Abstract
Potential promoters in the genome of Escherichia coli were searched by pattern recognition software PlatProm and classified on the basis of positions relative to gene borders. Beside the expected promoters located in front of the coding sequences we found a considerable amount of intragenic promoter-like signals with a putative ability to drive either antisense or alternative transcription and revealed unusual genomic regions with extremely high density of predicted transcription start points (promoter ‘islands’), some of which are located in coding sequences. PlatProm scores converted into probability of RNA polymerase binding demonstrated certain correlation with the enzyme retention registered by ChIP-on-chip technique; however, in ‘dense’ regions the value of correlation coefficient is lower than throughout the entire genome. Experimental verification confirmed the ability of RNA polymerase to interact and form multiple open complexes within promoter ‘island’ associated with appY, yet transcription efficiency was lower than might be expected. Analysis of expression data revealed the same tendency for other promoter ‘islands’, thus assuming functional relevance of non-productive RNA polymerase binding. Our data indicate that genomic DNA of E. coli is enriched by numerous unusual promoter-like sites with biological role yet to be understood.
INTRODUCTION
Even though expression data are now available for many organisms, a possibility to use them for tracking cell transcriptional output upon varying conditions requires a comprehensive map of genome regulatory elements. Currently there are two approaches opening a way to depict this map: computational search of promoter regions [most recent algorithms: (1–10)], and ChIP-on-chip spotting of RNA polymerase (RNAPol)-binding sites (11–14). In this study we tried to understand how predictable the data obtained in silico are and to what extent they correlate with the data obtained in vivo.
The genome-scale mapping of promoter sites has been performed by our pattern recognition software PlatProm (6,7). The same as other protocols operating with position-specific weight matrices, PlatProm has an advantage of predicting transcription start points (TSP) (the main promoter attribute, which is determined experimentally), rather than to define promoter regions as extensive genomic loci (9,10,15,16). Upon scanning PlatProm considers any nucleotide as a probable point of transcription initiation and scores this probability by searching for promoter-specific elements in proper positions. Rather high precision and low rate of false positives made our software suitable for genome scanning, providing a possibility to compare in silico and in vivo data. In the present work, we describe an overall map of TSPs predicted by PlatProm; evaluate the degree of correlation between PlatProm scores and RNAPol-binding efficiency registered by two independent research groups (12,13); focus attention on intragenic TSPs and promoter ‘islands’ having high density of transcription signals over long distance; and verify capacity of one ‘island’ to bind RNAPol and initiate RNA synthesis.
MATERIALS AND METHODS
An essence of computational approach
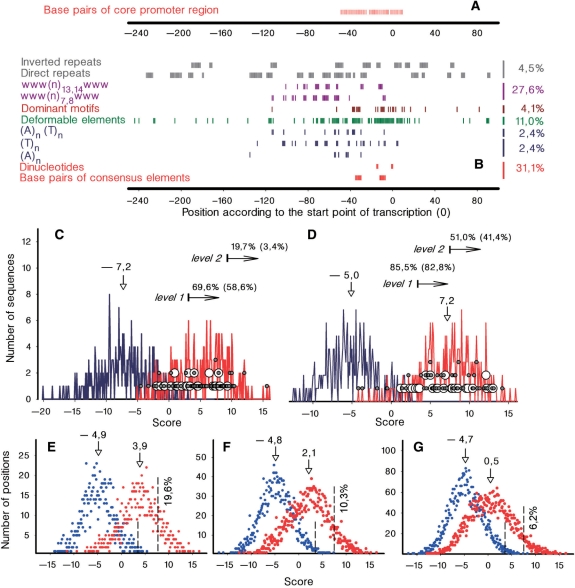
Our scoring system was designed as originally suggested by Hertz and Stormo (17). Sensitivity of this algorithm tested on our sets was the same as reported (Figure 1B). To increase the performance of this approach, we took into account sequence-dependent structural features in the genomic environment of promoter sites (6, 7). The final version of PlatProm (7) was refined on 271 experimentally identified non-homologous and non-overlapping σ70-promoters with a single TSP precisely pointed out by the previous version of our software (6). All sequences were 411-bp long (−255/+155 according to the start point nominated as 0). Allowed variations of the spacer, and the distance between the TSP and the element –10 were 14–21 and 2–9 bp, respectively. Variable length of the spacer was accounted by the matrix penalizing deviations from the optimal 17 bp in accordance with their frequencies in the training promoter set. The varying distance between element −10 and the TSP, ignored by previously published protocols, was taken into account the same way. Weight matrices corresponding to consensus elements −35 and −10 evaluated hexanucleotides, rather than 25 and 19 bp sequences, accounted by Hertz and Stormo (Figure 1A and B). They contain 6 × 4 scores equal to natural logarithms of normalized occurrence frequencies for each nucleotide at each position in preliminarily aligned promoters. Percentage of particular nucleotides in the genome was used for normalization. Sequence preference nearby the TSP previously accounted by a 12 × 4 weight matrix (17) was scored by the matrix precisely reflecting the distribution of dinucleotides in −1/0. Dinucleotides forming the ‘extended −10 element’ were accounted the same way. Since frequency coefficients depend on promoter alignment, optimal matrices were generated by the procedure of expectation–maximization.
Figure 1.
(A) Positions of base pairs accounted by Hertz and Stormo (17). (B) Promoter-specific elements scored by PlatProm (preferred positions are marked by vertical tics). Ciphers on the right indicate their contribution to S (percentage of promoters additionally recognized at levels 1 and 2). (C and D) Discriminative capacity of algorithm suggested by Hertz and Stormo (C) and PlatProm (D) tested on natural sequences. Blue plots represent distribution of scores for virtual TSPs ascribed to +256 position of CS1 sequences. These data were used to estimate the average value of S, standard deviation and threshold levels (indicated by horizontal arrows). Red curves show distribution of scores for experimentally mapped TSPs of testing compilation (±2 bp variations are allowed to pick out position with maximal score). These variations are aimed to compensate possible experimental or computational inaccuracy. In case of promoters with multiple TSPs, their scores were compared and the maximal one was plotted. These data are used to estimate percentage of recognized promoters (indicated above horizontal arrows) at two levels (S > 3.4 and 7.44). Distribution of scores for TSPs of promoters regulated by repressors and activators are shown by small gray and large open circles, respectively. Ciphers in parenthesis indicate sensitivity of PlatProm for positively regulated promoters. Vertical arrows and ciphers above indicate position and value of average S. (E–G) Distribution of S for all positions in the ±2 (E), ±5 (F) and ±10 (G) areas around virtual TSPs of CS1 (blue symbols) and real starts of testing compilation (red circles). Vertical lines delimit points with S exceeding levels 1 and 2. Percentage of positions with S > 7.44 is indicated nearby.
Additional sequence motifs were scored by the set of 34 cascade matrices exemplified in ref. (7). They include di-, tri- and tetranucleotides having occurrence frequencies at least 5 SD higher than the background level in the particular positions of 34 promoter subregions (Figure 1B). These matrices represent:
Sequence motifs potentially interacting with RNA polymerase α-subunits, including (A)n-tracts as perfect targets (18).
