vivo activity of ArdA
The methods employed for assaying ArdA activity in vivo used the efficiency of plating of phage lambda (virulent) λv on the various strains of E. coli have been previously described (27). All assays were performed in triplicate and at least 50 phage plaques per plate per experiment were counted. Experiments were performed on numerous days with fresh samples and control experiments performed each day. Little variation was observed during the replicate experiments. The standard deviations for the anti-restriction and anti-modification results are ±25% or less depending on the particular ardA investigated (Supplementary Table 1). The previously published results (27) for the activity of ardA from Tn916 are included here for completeness. Anti-restriction was defined as the efficiency of plating (eop) obtained with strains containing the plasmid expressing the ardA gene divided by the eop obtained from strains containing the vector plasmid. The degree of anti-modification was assessed by determining the eop of each phage stock on the restriction proficient strain relative to the non-restricting strain. Anti-modification was defined as 1/(eop).
Structural methods
The ArdA protein from transposon Tn916 was purified as described previously (27). Purified protein was concentrated to 15 mg/ml in 10 mM Tris, pH 7.5, 150 mM NaCl and 2 mM DTT in preparation for crystallization. Initial protein crystals were attained after screening purified ArdA against 384 different precipitants in 0.1 μl protein + 0.1 μl precipitant hanging drops set up on the Hamilton-Rhombix-Thermo integrated crystallization system at 293 K. Initial crystallization conditions were stochastically optimized and optimum crystals were found to grow in a precipitant range between 2.27 and 2.31 mM Na-K2-phosphate, 0.1 M sodium acetate, pH 4.0 at 293 K. To phase the structure a single ArdA crystal was soaked in 50 mM dipotassium tetrachloroplatinate (II) for ∼2 min before being back soaked in 2.4 mM Na-K2-phosphate, 0.1 M sodium acetate, pH 4.0, 25% ethylene glycol which acted as cryoprotectant. Native and derivative data were collected at beamline ID-29 at the ESRF, Grenoble, France. All data were processed with XDS (38), scaled in XSCALE and converted to scalepack format with XPREPX. Heavy atom sites (6 × Pt) were located using SHELXD from the SHELXC/D/E (39) suite of programs using native and derivative data with anomalous signal (SIRAS) to 3.5 Å. Located sites were entered into SOLVE/RESOLVE (40,41) and 2-fold averaging was automatically detected. Electron density maps were produced at 3.5 Å and a partial model built. The Cα coordinates from this model were used to automatically detect and employ 4-fold averaging at 3 Å in RESOLVE after which the asymmetric unit model was built by hand. The structure was refined using REFMAC5 (42). TLS parameters were introduced for each domain and non-crystallographic restraints applied throughout the refinement. XFIT (43) was used for manual model building. The quality of the structure was checked with MOLPROBITY (44). The final refined structure and data have been deposited with PDB accession code 2w82. Table 2 shows the data collection and refinement details.
Table 2.
The X-ray crystallography details (numbers in brackets refer to the highest-resolution shell)
| Data collection | ||
|---|---|---|
| Native | Platinum derivative | |
| Wavelength | ||
| Resolution (Å) | 12.5–2.8 (2.87–2.8) | 17.72–3.29 (3.38–3.29) |
| Space group | P212121 | P212121 |
| Temperature | 100K | 100K |
| Cell dimensions a, b, c (Å) | a = 63.8 b = 103.4 c = 173.0 | a = 63.8 b = 104.5 c = 171.5 |
| Vm(Å3/Da) | 3.73 | 3.74 |
| Solvent (4 mol/asu%) | 67 | 67 |
| Total number reflections | 68 208 | 140 499 |
| Unique reflections | 27 101 | 33 192 |
| I/σI | 12.6 (1.8) | 9.31 (2.31) |
| Redundancy | 2.5 (2.5) | 4.2 (4.1) |
| Completeness (%) | 93.3 (93.6) | 99.2 (95.4) |
| Anomalous correlation (%) | 40 | |
| Rmerge | 6.4 (56.5) | 10.6 (65.3) |
| Refinement | ||
| Rwork/Rfree | 20.7 (30.9)/24.9 (33.8) | |
| No. atoms | ||
| Protein | 10 203 | |
| Water | 51 | |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.006 | |
| Bond angles (°) | 0.817 | |
Phylogenetic analysis
Ninety-eight sequences of ArdA homologues were derived from the Pfam family PF07275 (45). A distance matrix was generated by Protdist (bootstrap analysis used a random seed number of 3 with 100 replicates) and analysed using Quicktree (46) before output in PhyloWidget (47).
RESULTS
Distribution of ArdA in sequenced genomes
Genes encoding putative ArdA homologues are widespread within the bacterial kingdom and can be identified by sequence homology in the Actinobacteria, the Cyanobacteria, the Proteobacteria and the Firmicutes (data not shown). This is consistent with the known promiscuity of conjugative plasmids and conjugative transposons such as Tn916. Interestingly the genus Bacillus does not, as yet, appear to contain any identifiable homologues of ArdA.
ArdA activity in vivo
Previously, in vivo assays have been performed for a few ArdA proteins from conjugative plasmids specific for E. coli against the IA, B and C families of Type I RM systems (13,21–24,26,48,49). Similar anti-restriction and anti-modification activities of the Ocr protein have been determined against the IA to ID families (34,35). Ocr was found to act as an anti-restriction protein against all four families but was only effective as an anti-modification protein against the IA and ID families (16,19,30).