(T)n- and (A)n(T)n-tracts able to promote additional protein–protein or DNA–protein contacts by inducing proper bending of the DNA helix (19).
Elements containing flexible YR steps (Y = T = C, R = A = G) favoring adaptive isomerization of the promoter DNA (20).
Other motifs previously revealed by cluster analysis (21).
Cascade matrices consider all sequence elements dominant in a particular promoter subregion, but only the most significant one found in a certain promoter is taken into account. Thus, for instance, there are 30 motifs dominating in the region −48/−41. The most frequent of them, hence the most significant one, is ATAA if located in position −44. It gives 0.91 to the overall score. Slightly less significant (contribution 0.82) is TATA if located in −45. Thus, for promoter containing TATAT in −45/−41 the overall score increases by 0.82, while for promoter, containing TATAA – by 0.91, rather than by 0.91+0.82. Lack of accounted motifs in a particular subregion of a promoter was penalized based on probability of their absence in this place in the training set.
Ten cascade matrices were used to formalize the presence of mixed w-tracts (w = A = T), highly typical for promoter DNA. They are regularly distributed with 1 or 1.5 helix turns periodicity (Figure 1B) and may participate in polymerase sliding along DNA (22). To focus attention on the observed regularity we search for (www(n)7,8www and (www(n)13,14www thus minimizing overlap with (A)n- and (T)n-specific matrices.
PlatProm also takes into account perfect direct and inverted repeats (5–11 bp long separated by 5 or 6 bp) as potential targets for interaction with regulatory proteins. They are scored as natural logarithms of lengths if centered in subregions of their frequent occurrence (Figure 1B).
The contributions of both types of repeats and elements accounted by cascade matrices were reduced by coefficients estimated as a ratio of the average information content [quantified as described in ref. (23)] in the particular subregion to the information content in the sixth position of the aligned element –35 (the least significant among conservative base pairs). This reduction was aimed to balance the endowment of additional elements with that of conservative base pairs. Inputs of all elements were summarized, giving the total score (S), which reflects the probability for any nucleotide to be a TSP and therefore the probability of its upstream region to bind RNAPol and fulfill promoter function.
Performance of the program was tested on a set of 290 known promoters and two control sets (CSs). All sequences were 411 bp long (−255/+155 according to real or virtual TSPs). The testing compilation was composed of experimentally characterized σ70 promoters not included in the training set. To increase the size of this set we allowed the presence of promoters (∼30%) with multiple TSPs (for instance discussed below dcuA), while homologous or overlapping promoters were excluded. CS1 was composed of 273 coding sequences taken from the fixed position of all convergent genes longer than 700 bp, separated by ≤50-bp intergenic space, which are not associated with promoter ‘islands’ (see below) and have 5′-ends similarly annotated in U00096 and U00096.2 E. coli gene maps. Sequences were taken so as virtual TSPs were 250 bp far from initiating codon (fall in position 254 inside the gene). This strategy minimizes the probability to pick out functional promoter for alternative transcription or for yet unidentified genes in intergenic regions. Having AT-content (48.3%) very close to that of the whole genome (49.2%) CS1 roughly represents natural sequences, which RNAPol has to bypass. CS2 was composed of 400 random sequences with the same AT-content as in chromosomal DNA. (All compilations are available by request.) Promoters were considered as recognized by PlatProm if experimentally mapped TSP(s) or a neighboring nucleotide (±2) had score higher than threshold level. Using this criteria and the last version of PlatProm we identified 85.5% of promoters from the test compilation at zero level of false positives (level 1, Figure 1) in both CSs (Smax = 3.4). Thus, at this level the combination of sensitivity (percentage of recognized promoters = 85.5%) and specificity (percentage of unrecognized non-promoter sequences = 100%) of PlatProm was better than that of previously suggested algorithms (68.7–82% and 82.2–99.1%, respectively) [(2,15,24) and Figure 1C]. Approximately 80% of recognized promoters possess experimentally mapped TSPs coinciding (in the range of ± 2 bp) with positions of local S maxima, i.e. nearly 70% of natural TSPs are accurately predicted by our software, which is also better than previously reported (<50%) (1,17).
Experimental procedures
The E. coli K12 strain was used as a source of genomic DNA and total RNA. DNA was isolated by phenol–chloroform extraction. Total RNA was purified as described in ref. (25).
Three DNA fragments containing part of promoter ‘island’ associated with appY were obtained by PCR. A 406-bp fragment spanning over the region −260/+146 relative to the initiating codon of appY was amplified using the primers 5′-GCAAGAGGTTTCAGGTGCGTTGTAGTGAG-3′(F1) and 5′-CTTAGTTTAGAGGGGCAT-3′(R1). Shortened (378 bp) and elongated (471 bp) templates were obtained with F1 + 5′-CCCTTCTAGATTTGTCGCTTACAATAAA-3′(R2) (−260/+118) and 5′-GATAAGATCTGCAAGTAAAAATGATACTC-3′(F2) + R1 (−325/+146), respectively.
RNAPol-σ70-binding ability was tested by gel-retardation and potassium permanganate footprinting. Complexes were formed at 37°C in buffer containing 50 mM Tris–HCl (pH 8.0), 0.1 mM EDTA, 0.1 mM DTT, 10 mM MgCl2, 50 mM NaCl, 250 mkg/ml BSA, 1 pm of 406-bp DNA-fragment and 2–8 pm of RNAPol. Templates for permanganate footprinting were 32P-labeled. Interaction was allowed for 30 min. In gel-shift experiments, 20 mkg/ml of heparin was added before loading the sample on 4% polyacrylamide gel pre-warmed to 37°C. Gels were run at the same temperature and stained by ethydium bromide. Footprinting assays were performed according to (26). Unpaired thymines in open transcription complexes were modified by potassium permanganate and revealed after piperidine treatment. The products of chemical hydrolysis were separated in 8% polyacrylamide gel in the presence of 8 M urea and visualized by radioautography.
Primer extension was carried out using RevertAid™ M-MulV reverse transcriptase (Fermentas), 4 pm of 32P-labeled primer and variable amounts of total RNA (1–6 mkg) purified from E. coli cells grown either in LB or M9 media and harvested at logarithmic or stationary phases. Before the reaction the mixture of RNA and primer was heated at 72°C for 10 min; cooled down to 58°C, and primer annealing was allowed for 3 min. Then the probe was chilled on ice, mixed with the buffer provided by manufacturer and incubated with the enzyme at 42°C for 45 min. Reverse transcription was terminated by heating. cDNA was precipitated by N-butanol and analyzed the same way as the products of permanganate footprinting.
Single round transcription was carried out as described previously (27).