We have tested the in vivo activity of ArdA proteins from six bacterial species (25,50–53,36) (Table 1) against representatives of the four major Type I RM families (Table 3, Supplementary Table 1). We arbitrarily define that the anti-restriction and anti-modification activities should have a value >2 for the ArdA protein to be considered truly active in vivo. The effectiveness varies greatly between different ArdA proteins and between different Type I RM families. All of the ArdA proteins were active as both anti-restriction and anti-modification proteins against the IA family, the IB family and the ID family. ArdA showed anti-restriction activity against the IC family, although the anti-modification activity of the ArdA from S. agalactiae and from B. fragilis was unmeasurable against the IC family. Despite these variations, the fact that all the ArdA were active in blocking restriction to a measurable extent in an unnatural host without induction of high levels of protein expression suggests that these ArdA will function efficiently as anti-restriction proteins if expressed in their normal hosts and be capable of modulating HGT.
Table 3.
Effect of expression of 6 different ardA on the restriction and modification of bacteriophage λv by 4 different Type I RM systems, each representing one of the 4 families (IA to ID) of Type I systems
| Source organism | Gene expressed in host E. coli | Chromosomal restriction system in host E. coli |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| NM1049 Type IA EcoKI |
NK354 Type IB EcoAI |
NK402 Type IC EcoR124I |
NM1009 Type ID StySBLI |
||||||
| Anti-restriction | Anti-modification | Anti-restriction | Anti-modification | Anti-restriction | Anti-modification | Anti-restriction | Anti-modification | ||
| Enterococcus faecalis | Tn916 (ORF18) | 30 000 | 13 | 130 | 17 | 1500 | 390 | 1700 | 4200 |
| Enterococcus faecalis V583 | EF2335 | 6200 | 12 | 100 | 11 | 430 | 3.4 | 14 000 | 160 |
| Staphylococcus aureus Mu50 | SAV0405 | 4600 | 4.8 | 100 | 2.8 | 430 | 6600 | 11 000 | 200 |
| Clostridium difficile 630 | CD0376 | 5000 | 360 | 100 | 60 | 1900 | 2.5 | 8500 | 52 |
| Streptococcus agalactiae 2603V/R | SAG2011 | 3400 | 11 | 40 | 2.6 | 6.9 | 1.2 | 12 000 | 1600 |
| Bacteroides fragilis NCTC 9343 | BF1222 | 68 000 | 10 000 | 140 | 13 | 1200 | 1.0 | 2100 | 1500 |
| Plasmid vector alone | None | 1.0 | 0.9 | 1.0 | 1.7 | 1.0 | 1.7 | 1.0 | 1.1 |
The host E. coli contained a chromosomal copy of the RM system and a plasmid expressing ardA at low level. An anti-restriction or anti-modification value >2 was taken to indicate ArdA activity in vivo and the standard deviation in these values is no more than +/– 25%.
ArdA crystal structure
The monomer structure
The asymmetric unit contains four monomers of the protein forming two independent dimers. Each monomer can be decomposed into three domains: the N-terminal domain 1 (residues 3–61), the central domain 2 (62–103) and the C-terminal domain 3 (residues 104–165) (Figure 1). The three domains of the ArdA monomer are arranged in an approximately linear manner giving a very elongated molecular shape (70 Å × 20 Å) with a definite curve. The arrangement of the three domains has no counterpart in any known structure. As a consequence of the elongated arrangement, only eight residues are buried from solvent. In effect the protein lacks the conventional hydrophobic core. Superimposing the four monomers in the asymmetric unit reveals very small variations in the precise arrangement of the domains with respect to each other despite the limited contact area between the domains.
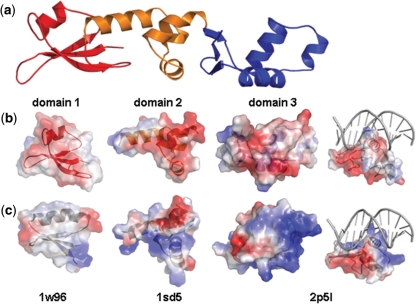
Figure 1.
Domains in the Tn916 Orf18 ArdA monomer. (a) The N-terminal domain 1 is red, the central domain 2 is orange and the C-terminal domain is blue. (b) An electrostatic representation of each ArdA domain demonstrating their negatively charged surface. The panel on the extreme right has DNA from the structure 2p5l (DNA bound to the winged helix–turn–helix domain) modelled on to the ArdA domain 3. As can be seen, the charges on ArdA are not consistent with binding of DNA despite its fold being similar to that of 2p51. (c) Electrostatic representations of domains homologous to ArdA, including the DNA binding motif of 2p5l. Although the overall folds of the biotin carboxylase B domain (1w96), the ANTAR domain (1sd5) and the winged helix–turn–helix domain (2p51) are similar to those in ArdA, the charge distributions are very different.
The N-terminal domain consists of a three-stranded anti-parallel β sheet and one short α helix interspersed with three large loops of 10 or more residues. The arrangement of the secondary structure elements is reminiscent of, but distinct from, the B domain in biotin carboxylase (54) (PDB 1w96), residues 248–293, from yeast (rmsd 2.1 Å over 36 residues). The central domain of ArdA is a four-helix bundle and shows weak similarity to the anti-termination ANTAR domain found in RNA-binding proteins (55). A superposition of the central domain with the closest structural match (PDB 1sd5) has rmsd of 3.2 Å for 40 Cα atoms. The N-terminal and central domain pack against each other burying around 1700 Å2 of surface area. The junction of the two domains creates a cleft with approximate dimensions 15 × 10 Å, which is partially occluded by the 12-residue loop connecting the two domains. The C-terminal domain has a three-stranded β sheet and three α helices packed together in a manner that creates a groove in the structure ∼11 Å wide. The C-terminal residue of the protein (Y165) is at the very end of the middle β strand. The helical arrangement in the C-terminal domain contains a component of the winged helix–turn–helix motif. Superposition of the C-terminal domain with arginine repressor protein (56) (PDB code 2p5l) reveals 42 Cα atoms superimpose with an rmsd of 2.1 Å. The arrangement of domains 2 and 3 occludes only 600 Å2 and no cleft is observed at the juncture of domains 2 and 3.