RESULTS
Figure 1 demonstrates advantage given by each additional element (Figure 1B) and increased sensitivity of the program (Figure 1C and D) for the whole testing compilation and positively regulated promoters. All additional elements improve the resolution but the largest contribution is given by w-tracts increasing sensitivity for 27.6% (Figure 1B). This high significance of additional elements provides a chance to detect regulatory regions in newly sequenced genomes if information about their own promoters is insufficient.
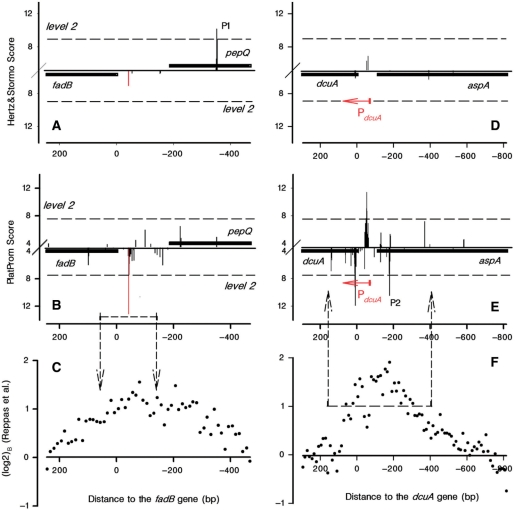
Figure 2 exemplifies promoters accurately recognized (fadB) and unrecognized (dcuA) by PlatProm. The start point of fadB-transcription exactly fits the experimentally mapped TSP (Figure 2B), while the original program (17) suggests intragenic P1 as a dominant transcription signal (Figure 2A) and algorithm developed by Huerta and Collado-Vides (1) predicts fadB promoter ∼150 bp upstream of the real TSP [see Figure 4 in (1)].
Figure 2.
Distribution of potential TSPs within regulatory regions of fadB/pepQ (A–C) and dcuA/aspA (D–F) as predicted by Hertz and Stormo algorithm (top panels) and PlatProm (middle panels). Black lines mark the coding sequences. Upward and downward bars reflect promoter scores quantified for the ‘+’ and ‘−’ strands, respectively. X-axis is placed at ‘level 1’ (Figure 1). Dashed lines indicate ‘level 2’. Red bars and arrows correspond to the experimentally mapped TSPs. Bottom plots show ChIP-on-chip data (13) for analyzed regions. Dashed arrows in plots (B) and (C) confine genomic area around the predicted or experimentally mapped TSP(s), where interaction with RNAPol was verified. Dashed arrows in plots (E) and (F) outline area nearby registered RNAPol-binding site, where predicted TSPs were searched.
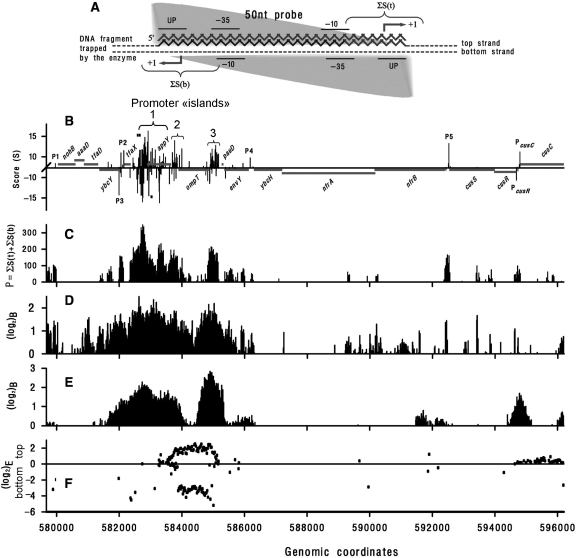
Figure 4.
(A) Scheme illustrating the strategy used to convert PlatProm scores into probability of RNAPol binding (P). Fifty-nucleotide microarray probe is shown as a gray zigzag line. Dashed and composite lines represent two strands of DNA trapped by RNAPol. To take into account both orientations of the enzyme (gray triangle) P was calculated as the sum of S near the expected TSPs on both strands (indicated by braces). The most adequate values of P are expected if a given probe exactly fits to a promoter but allowed (±8 bp) variation in position of the potential start ensures capturing scores of promoters shifted by 1–6 bp as well (microarray probes are distributed along DNA with 12 bp periodicity). Plot (C) shows the result of this converting procedure for PlatProm scores predicted in 57 9670–596 200 bp genomic region (B). Horizontal gray lines indicate gene positioning. The X-axis is placed at ‘level 2’. Known TSPs of PcusR and PcusC are indicated. Three braces in plot (B) confine promoter ‘islands’. Tick marks indicate positions of primers used for PCR. (D) RNAPol-binding efficiency represented as log2 of the ratio of hybridization signal obtained with DNA co-immunoprecipitated with the enzyme by β′-specific antibody to control DNA recovered from complexes without specific immunoprecipitation (12). (E) The same as (D) but σ-specific antibody was used to collect RNAPol–DNA complexes (13). (F) Transcription efficiency from the top and the bottom strands, respectively, represented as log2 of the ratio of fluorescence signals originated from hybridized cDNA to signals obtained from hybridized sonicated genomic DNA (13).
Unrecognized promoter PdcuA (Figure 2D–F) has four TSPs, experimentally mapped 69–72 bp upstream of the initiating codon (28). S values at these positions vary in the range −2.1/−5.7, i.e. far below the level required for recognition. All promoter-specific elements in PdcuA are weakly pronounced, thus no refinement of the current scoring system can compensate for these negative values. Although it can not be excluded that PdcuA has some important determinants, which are not formalized by PlatProm, limitations of experimental mapping performed by primer extension (28) also may result in some difference between apparent and actual TSPs. Reverse transcriptase, for example, can stop at sites with stable hairpins in mRNA structure. In this case, the registered site of transcription initiation would be pointed out downstream from the real one. PlatProm predicts a good candidate (P2) perfectly fitting the RNAPol-binding site registered by ChIP-on-chip assay (Figure 2F). Thus, comparing in silico and in vivo data one should take into account finite sensitivity and selectivity of computer model as well as limited accuracy of experimental approaches. Even though PlatProm may be further improved on the basis of additional features, current version predicts TSPs with rather high accuracy and was used to depict the genome-wide distribution of potentially transcribed regions.
Genome scanning revealed unexpected intragenic promoters
The genome sequence of E. coli K12 (NCBI, GenBank entry U00096.2) was used for large-scale promoter prediction (see Supplementary Table 1 for all PlatProm scores on both strands). Only signals with S >7.44 (level 2 in Figure 1D) were considered reliable. This cut-off level provides a possibility to identify only 51.0% of known promoters (Figure 1D) but ensures p < 0.000043 (Student's T-test statistics) if average S (−5.0) and SD (3.1) were estimated using scores of CS1. Artificially generated sequences of CS2 showed the same variability of S (SD = 3.1) but average score was higher (−4.2), thus assuming ∼2.5-fold higher p-value (p < 0.00011). A total of 30 188 individual or clustered TSPs (Figure 2) forming ∼5000 promoter-like regions were found. Since only half of known promoters have TSPs with S >7.44, the data obtained presume existence of more than 9000 of separate promoter-like sites, which is twice greater than the number of annotated genes in the genome of E. coli. To what extent this excess could be explained by the presence of false positives?