Although domains 2 and 3 from ArdA have folds reminiscent of nucleotide binding proteins, a comparison of the electrostatic surface of ArdA shows these domains have a profoundly negative potential (the pI of ArdA is 4), the opposite of the profoundly positive potential on nucleotide binding proteins. This reversal in surface potential would seem to rule out ArdA as a DNA- (or RNA)-binding protein. It remains an open question whether the similarity is due to evolutionary divergence or simply that the robust structural frameworks of DNA-binding proteins and ArdA have been arrived at by convergent evolution.
The dimer structure and its mimicry of DNA
Analysis of the structure with the program PISA (57) identifies a head-to-head dimeric arrangement as the functional biological unit (Figure 2a and b). The presence of two such independent dimers in the asymmetric unit is additional evidence for the biological significance of the arrangement. A small α-helix in one monomer (residues L11 to E16) is replaced with a short loop in the other subunit. The dimer, like the monomer, is highly elongated and curved. The chord that connects the extreme ends has a length of 140 Å. There is no evidence in the crystal structure for higher order oligomers such as those observed for a DNA mimic encoded by a eukaryotic virus (58). This would appear to be inconsistent with our previous gel filtration analysis in which the molecular weight of the protein increased on going from low concentration (∼0.1 μM) to high concentration (∼10 μM) (27). The chromatography calibration relied upon globular protein standards. As the structure of ArdA is now revealed to be highly elongated such a calibration is not valid and the gel filtration data, originally interpreted as a change from a dimer at low concentration to possibly a hexamer at high concentration, should now perhaps be interpreted as a change from monomer to dimer. This would be consistent with the observation (27) that the change in molecular weight as a function of concentration followed a simple one step binding equilibrium equation. The high protein concentration used for crystallization is near the upper limit of the concentration range studied by the gel filtration experiment and the crystallization sample eluted as a single peak as expected (27).
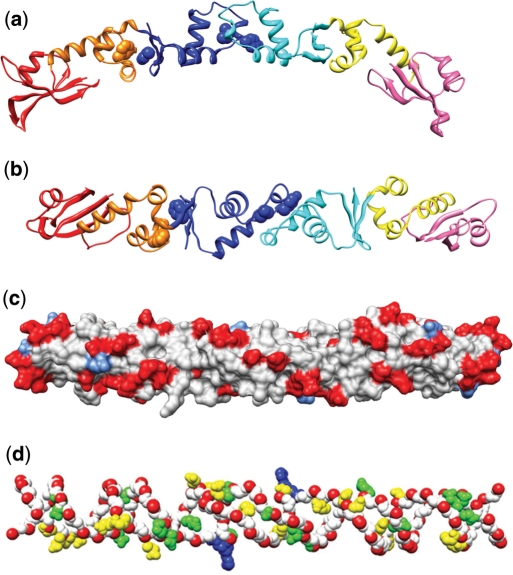
Figure 2.
The dimeric structure of ArdA and its distribution of negative charge clearly show mimicry of the DNA double helix. (a) The ArdA dimer with domain 1 coloured red and salmon, domain 2 coloured orange and yellow and domain 3 coloured blue and cyan. Residues F91, Y137, I138 and F161 are shown space-filled. F91 is orange, Y137 and I138 are blue and located at the dimer interface and F161 is blue and at the interface with domain 2. (b) The ArdA dimer rotated 90° from (a) looking at the convex face. The residues F91, Y137, I138 and F161 are highlighted as above. (c) The surface of ArdA in the same orientation as in (b) with acidic residues coloured red and basic residues coloured light blue. The negatively charged residues form a helical pattern across the surface. (d) An overlay of a selection of the acidic residues in the dimer, viewed from the same angle as in (b) and (c), onto the DNA duplex from the model structure of the MTase bound to DNA (32). Only the phosphate backbone of DNA is shown (red and white but with the nucleotides flipped out by the MTase for methylation shown in blue) with acidic residues from ArdA matching one strand shown in yellow and matching the other strand shown in green.
The two ArdA dimers in the unit cell when compared to each other show minor variation along the molecular axis consistent with a flexible dimer interface. The highly elongated dimer has a very large solvent surface area of 20 000 Å2. Examining the protein surface with a 1.5-Å probe shows a wide and narrow groove running the length of the structure, akin to the major groove in DNA. In addition to the general negative potential of the surface, there are two ribbons of negative charge on a raised surface that entwine around the entire length of the molecule (Figure 2c). This striking pattern is reminiscent of the phosphate backbone of a polynucleotide. Many of the carboxyl groups can be superimposed upon the DNA (32) structure derived from the structural model of a Type I MTase core bound to DNA (Figure 2d). The pattern of negative charge even extends across the dimer interface through the conserved residues D109, D111, D112, D115, E122, E123 and E129. This distribution and conservation of charged residues is evidence for the necessity of dimer formation for protein function and suggests that ArdA across all species will have similar structural requirements.