Relying on p-values only 399–928 of 30 188 predicted TSPs (1–3%) may be false signals (4 639 675 bp × 2 × p). However, intragenic and artificial sequences of CS1 and CS2 may be deficient in some promoter-specific features. That is why PlatProm performance was further tested in intergenic regions separating convergent genes, even though each of them may contain yet unidentified genes. Predicted TSPs were searched in the central part of intergenic space (≥250 bp from the ends of flanking genes) so as to ensure that only non-coding sequences undergo scoring. Only 15 regions between convergent genes are longer than 500 bp but two of them overlap with promoter ‘islands’ (see below) and were eliminated. The total length of the remaining ones is 10 226 bp (both strands). As expected, AT-content of these regions is slightly higher (49.2%) than that of CS1 (48.3%). Altogether they contain four predicted TSPs (one per ∼2500 bp). If all of them are spurious, the probability to come across a false positive is 0.00039, i.e. higher than p-values mentioned above. However, two predicted TSPs are perfectly suitable for small regulatory RNAs HB_48 and HB_139 predicted by Carter et al. (29) and one is located in front of IS yi21_6. Supposing that they belong to real promoters, false positive rate given by statistic analysis and scanning procedure correspond to each other. Even if p = 0.00039 is taken as a real frequency of false signals, still ∼90% of TSPs predicted by PlatProm should be considered as promoter-like signals deserving more detailed analysis.
An important question is how to perceive PlatProm signals forming clusters near real and accurately recognized by PlatProm TSPs (see Figure 2B for example). Should they be considered as false positives or not? To address this question we compared distribution of scores in positions surrounding real (test compilation) and virtual (CS1) transcription starts (Figure 1E–G). An average S quantified for CS1 remains almost constant if the size of analyzed area increases. The number of signals exceeding ‘level 1’ in ± 2 and ± 5 bp area (11 and 24, respectively) corresponds to the expected values (11 and 26, respectively) if CS2 statistic is used but is twice larger than expected from p-value (<0.0038) calculated on the basis of CS1 scores (6 and 12). The rate of false positives given by CS2 statistic is, therefore, quite realistic. It assumes 0.63 probability for the presence of highly scoring position in the ± 10 area around virtual TSPs of CS1 (total number of positions is 5733). One signal with S >7.44 (‘level 2’) was really registered in this region (Figure 1G).
Portion of positions with S exceeding ‘level 2’ and the average S decrease upon widening of the analyzed area around real promoters, however, distribution of their scores does not reveal expected maxima typical for CS1 (Figure 1E and F). Only in ±10 region S-values show noticeable divergence. That means that PlatProm scores around real TSPs are higher than background values. Thus, the total number of positions with S >3.4 for ±2 bp area (1681 points) is significantly larger (852) than the total number of experimentally mapped starts including multiple ones (480). Detailed examination showed that ∼92% of additional signals are generated by PlatProm using the same frame for −35 and −10 hexanucleotides as position with maximal S. Remaining ones correspond to alternative −10 element (4.4%) or −10 and −35 elements (3.6%), suggesting the presence of overlapping promoter. Promoters with multiple TSPs tend to have additional signals corresponding to alternative pair of core elements. Thus, in all cases PlatProm signals proximal to known TSPs denote the presence of either the same or alternative promoter and should not be considered as false positives.
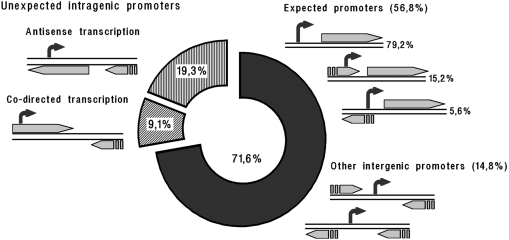
Predicted TSPs were classified in respect to the borders of mapped genes considering 750 bp as the allowed distance between the predicted transcription starts and the coding sequences of downstream genes [∼10% of known promoters are 250–700 bp far from the coding sequences (6)]. Most predicted TSPs were ranked by this strategy as ‘expected’ promoters (Figure 3), although some of them fall into upstream genes (see, for instance P2 in Figure 2E).
Figure 3.
Classification of predicted TSPs in respect to their genomic positioning. The main group contains anticipated transcription starts located either within intergenic regions or <750 bp upstream of open reading frames. Schemes on the right illustrate all possible variants of their genomic environment. Schemes on the left exemplify positions of unexpected intragenic promoter-like sites.
Approximately 19% of the remaining TSPs were found on the opposite strand of coding sequences (Figure 3). Antisense RNAs, if initiated therein, may block translation by base pairing with mRNAs, regulate their stability or interfere in other molecular processes. Such RNAs control expression of many plasmid and transposon genes, but until now they were not considered typical for bacterial regulatory networks. Nonetheless several lines of evidence support our predictions:
Microarray probes that query expression of antisense strands gave positive hybridization signals for all genes possessing predicted antisense promoters (30).
Among RNAs matching antisense strands and found by Vogel et al. (31) and Kawano et al. (32), 10 species may be transcribed from intragenic promoters predicted by PlatProm.
aRNAs I002 (33) and GadY (34) have 5′-ends exactly pointed out by PlatProm.
Two intragenic promoters found by PlatProm within the coding sequences of hns and htgA/yaaW appeared to be active in vitro (35).
Thus, the first amusing outcome obtained from the genome scanning was a large amount of genes containing promoters with antisense orientation.
Unexpected intragenic promoters of another group have sense orientation (Figure 3). About 23% of corresponding TSPs lie <50 bp downstream from the initiating codon. They were not considered as candidates for independent transcription initiation, since corresponding promoter-like regions may participate in polymerase trapping near real promoters (1). Some intragenic promoters may intensify transcription of properly oriented downstream genes. If they are also ignored, there are still ∼400 potential TSPs with a less comprehensible destination. Some of them may produce RNAs antisense to mRNAs of the convergently oriented downstream genes. Synthesis of shortened mRNAs seems to be the most exotic assumption to date. At the same time, ORF Finder (NCBI) reveals alternative ORFs supplied with a suitable Shine-Dalgarno sequence downstream of ∼50% of intragenic co-directed promoters.
The genome of E. coli has promoter ‘islands’ with high density of promoter-like signals
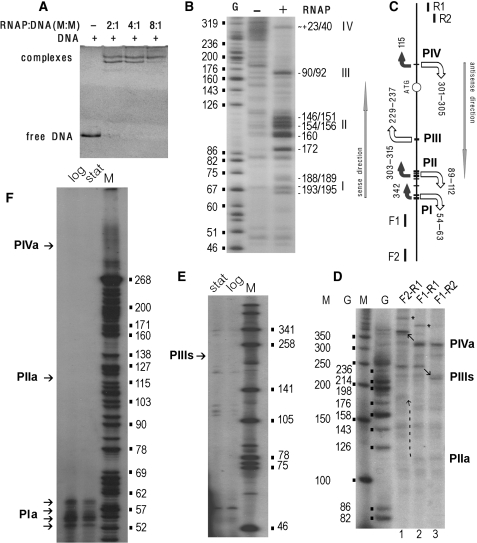
In silico promoter mapping revealed 78 unusual genomic regions spanning for at least 300 bp and containing eight or more potential TSPs on any strand in the running window of 100 bp. Figure 4B exemplifies this phenomenon. We call these regions ‘promoter islands’ (PI) so as to distinguish them from clustered ‘promoter-like signals’ (Figure 2B) found in front of almost all (92%) genes of E. coli (1,36). ChIP-on-chip assays registered complexes with RNAPol for all three PIs in Figure 4 (D and E). However, transcription efficiency from appY-associated PI1 seemed to be much weaker than that from PI3, containing promoters for ompT and pauD and PI2 carrying out antisense transcription from ompT (Figure 4F). This curious observation prompted us to verify promoter activity within the appY genomic locus.
In perfect conformity with ChIP-on-chip data (Figure 4D and E), the 406-bp DNA fragment, containing both the expected intergenic promoters and unusual intragenic promoter-like sites within appY, formed two major heparin-resistant complexes with RNAPol (Figure 5A). Their relative abundance depended on enzyme:DNA ratio, supposing two or more sites for simultaneous interaction with the enzyme. Permanganate footprinting revealed four transcription bubbles, assuming formation of transcriptionally competent open complexes (Figure 5B). Three of them lie ∼190 (I), ∼155 (II) and ∼90 bp (III) upstream from the appY ATG codon, and are suitable candidates for appY promoters, while the fourth one is located within the coding sequence (Figure 5C). Single round transcription assay performed with the same DNA fragment (Figure 5D, lane 2) testified synthesis of two major RNA-products: ∼305 and ∼237 nt. Two additional templates were used to ascribe them to particular transcription bubbles. The longer product may be initiated at either PII, or PIV (Figure 5C). It remains unchanged when template shortened in the coding region was used (lane 3), while becomes ∼65 nt longer when DNA fragment elongated in opposite direction was taken for transcription assay (lane 1). Thus we conclude that ∼305 nt-RNA is initiated at PIV and is transcribed in antisense direction. Since the ∼237 nt-product, vice versa, remains unchanged when the longer DNA fragment is used but becomes shorter in the case of the shortened template, it can be transcribed only from PIII towards the coding sequence of appY and, therefore, may represent appY-mRNA. However, reverse transcription from total RNA failed to reveal the expected cDNAs in any of five experiments performed in different conditions (Figure 5E).
Figure 5.
Experimental verification of promoter activity within PI1 (Figure 4B). All procedures are described in ‘Materials and methods’ section. (A) Gel-shift assays were performed at different RNAPol–DNA ratios as indicated above the gel. (B) Local DNA melting as revealed by potassium permanganate footprinting. The 5′-end of F1 (C) was 32P-labeled. ‘G’-sequencing ladder. Arabic ciphers on the right indicate positions of unpaired thymines respective to the appY ATG codon. Transcription bubbles are denoted by Roman numerals. (C) Positioning of transcription bubbles (I–IV) in the analyzed DNA fragment. Horizontal lines mark unpaired thymines. Bent arrows with size roughly reflecting the value of S show expected directions of transcription. The lengths of the expected RNA products to the end of DNA fragment amplified with F1 and R1 are indicated nearby. Arrows are white if corresponding RNAs were registered either in vitro (D) or in vivo (F, E). Straight arrows show directions of sense and antisense transcription according to appY. (D) Single-round transcription assay. Arrows show transcripts with migration rate altered due to changed length of the templates. End-to-end products are indicated by asterisks (‘a’ and ‘s’ denote antisense and sense directions). (E) and (F) Products of reverse transcription from the total RNA using R1 (E) and F1 (F). Bacterial cells were harvested at logarithmic and stationary phases as indicated. Lanes M and ciphers nearby calibrate gels on D–F plots.
Low-transcription efficiency of appY in vivo is consistent with the expression data [Figure 4F, (13)] and the data obtained by Isalan et al. (37), who tested nearly 600 cross-combinations of genes encoding transcription regulators or σ-factors and their promoters. Reporter gene expression [Supplementary 1_GFP; (37)] indicated increased transcription in 17 out of 22 constructions when cognate promoters were replaced by appY regulatory region, thus assuming very high activity of appY promoter. The effect, however, was quite opposite if the appY coding sequence was fused to different promoters. Transcription efficiency of the reporter gene significantly decreased practically for all promoters. Thus, it appears to be that strong interaction with RNAPol and even ability to form open complexes do not necessarily result in efficient RNA synthesis. In the case of appY transcription depends on impediments buried in its coding sequence. Intragenic promoter-like sites certainly may play such a role. Genomic DNA of E. coli, therefore, contains several types of promoter-like sites with poorly understood function. ChIP-on-chip data were used to evaluate the ability of predicted promoters to bind RNAPol in the genome-wide scale.
Coordinates of predicted promoters overlap with RNAPol-binding sites found in vivo
Three questions were addressed to compare in silico and in vivo data:
how many predicted TSPs are associated with the registered sites of RNAPol binding;
how many RNAPol-binding sites are associated with the predicted TSPs;
how many PIs form complexes with RNAPol in vivo?
To answer the first question, we used the whole set of 30 188 predicted TSPs. They form 4810 clusters or single promoter-like sites separated from each other by at least 100 bp (overlapping clusters on opposite DNA strands were combined). Predicted TSPs or TSP clusters were considered as confirmed by in vivo data, if at least one positive chip probe was found by Reppas et al. [(13); experiment B] within a ± 100 bp surrounding region (Figure 2B and C illustrate scrutinized area). Ninety-four percent of individual TSPs (or 77% of promoter clusters) fall into genomic loci interacting with RNAPol in vivo or are located within the allowed distance. Thus, most predicted promoters of all types perform at least one promoter function.
To address the second question, we used the set of microarray probes efficiently hybridized with enzyme-trapped DNAs. A total of 51 922 probes show fluorescent signal at least 2-fold higher if RNAPol–DNA complexes were extracted using specific antibodies (Figure 2F). They represent 1209 continuous genomic regions. RNAPol residence sites observed in vivo were considered related to potential promoters if at least one TSP with S > 7.44 was found within region covered by oligonucleotide probes or in ±100 bp surrounding area. Ninety-three percent of individual probes (86.8% of continuous regions) appeared to be associated with predicted promoter sites.
All 78 promoter ‘islands’ form extensive complexes with RNAPol (Supplementary Table 2). This coincident disposition of TSPs found in silico and polymerase-binding sites registered in vivo allows considering coordinates of predicted TSPs as a valid map of transcription signals.