The residues, F91, Y137, I138 and F161 were previously identified as essential (26). The crystallographic data suggest their role is not functional per se but rather they maintain the fold of the protein at the dimer interface (Y137 and I138) and at the interface of domains 2 and 3 (F91 and F161) (Figure 2a and b).
The essential VF-motif (I160 F161 in the Tn916 ArdA protein) is located in the domain 3 of ArdA (26) and is part of the middle β-strand in this domain. Removal of these residues (26) would cause the β-sheet to collapse destroying the structural integrity of domain 3 and the interface with domain 2. F161 and I163, on the interface between domains 2 and 3, contact E79, L80, E83 and F91 (another essential residue) in domain 2.
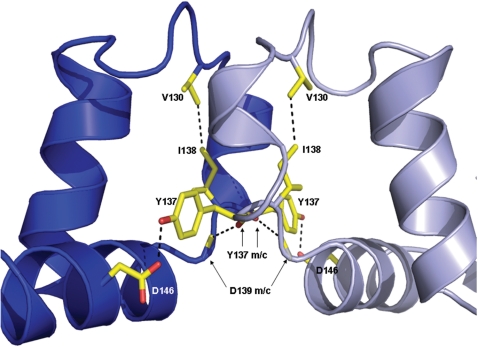
The dimer interface (Figure 3) contains the anti-restriction motif (amino acids 126–140 in the Tn916 ArdA protein) identified by Belogurov and Delver (59) and the two essential residues Y137 and I138 identified by molecular genetics (26). The structure makes it clear that this motif serves a structural purpose rather than being involved directly in the anti-restriction activity. The dimer buries just under 600 Å2 per monomer (<6% of the total surface area) but is predominantly hydrophobic with L127, L134 and Y137 providing the bulk of the contacts. There are four hydrogen bonds; two involving main chain to main chain contacts (Y137 to D139) and two side chain to side chain contacts (Y137 to D146). Y137 and I138 are at the core of the dimer interface with Y137 making two H-bond contacts with D139 and D146 of the opposing monomer and I138 acting to stabilize the loop regions either side of the interface helix through interaction with V130 on the same monomer.
Figure 3.
The ArdA dimer interface showing the residues involved in H-bonding (m/c indicates a main chain H-bond).
The structure of ArdA compared to other ArdA sequences
Figure 4 shows an alignment of the amino acid sequences of the ArdA investigated in this study and by others (13,21–27). As can be seen, there is considerable variability in sequence in this set but the pattern of charged residues and the two motifs identified previously (26,59) are well conserved, indicating that all ArdA are very likely to have the same structure and to operate in the same manner. Biochemical data on the ArdA listed in Table 1 (Roberts,G.A. and Sanghvi,B., unpublished data) when compared to that from Tn916 ArdA (27), support this assertion.
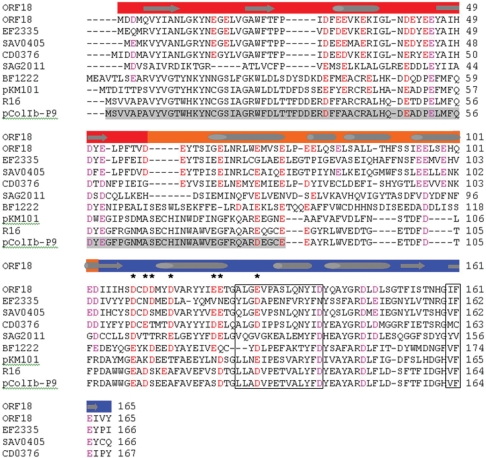
Figure 4.
CLUSTALW multiple sequence alignment of ArdA proteins in Table 1 plus ArdA from several previously studied conjugative plasmids from different incompatibility groups. The colours above each residue in the sequence alignment indicate the domain of the Tn916 Orf18 ArdA; red is domain 1, orange is domain 2 and blue is domain 3. Secondary structure from the crystallographic analysis is shown above the sequence as cylinders for helix and block arrows for strands. The secondary structure assignment is from the first monomer of the dimer. The second monomer lacks the first helix. Acidic residues that entwine ArdA are highlighted in red and magenta. Those conserved across the dimer interface are highlighted with asterisk. Highlighted in an open box are the ‘anti-restriction motif’ (59) and the VF-motif both essential for activity (26). The region of ArdA from the conjugative plasmid pColIb-P9 that can be deleted without loss of anti-restriction activity is highlighted in grey (26).