Scores of predicted promoters correlate with efficiency of RNA polymerase binding
At the last step we estimated the degree of correlation between the values of PlatProm scores, and efficiencies of RNAPol binding. Since S reflects probability of any single nucleotide in the genome to be a TSP, while ChIP-on-chip method estimates efficiency of RNAPol binding to a particular genomic region represented by 50 nt probes on high-density tiled microarrays, the pattern of S should be converted into a comparable mode. Converting procedure should take into account that the geometrical center of RNAPol-binding site does not coincide with the TSP. Moreover, as RNAPol in vivo interacts with double-stranded DNA, samples prepared for chip-hybridization contained both strands of enzyme-trapped fragments. Thus, fluorescent signal registered for a particular probe may result from two types of complexes differing in enzyme orientation (Figure 4A). Supposing each 50 nt probe as a target for interaction with polymerase we thus considered the sum of S corresponding to the virtual TSPs on both strands as a probability of enzyme binding (P):
| 1 |
As the spacer length and the distance between the TSP and −10 element differ in real promoters, ±8 nt variation in +1 positioning has been allowed. Location of these regions relative to 5′-ends was selected experimentally as position giving maximal correlation coefficient (R) between P and hybridization signals. Figure 4C shows the predicted TSPs of the Figure 4B converted in such a way. X-axis is shifted 1.96 SD (1 SD = 55.2) above the background level of P (average value of all negative P), providing p < 0.05 for positive values.
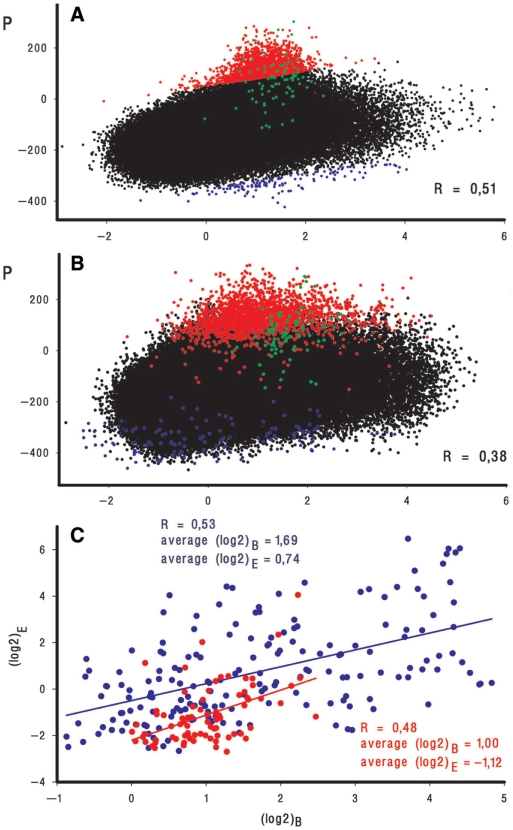
Log2 ratios [(log2)B] reflecting efficiencies of RNAPol binding in vivo, obtained by Herring et al. in experiment 2 (12) and by Reppas et al. in experiment B (13) were used to estimate correlation with P. Even though hybridization signals registered by two experimental assays differ in amplitude (Figure 4D and E), their profiles resemble each other and the pattern of P (Figure 4C). Quantified values of R are 0.51 (RH) and 0.38 (RR), respectively (Figure 6). RH is comparable with R values estimated for three replicates performed in similar conditions by the same research group (R = 0.46; 0.48 and 0.52) (12).
Figure 6.
Correlation between P and efficiency of enzyme binding (log2)B as registered by ChIP-on-chip assays in experiment 2 carried out by Herring et al. (12) (A) and experiment B performed by Reppas et al. (13) (B). Only the ‘+’ strand probes [185 519 and 191 088 for (A) and (B), respectively] are plotted. Outlying points on plot (A) are dissected by invisible lines equally shifted above and below the median and colored. Red and blue symbols on plot (B) represent the same genomic regions in the Reppas et al. (13) data set. Since blue outliers in the second data set tend to fit better to the expected range of log2 ratios, this slight deviation was not considered as regular and was not further analyzed. Points representing promoter ‘islands’ 1–3 (Figure 4B) are shown in green. The indicated values of R were quantified for the whole set of microarray probes using (log2)B and P averaged within running window 3. (C) Correlation between RNAPol binding (log2)B and expression (log2)E efficiencies plotted for 78 PIs (red symbols) and 181 single promoters (blue symbols). For PIs (log2)B were quantified as average values of ChIP-on-chip signals for all probes covering corresponding genomic areas (Supplementary Table 2). (Log2)E was first quantified as an average log2 ratio of chip signals obtained upon cDNA hybridization with 10 probes representing the 250-bp region downstream from the last predicted TSP on each strand. Two values characterizing transcription in both directions were then compared and the largest one was considered as the measure of expression efficiency. Other variants of (log2)E calculation see in Supplementary Table 2. (Log2)B and (log2)E for ‘normal’ promoters were quantified within 250 bp region upstream and downstream from the position +1, respectively (Supplementary Table 3). Both (log2)B and (log2)E were averaged for two replicates of Reppas et al. (13) data set. First-order regression lines show apparent dependence between (log2)B and (log2)E for both promoter ‘islands’ and ‘normal’ promoters.
Are these RH and RR values high or not? Although the data obtained in silico and in vivo reflect the same phenomena: anticipated and registered interaction with RNAPol, there are some peculiarities decreasing evaluated correlation. PlatProm, for instance, accounts putative modules for interaction with regulatory proteins as positive elements, though repressors decrease polymerase binding. Many weak RNAPol-binding sites are not detected in vivo due to low signal-to-noise ratio (12) but PlatProm discriminates them. Intensity of ChIP signals from some promoters may disproportionately decrease due to possibility of epitope occlusion by other interactions (12). Finally, PlatProm is currently attuned to σ70-specific promoters, while σ70-specific antibodies precipitate σ28 as well (13). Usage of β′-specific antibodies in combination with rifampicin-chase selection allows promoters of all types to be extracted. Considering all these realities, we conclude that both values of R are rather high.
Promoter ‘islands’ have unusual functional features
The shape of Figure 6A reveals 2106 outliers with lower than expected hybridization signals (red symbols), which can not be attributed to experimental or technical errors, since oligonucleotide probes to the same genomic regions in the other ChIP-on-chip data set also tend to weaker hybridization (Figure 6B). This phenomenon can not be also explained by repressor functioning since the percentage of probes representing genes with inhibited transcription (22%) among these outlying probes approximately equals the percentage (25%) of such genes in the genome [RegulonDB 5.5 (38)]. Further analysis showed that almost all out-of-ordered probes correspond to regions with multiple promoter-like sites and every one of 78 promoter ‘islands’ turned out to be represented in the prominent part of Figure 6A. They comprise 32% of all the deviating probes, rather than 0.7% expected by chance. RH and RR for promoter ‘islands’ are low: 0.09 and 0.17, respectively. Thus, it appears that polymerase recognizes any single promoter-like site within the region of multiple competing promoters with less predictable efficiency than across the whole genome. High density of promoter-like sites tends to weaken rather than strengthen RNAPol binding.