DISCUSSION
The interaction of ArdA with the EcoKI Type I RM enzyme: comparison of ArdA with the DNA footprint of the enzyme
The ArdA dimer appears to mimic ∼42 bp of bent B-form DNA. This is comparable in length to the footprint of the EcoKI Type IA RM enzyme, without its cofactors, on DNA (60,61). In comparison, the Ocr dimer from phage T7 mimics only ∼24 bp, similar in length to the ∼30-bp footprint of the Type I RM enzyme in the presence of its cofactors and to the footprint of the MTase core, M.EcoKI, of the Type I RM enzyme (60–62). Binding experiments for the interaction of Ocr with M.EcoKI indicated that Ocr was engulfed by the MTase core (63) and this has recently been confirmed by image reconstruction from electron micrographs of an M.EcoKI–Ocr complex (32). The typical DNA target for a Type I RM enzyme is ∼14-bp long and bipartite, e.g. EcoKI recognizes 5′-AACNNNNNNGTGC-3′, and lies centrally in the experimental DNA footprints. Figure 5a shows the M.EcoKI–DNA model built using the EM reconstruction (32) with the DNA extended in length to 42 bp. Figure 5b shows the ArdA structure within the M.EcoKI structure. This is derived by taking advantage of the superposition of carboxylates in ArdA and phosphates in DNA (Figure 2d). A correspondence between the domains of ArdA and the regions of DNA is obvious: domain 3 overlaps the EcoKI target sequence, domain 2 contacts the extremities of the DNA-binding groove in M.EcoKI and domain 1 projects beyond the M.EcoKI structure. The footprint of the entire EcoKI RM enzyme is ∼42 bp (61), hence it would appear that domain 1 mimics the region of DNA in contact with the HsdR restriction subunits of EcoKI (these HsdR subunits are of course absent from the MTase structure). This would also be consistent with the model proposed recently from the structure of HsdR (33). This model approximately placed the HsdR against the ends of the DNA-binding groove in the MTase. Their placement would be consistent with the ArdA-MTase model in Figure 5b and earlier footprinting data. However, the exact orientation of the two HsdR with respect to the MTase is still unknown and large changes in the DNA footprint of the EcoKI nuclease (61) suggest large conformational changes. In addition, the 12-residue loop linking domains 1 and 2 may indicate that domain 1 can move relative to the rest of the protein to better contact the HsdR subunits. Domain 1 is not essential for anti-restriction as it can be deleted (26) indicating that the key aspect of anti-restriction by ArdA is the binding to the MTase core using domains 2 and 3.
Figure 5.
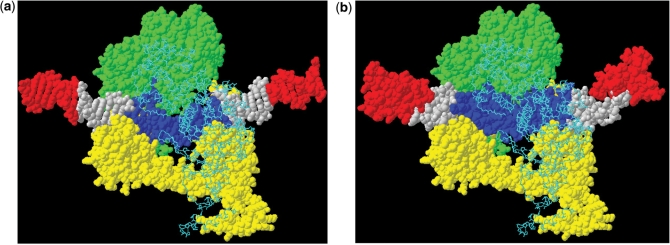
The domains of ArdA correspond to different parts of the DNA bound by EcoKI. (a) Forty-two base pairs of DNA within the EcoKI MTase (HsdS subunit yellow, HsdM subunits in green space-fill and cyan backbone trace) (32). The DNA contains a substantial bend matching the bend in ArdA. The DNA target sequence (central 14 bp) in blue plus the regions shown in white (7 bp on either side of the target sequence), totalling 28 bp, are protected in footprinting assays by EcoKI MTase or the EcoKI nuclease in the presence of ATP (46–48). The regions of DNA shown in red are the additional 7-bp regions protected in the footprinting assay by the EcoKI nuclease when no cofactors are present. The protection of these red regions of the DNA is attributed to the presence of the additional two HsdR subunits in the nuclease. (b) The ArdA domains of the dimer coloured red, white and blue for domains 1, 2 and 3 (each is approximately equivalent in length to 7 bp of DNA) replacing the DNA in the EcoKI MTase structural model. A clear correspondence between the domains of ArdA and the regions of DNA defined by footprinting experiments is visible.
The degree of anti-restriction observed in vivo varied considerably between different ArdA and different Type I families (from 7 to 70 000). The Type I RM systems are all expressed from a single chromosomal copy of their genes but because of the different stabilities of the Type I enzymes (63–65), the number of active enzyme molecules in the different host strains may vary considerably. The number of EcoKI Type I enzyme has been estimated as <100 per cell (66,67). The number of target sequences on phage lambda for the RM enzymes also varies with 5, 1, 14 and 3 targets for the IA, IB, IC and ID systems, respectively. In addition, the expression levels of the different ardA genes may vary. These factors may account for the unpredictable differences in anti-restriction. The IA, ID and, taking into account the single target site, IB systems seem most susceptible to anti-restriction by ArdA. Taking into account the number of potential target sequences for the IC system, the IC system seems rather resistant to ArdA. This is perhaps because even a single interaction of the IC enzyme with one of the numerous target sequences on the phage DNA would lead to restriction and there is insufficient ArdA to knock out every enzyme molecule. However, it is also noticeable that the IC enzyme is most able to carry out its methylation function as shown by a low anti-modification value for most of the ArdA (anti-modification varies between 1 and 10 000). This difference in the ability of ArdA to inhibit methylation of phage DNA by Type I families may reflect different binding affinities of the ArdA proteins for the Type I enzymes. At present, binding affinity is not well characterized but for the ArdA–EcoKI interaction it has been estimated that the dissociation constant, Kd, is <1 nM in vitro (27) and ∼170 nM in vivo (68). This is not as strong as the interaction between Ocr and EcoKI, where the Kd is ∼50 pM (69); therefore, perhaps ArdA is just not such a strong competitive inhibitor as Ocr. A weaker equilibrium binding affinity would allow a greater fraction of the RM enzymes to remain active in vivo. These questions will only be addressable with extensive in vitro analysis of different ArdA/RM enzyme combinations.
The ArdA structure and that of other DNA mimics, e.g. the phage lambda gam protein which inhibits the RecBCD exonuclease (70), has implications for HGT. The ArdA structure is a novel fold comprising three small domains, each of a known fold, arranged in a row in each monomer. The dimer interface is very small and the overall protein is extremely elongated and decorated with negative side chains arranged to mimic the phosphate backbone of DNA. The evolution of ArdA is curious as two of its domains show a related fold but the opposite charge to oligonucleotide binding proteins. The sequential acquisition of single-point mutations to code for a predominance of acidic amino acids and DNA mimicry is certain to be a rare event. This may be the reason that few mimics of extended DNA structures have been found. This rarity may also mean that whenever the mimics do arise, they spread rapidly throughout their ecological niche, for example, ArdA is now widely spread in the eubacteria on plasmids and transposons (24,26,27,59), phage T7 Ocr is found in other related phage (28,29) and the MfpA pentapeptide repeat protein (71) involved in quinolone resistance is spreading rapidly (4). Recently, a DNA mimic has been found in a highly infectious eukaryotic virus of crustaceans (58). One may anticipate that this mimic will also spread rapidly.
All ArdA tested here are active against restriction by Type I RM systems, so if they are expressed in their natural host then they will certainly function as anti-restriction proteins to assist HGT allowing, for example, the acquisition of resistance genes. HGT would be rampant without the existence of RM systems resulting in disruption of the host genome. An interplay between anti-restriction systems and RM systems is clearly beneficial for the evolution of bacterial species.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Biotechnology and Biological Sciences Research Council (BB/D001870/1 to D.T.F.D.); the Wellcome Trust (GR080463MA to D.T.F.D., M.D.W., J.H.N. and G.B.); the Darwin Trust of Edinburgh (a PhD studentship to B.S.); and the BBSRC (BB/S/B14450) and the Scottish Funding Council (SULSA and SSPF). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professor Noreen Murray and numerous colleagues in her laboratory for countless discussions and Dimitra Serfiotis-Mitsa for the expression plasmid and assistance with early crystallization trials.
Footnotes
The first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Bennett PM. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br. J. Pharmacol. 2008;153:S347–S357. doi: 10.1038/sj.bjp.0707607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindsay JA, Holden MT. Understanding the rise of the superbug: investigation of the evolution and genomic variation of Staphylococcus aureus. Funct. Integr. Genomics. 2006;6:186–201. doi: 10.1007/s10142-005-0019-7. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy AD, Otto M, Braughton KR, Whitney AR, Chen L, Mathema B, Mediavilla JR, Byrne KA, Parkins LD, Tenover FC, et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc. Natl Acad. Sci. USA. 2008;105:1327–1332. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 2006;6:629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 5.Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 2007;5:175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- 6.Vedler E, Vahter M, Heinaru A. The completely sequenced plasmid pEST4011 contains a novel IncP1 backbone and a catabolic transposon harboring tfd genes for 2,4-dichlorophenoxyacetic acid degradation. J. Bacteriol. 2004;186:7161–7174. doi: 10.1128/JB.186.21.7161-7174.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts RJ. How restriction enzymes became the workhorses of molecular biology. Proc. Natl Acad. Sci. USA. 2005;102:5905–5908. doi: 10.1073/pnas.0500923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev SKh, Dryden DTF, Dybvig K, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tock MR, Dryden DTF. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 2005;8:466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 12.Bickle TA, Krüger DH. Biology of DNA restriction. Microbiol. Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkins BM. Plasmid promiscuity: meeting the challenge of DNA immigration control. Env. Microbiol. 2002;4:495–500. doi: 10.1046/j.1462-2920.2002.00332.x. [DOI] [PubMed] [Google Scholar]
- 14.King G, Murray NE. Restriction alleviation and modification enhancement by the Rac prophage of Escherichia coli K-12. Mol. Microbiol. 1995;16:769–777. doi: 10.1111/j.1365-2958.1995.tb02438.x. [DOI] [PubMed] [Google Scholar]
- 15.Webb JL, King G, Ternent D, Titheradge AJ, Murray NE. Restriction by EcoKI is enhanced by co-operative interactions between target sequences and is dependent on DEAD box motifs. EMBO J. 1996;15:2003–2009. [PMC free article] [PubMed] [Google Scholar]
- 16.Bandyopadhyay PK, Studier FW, Hamilton DL, Yuan R. Inhibition of the Type I restriction-modification enzymes EcoB and EcoK by the gene 0.3 protein of bacteriophage T7. J. Mol. Biol. 1985;182:567–578. doi: 10.1016/0022-2836(85)90242-6. [DOI] [PubMed] [Google Scholar]
- 17.Dryden DTF. DNA mimicry by proteins and the control of enzymatic activity on DNA. Trends Biotechnol. 2006;24:378–382. doi: 10.1016/j.tibtech.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Dryden DTF, Tock MR. DNA mimicry by proteins. Biochem. Soc. Trans. 2006;34:317–319. doi: 10.1042/BST20060317. [DOI] [PubMed] [Google Scholar]