To explore further this phenomenon a set of 181 known promoters accurately recognized in silico and possessing no more than two additional PlatProm signal(s) on either strand in a ±50 bp area was compiled (Supplementary Table 3). Amid others it contains promoters PfadB, PcusC, and PcusR (Figures 2 and 4). Only 21 promoters from this set contribute to the population of 2106 outlying probes by 42 representatives, which is exactly as much as expected by chance (∼2%). The set of ‘normal’ promoters was compared with 78 PIs in terms of RNAPol binding and transcription efficiency calculated as described in Figure 6 legend (Supplementary Tables 2 and 3, and Figure 6C). All PIs and 87% of ‘normal’ promoters (Supplementary Tables 2 and 3) form complexes with RNAPol. At the same time RNA synthesis has been detected for 107 ‘normal’ promoters (59%) but only for 20 out of 156 (78 × 2) genomic loci flanking promoter ‘islands’ (13%). Figure 6C illustrates expression efficiency and RNAPol binding for two compared groups. Points representing promoter ‘islands’ are mainly concentrated in the bottom part of the plot. The yield of particular RNAs in the cell may, therefore, depend on the presence of competing promoters, which makes genomic regions differing in abundance of transcription signals dissimilar in terms of functional characteristics.
DISCUSSION
Comprehensive annotation of newly sequenced genomes implying mapping of genes along with their regulatory elements is currently becoming a long-term strategy appealing for effective promoter-finders. In the case of genes encoding proteins, rRNAs and tRNAs, which by itself are accurately identified by almost perfectly attuned computer programs, promoter-finders may assist in localization of regulatory regions. In the case of genes encoding small untranslated RNAs they are used to point out positions where sRNAs may be initiated (29,39), whereas other programs classify potential products as putative sRNAs based on their thermodynamic features and folding propensity. In the case of aRNAs promoter-finders may provide unique indicators of potential antisense transcription. So far only two dozens of aRNAs at least partly synthesized from the antisense strands of other genes have been found in E. coli. This rather small number apparently does not reflect their multiplicity, as genome-wide expression studies registered a huge amount of RNAs generated from antisense strands (30); PlatProm predicted many potential promoters for antisense transcription, while ChIP-on-chip technique detected suitable intragenic RNAPol-binding sites (12,13). Even though many signals registered by microarrays may result from run-through transcripts unterminated at 3′-ends of neighboring genes, some promoters predicted in silico may be false positives while intragenic RNAPol-binding sites may represent paused elongation complexes, combined data vote for antisense transcription in bacteria.
Along with potential TSPs for antisense transcription our screen revealed a large number of intragenic promoters with sense orientation and preferred location near the initiating codon. Though expediency of alternative transcription remains vague, very similar phenomenon has been recently registered in eukaryotic genomes [see, e.g. ref. (40)]. In the chromosome of E. coli, there are several pairs of extensively overlapping genes encoding two proteins or a protein and an untranslated RNA. Many of them may be transcribed from common promoters. However genes hokC, tpr and ygeN with 5′-ends mapped within mokC, rttR and ygeO, respectively, as well as 46 transcripts partly matching sense strand and detected in the fraction of short RNAs by Vogel et al. (31) may be also initiated from their own promoters predicted by PlatProm. Internal promoters with sense orientation may, thus, mark candidates for alternative transcription.
The data obtained drew attention to 78 regions with high density of TSPs over long distance. Fifty-five of them at least partly overlap with intergenic loci presumed to contain active promoters, nine of which are mapped experimentally. They initiate RNA synthesis from single or several (up to eight) positions. Multiplicity of the TSPs might, therefore, be exploited by transcription machinery. Four promoter-rich clusters lie between convergently transcribed genes like in the case of appY/ompT (Figure 4B). Their transcription activity, if detected, assumes either presence of yet unidentified genes in intergenic regions or antisense transcription in one or both directions. Putative sRNA IS021 (39), for instance, may be synthesized between appY and ompT, while gene for sRNA HB_171 is predicted between ygcE and ygcF (29). The most interesting are the remaining 19 PIs either immersed into one coding sequence or covering the junction point between two operonic genes, where promoter activity is not required.
Even though all 78 promoter ‘islands’ bind RNAPol in vivo, transcription has been detected for only 13 genomic loci (Figure 6C). In all but one case (PI submerged into the wbbK coding sequence) they occupy intergenic regions. PI1 (Figure 4B) exemplifies cluster with weak expression observed in only one experiment (Supplementary Table 2). Our data show that RNAPol can interact with PI1 and initiate RNA synthesis in both directions in vitro but primer extension (Figure 5E) failed to reveal the presence of appY-mRNA among RNAs produced in vivo. This transcriptional silence makes the value of R between promoter scores within PIs and corresponding (log2)E statistically insignificant, whereas faint correlation between these parameters was registered for the subset of ‘normal’ promoters (0.16) and even the whole set of predicted promoter-like regions (0.075). That means that the overall transcription from multiple TSPs does not represent the sum of expected outputs from individual ones. Some sites seem to bind RNAPol without initiating productive synthesis, thus performing only part of functions commonly ascribed to promoter regions. If so, they may be considered as a kind of suppressor elements.
Currently it is not clear whether promoter ‘islands’ found in this study and ‘clusters with high density of promoter-like signals’ (1,36) reflect one and the same phenomenon. Previously discovered clusters are defined as accessories of intergenic loci, surrounding functional promoters and participating in their proper regulation, while 23 PIs (∼30%) are located far from expected promoters. Ninety-two percent of E. coli genes have promoters surrounded by other promoter-like signals (36). This clustering, thus, should be considered as a feature typical for E. coli genes, while ‘single’ promoters should be ranked as a peculiarity. At the same time only 78 regions have extremely high density of potential TSPs assuming their special role in genome organization and function. Since DNA of all but three PIs has high potentiality to undergo stress-induced deformations [(10); http://www.genomecenter.ucdavis.edu/benham/sidd_database/], there is a reason to suggest a possibility that PIs participate in structural remodeling of bacterial chromosome.
Our conclusions were drawn on the basis of comparative analysis of data sets obtained in silico and in vivo. Although both methods have certain limitations if used separately, together they provide valid information on the genome positioning of RNAPol-binding sites and allow spotting a larger set of potential promoters. Thus, even weak binding signals associated with a predicted TSP may indicate the presence of an active promoter, whereas complex formation with the enzyme nearby low scoring promoters supports their functionality. Moreover, the combined use of two approaches allows mapping of all types of transcribed regions including those encoding untranslated RNA species, hardly identifiable by other computational screens.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Foundation for Basic Research (07-04-01066).
Conflict of interest statement. None declared.