- 19.Krüger DH, Schroeder C, Hansen S, Rosenthal HA. Active protection by bacteriophages T3 and T7 against E. coli B- and K-specific restriction of their DNA. Mol. Gen. Genet. 1977;153:99–106. doi: 10.1007/BF01036001. [DOI] [PubMed] [Google Scholar]
- 20.Stephanou AS, Roberts GA, Tock MR, Pritchard EH, Turkington R, Nutley M, Cooper A, Dryden DTF. A mutational analysis of DNA mimicry by Ocr, the gene 0.3 antirestriction protein of bacteriophage T7. Biochem. Biophys. Res. Commun. 2009;378:129–132. doi: 10.1016/j.bbrc.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 21.Belogurov AA, Yussifov TN, Kotova VU, Zavilgelsky GB. The novel gene(s) ARD of plasmid pKM101: alleviation of EcoK restriction. Mol. Gen. Genet. 1985;198:509–513. doi: 10.1007/BF00332948. [DOI] [PubMed] [Google Scholar]
- 22.Belogurov AA, Delver EP, Rodzevich OV. Plasmid pKM101 encodes two nonhomologous antirestriction proteins (ArdA and ArdB) whose expression is controlled by homologous regulatory sequences. J. Bacteriol. 1993;175:4843–4850. doi: 10.1128/jb.175.15.4843-4850.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belogurov AA, Delver EP, Agafononva OV, Belogurova NG, Lee L, Kado C. Antirestriction protein Ard (Type C) encoded by the IncW plasmid pSa has a high similarity to the “protein transport” domain of TraC1 primase of promiscuous plasmid RP4. J. Mol. Biol. 2000;296:969–977. doi: 10.1006/jmbi.1999.3493. [DOI] [PubMed] [Google Scholar]
- 24.Chilley PM, Wilkins BM. Distribution of the ardA family of antirestriction genes on conjugative plasmids. Microbiology. 1995;141:2157–2164. doi: 10.1099/13500872-141-9-2157. [DOI] [PubMed] [Google Scholar]
- 25.Flannagan SE, Zitzow LA, Su YA, Clewell DB. Nucleotide sequence of the 18-kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid. 1994;32:350–354. doi: 10.1006/plas.1994.1077. [DOI] [PubMed] [Google Scholar]
- 26.Nekrasov SV, Agafonova OV, Belogurova NG, Delver EP, Belogurov AA. Plasmid-encoded antirestriction protein ArdA can discriminate between type I methyltransferase and complete restriction-modification system. J. Mol. Biol. 2007;365:284–297. doi: 10.1016/j.jmb.2006.09.087. [DOI] [PubMed] [Google Scholar]
- 27.Serfiotis-Mitsa D, Roberts GA, Cooper LP, White JH, Nutley M, Cooper A, Blakely GW, Dryden DTF. The ORF18 gene product from conjugative transposon Tn916 is an ArdA antirestriction protein that inhibits Type I DNA restriction-modification systems. J. Mol. Biol. 2008;383:970–981. doi: 10.1016/j.jmb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Garcia E, Elliott JM, Ramanculov E, Chain PS, Chu MC, Molineux IJ. The genome sequence of Yersinia pestis bacteriophage phiA1122 reveals an intimate history with the coliphage T3 and T7 genomes. J. Bacteriol. 2003;185:5248–5262. doi: 10.1128/JB.185.17.5248-5262.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scholl D, Kieleczawa J, Kemp P, Rush J, Richardson CC, Merril C, Adhya S, Molineux IJ. Genomic analysis of bacteriophages SP6 and K1-5, an estranged subgroup of the T7 supergroup. J. Mol. Biol. 2004;335:1151–1171. doi: 10.1016/j.jmb.2003.11.035. [DOI] [PubMed] [Google Scholar]
- 30.Walkinshaw MD, Taylor P, Sturrock SS, Atanasiu C, Berge T, Henderson RM, Edwardson JM, Dryden DTF. Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol. Cell. 2002;9:187–194. doi: 10.1016/s1097-2765(02)00435-5. [DOI] [PubMed] [Google Scholar]
- 31.Murray NE. 2001 Fred Griffith review lecture. Immigration control of DNA in bacteria: self versus non-self. Microbiology. 2002;148:3–20. doi: 10.1099/00221287-148-1-3. [DOI] [PubMed] [Google Scholar]
- 32.Kennaway CK, Obarska-Kosinska A, White JH, Tuszynska I, Cooper LP, Bujnicki JM, Trinick J, Dryden DTF. The structure of M.EcoKI Type I DNA methyltransferase with a DNA mimic antirestriction protein. Nucleic Acids Res. 2009;37:762–770. doi: 10.1093/nar/gkn988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lapkouski M, Panjikar S, Janscak P, Smatanova IK, Carey J, Ettrich R, Csefalvay E. Structure of the motor subunit of type I restriction-modification complex EcoR124I. Nat. Struct. Mol. Biol. 2009;16:94–95. doi: 10.1038/nsmb.1523. [DOI] [PubMed] [Google Scholar]
- 34.Titheradge AJ, King J, Ryu J, Murray NE. Families of restriction enzymes: an analysis prompted by molecular and genetic data for type ID restriction and modification systems. Nucleic Acids Res. 2001;20:4195–4205. doi: 10.1093/nar/29.20.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chin V, Valinluck V, Magaki S, Ryu J. KpnBI is the prototype of a new family (IE) of bacterial type I restriction-modification system. Nucleic Acids Res. 2004;32:e138. doi: 10.1093/nar/gnh134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cerdeno-Tarraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, et al. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 37.Amann E, Ochs B, Abel KJ. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- 39.Sheldrick GM. A short history of SHELX. Acta Cryst. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 40.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Cryst. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Cryst. D. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.CCP4. Collaborative computational project number 4. Acta Cryst. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 43.McRee DE. XtalView/Xfit – a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 44.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finn RD, Tate J, Mistry J, Coggill PC, Sammut JS, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Howe K, Bateman A, Durbin R. QuickTree: building huge Neighbour-Joining trees of protein sequences. Bioinformatics. 2002;18:1546–1547. doi: 10.1093/bioinformatics/18.11.1546. [DOI] [PubMed] [Google Scholar]
- 47.Jordan GE, Piel WH. PhyloWidget: web-based visualizations for the tree of life. Bioinformatics. 2008;24:1641–1642. doi: 10.1093/bioinformatics/btn235. [DOI] [PubMed] [Google Scholar]
- 48.Thomas AT, Brammar WJ, Wilkins BM. Plasmid R16 ArdA protein preferentially targets restriction activity of the Type I restriction-modification system EcoKI. J. Bacteriol. 2003;185:2022–2025. doi: 10.1128/JB.185.6.2022-2025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zavil'gel'skiĭ GB, Letuchaia TA, Rastorguev SM. Antirestriction and antimodification activities of the ArdA protein encoded by the IncI1 transmissive plasmids R-64 and ColIb-P9. Russ. J. Genet. 2006;42:252–258. [PubMed] [Google Scholar]
- 50.Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 51.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 52.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Tárraga AM, Wang H, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 2006;38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 53.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl Acad. Sci. USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen Y, Volrath SL, Weatherly SC, Elich TD, Tong L. A mechanism for the potent inhibition of eukaryotic acetyl-coenzyme A carboxylase by soraphen A, a macrocyclic polyketide natural product. Mol. Cell. 2004;16:881–891. doi: 10.1016/j.molcel.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 55.Morth JP, Feng V, Perry LJ, Svergun DI, Tucker PA. The crystal and solution structure of a putative transcriptional antiterminator from Mycobacterium tuberculosis. Structure. 2004;12:1595–1605. doi: 10.1016/j.str.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 56.Garnett JA, Marincs F, Baumberg S, Stockley PG, Phillips SE. Structure and function of the arginine repressor-operator complex from Bacillus subtilis. J. Mol. Biol. 2008;379:284–298. doi: 10.1016/j.jmb.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 57.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 58.Wang HC, Wang HC, Ko TP, Lee YM, Leu JH, Ho CH, Huang WP, Lo CF, Wang AH. White spot syndrome virus protein ICP11: A histone-binding DNA mimic that disrupts nucleosome assembly. Proc. Natl Acad. Sci. USA. 2008;105:20758–20763. doi: 10.1073/pnas.0811233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Belogurov AA, Delver EP. A motif conserved among the Type I restriction-modification enzymes and antirestriction proteins: a possible basis for mechanism of action of plasmid-encoded anti-restriction functions. Nucleic Acids Res. 1995;23:785–787. doi: 10.1093/nar/23.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davies GP, Martin IV, Sturrock SS, Cronshaw A, Murray NE, Dryden DTF. On the structure and operation of type I DNA restriction enzymes. J. Mol. Biol. 1999;290:565–579. doi: 10.1006/jmbi.1999.2908. [DOI] [PubMed] [Google Scholar]
- 61.Powell LM, Dryden DTF, Murray NE. Sequence-specific DNA binding by EcoKI, a type IA DNA restriction enzyme. J. Mol. Biol. 1998;283:963–976. doi: 10.1006/jmbi.1998.2143. [DOI] [PubMed] [Google Scholar]
- 62.Powell LM, Dryden DTF, Willcock DF, Pain RH, Murray NE. DNA recognition by the EcoK methyltransferase. The influence of DNA methylation and the cofactor S-adenosyl-L-methionine. J. Mol. Biol. 1993;234:60–71. doi: 10.1006/jmbi.1993.1563. [DOI] [PubMed] [Google Scholar]
- 63.Suri B, Shepherd JC, Bickle TA. The EcoA restriction and modification system of Escherichia coli 15T−: enzyme structure and DNA recognition sequence. EMBO J. 1984;3:575–579. doi: 10.1002/j.1460-2075.1984.tb01850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dryden DTF, Cooper LP, Thorpe PH, Byron O. The in vitro assembly of the EcoKI type I DNA restriction/modification enzyme and its in vivo implications. Biochemistry. 1997;36:1065–1076. doi: 10.1021/bi9619435. [DOI] [PubMed] [Google Scholar]
- 65.Janscak P, Dryden DTF, Firman K. Analysis of the subunit assembly of the typeIC restriction-modification enzyme EcoR124I. Nucleic Acids Res. 1998;26:4439–4445. doi: 10.1093/nar/26.19.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heip J, Rolfe B, Schell J. Abolition of host cell restriction by high multiplicity of phage infection. Virology. 1974;59:356–370. doi: 10.1016/0042-6822(74)90450-4. [DOI] [PubMed] [Google Scholar]
- 67.Kelleher JE, Raleigh EA. Response to UV damage by four Escherichia coli K-12 restriction systems. J. Bacteriol. 1994;176:5888–5896. doi: 10.1128/jb.176.19.5888-5896.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zavilgelsky GB, Kotova VY, Rastorguev SM. Comparative analysis of anti-restriction activities of ArdA (ColIb-P9) and Ocr (T7) proteins. Biochemistry. 2008;73:906–911. doi: 10.1134/s0006297908080087. [DOI] [PubMed] [Google Scholar]
- 69.Atanasiu C, Su T-J, Sturrock SS, Dryden DTF. Interaction of the Ocr gene 0.3 protein of bacteriophage T7 with EcoKI restriction/modification enzyme. Nucleic Acids Res. 2002;30:3936–3944. doi: 10.1093/nar/gkf518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Court R, Cook N, Saikrishnan K, Wigley D. The crystal structure of lambda-Gam protein suggests a model for RecBCD inhibition. J. Mol. Biol. 2007;371:25–33. doi: 10.1016/j.jmb.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 71.Hegde SS, Vetting MW, Roderick SL, Mitchenall LA, Maxwell A, Takiff HE, Blanchard JS. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science. 2005;308:1480–1483. doi: 10.1126/science.1110699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.