REFERENCES
- 1.Huerta AM, Collado-Vides J. Sigma70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J. Mol. Biol. 2003;333:261–278. doi: 10.1016/j.jmb.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 2.Gordon L, Chervonenkis AY, Gammerman AJ, Shahmuradov IA, Solovyev VV. Sequence alignment kernel for recognition of promoter regions. Bioinformatics. 2003;19:1964–1971. doi: 10.1093/bioinformatics/btg265. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell JE, Zheng D, Busby SJW, Minchin SD. Identification and analysis of “extended -10” promoters in Escherichia coli. Nucleic Acids Res. 2003;31:4689–4695. doi: 10.1093/nar/gkg694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. Anatomy of Escherichia coli sigma70 promoters. Nucleic Acids Res. 2007;35:771–788. doi: 10.1093/nar/gkl956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacques P-E, Rodrigue S, Gaudreau L, Goulet J, Brzezinski R. Detection of prokaryotic promoters from the genomic distribution of hexanucleotide pairs. BMC Bioinformatics. 2006;7:423–437. doi: 10.1186/1471-2105-7-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brok-Volchanski AS, Masulis IS, Shavkunov KS, Lukyanov VI, Purtov YuA, Kostyanicina EG, Deev AA, Ozoline ON. Predicting sRNA genes in the genome of E. coli by the promoter-search algorithm PlatProm. In: Kolchanov N, Hofestaedt R, Milanesi L, editors. Bioinformatics of Genome Regulation and Structure. II. New York, NY, USA: Springer Science and Business Media Inc.; 2006. pp. 11–20. [Google Scholar]
- 7.Ozoline ON, Deev AA. Predicting antisense RNAs in the genomes of Escherichia coli and Salmonella typhimurium using promoter-search algorithm PlatProm. J. Bioinf. Comput. Biol. 2006;4:443–454. doi: 10.1142/s0219720006001916. [DOI] [PubMed] [Google Scholar]
- 8.Dekhtyar M, Morin A, Sakanyan V. Triad pattern algorithm for predicting strong promoter candidates in bacterial genomes. BMC Bioinormatics. 2008;9:233. doi: 10.1186/1471-2105-9-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanhere A, Bansal M. A novel method for prokaryotic promoter prediction based on DNA stability. BMC Bioinformatics. 2005;6:1. doi: 10.1186/1471-2105-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Benham CJ. Promoter prediction and annotation of microbial genomes based on DNA sequence and structural responses to superhelical stress. BMC Bioinformatics. 2006;7:248–263. doi: 10.1186/1471-2105-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wade JT, Struhl K, Busby SJ, Grainger DC. Genomic analysis of protein–DNA interactions in bacteria: insights into transcription and chromosome organization. Mol. Microbiol. 2007;65:21–26. doi: 10.1111/j.1365-2958.2007.05781.x. [DOI] [PubMed] [Google Scholar]
- 12.Herring CD, Raffaelle M, Allen TE, Kanin EI, Landick R, Ansari AZ, Palsson BO. Immobilization of Escherichia coli RNA polymerase and location of binding sites by use of chromatin immunoprecipitation and microarrays. J. Bacteriol. 2005;187:6166–6174. doi: 10.1128/JB.187.17.6166-6174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reppas NB, Wade JT, Church GM, Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol. Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 14.Grainger DC, Hurd D, Harrison M, Holdstock J, Busby SJW. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl Acad. Sci. USA. 2005;102:17693–17698. doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horton PB, Kanehisa M. An assessment of neural network and statistical approaches for prediction of E.coli promoter sites. Nucleic Acids Res. 1992;20:4331–4338. doi: 10.1093/nar/20.16.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedersen AG, Engelbrecht J. Investigations of Escherichia coli promoter sequences with artificial neural networks: new signals discovered upstream of the transcriptional start point. In: Rawlings C, editor. CA, USA: AAAI Press, Menlo Park; 1995. pp. 292–299. In Proceedings of the Third International Conference on Intelligent Systems for Molecular Biology. [PubMed] [Google Scholar]
- 17.Hertz GZ, Stormo GD. Escherichia coli promoter sequences: analysis and prediction. Methods Enzymol. 1996;273:30–42. doi: 10.1016/s0076-6879(96)73004-5. [DOI] [PubMed] [Google Scholar]
- 18.Estrem ST, Gaal T, Ross W, Gourse RL. Identification of an UP element consensus sequence for bacterial promoters. Proc. Natl Acad. Sci. USA. 1998;95:9761–9766. doi: 10.1073/pnas.95.17.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hivzer J, Rozenberg H, Frolow F, Rabinovich D, Shakked Z. DNA bending by an adenine-thymine tract and its role in gene regulation. Proc. Natl Acad. Sci. USA. 2001;98:8490–8495. doi: 10.1073/pnas.151247298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ozoline ON, Deev AA, Trifonov EN. DNA bendability - a novel feature in E. coli promoter recognition. J. Biomol. Struct. Dynam. 1999;16:825–831. doi: 10.1080/07391102.1999.10508295. [DOI] [PubMed] [Google Scholar]
- 21.Ozoline ON, Deev AA, Arkhipova MV. Non-canonical sequence elements in the promoter structure. Cluster analysis of promoters recognized by Escherichia coli RNA polymerase. Nucleic Acids Res. 1997;25:4703–4709. doi: 10.1093/nar/25.23.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozoline ON, Deev AA, Arkhipova MV, Chasov VV, Travers A. Proximal transcribed regions of bacterial promoters have a non-random distribution of A/T tracts. Nucleic Acids Res. 1999;27:4768–4774. doi: 10.1093/nar/27.24.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider TD, Stormo GD, Gold L, Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J. Mol. Biol. 1986;188:415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- 24.Leung S, Mellish C, Robertson D. Basic gene grammars and DNA-ChartParser for language processing of Escherichia coli promoter DNA sequences. Bioinformatics. 2001;17:226–236. doi: 10.1093/bioinformatics/17.3.226. [DOI] [PubMed] [Google Scholar]
- 25.Favre-Bonte S, Joly B, Forestier Ch. Consequences of reduction of Klebsiella pneumoniae capsule expression on interactions of this bacterium with epithelial cells. Infect. Immun. 1999;67:554–561. doi: 10.1128/iai.67.2.554-561.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaychikov E, Denissova L, Meier T, Gotte M, Heumann H. Influence of Mg2+ and temperature on formation of the transcription bubble. J. Biol. Chem. 1997;272:2259–2267. doi: 10.1074/jbc.272.4.2259. [DOI] [PubMed] [Google Scholar]
- 27.Ozoline ON, Fujita N, Ishihama A. Mode of DNA–protein interaction between the C-terminal domain of Escherichia coli RNA polymerase alpha-subunit and T7D promoter UP element. Nucleic Acids Res. 2001;29:4909–4919. doi: 10.1093/nar/29.24.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golby P, Kelly DJ, Guest JR, Andrews SC. Transcriptional regulation and organization of the dcuA and dcuB genes, encoding homologous anaerobic C4-dicarboxylate transporters in Escherichia coli. J. Bacteriol. 1998;180:6586–6596. doi: 10.1128/jb.180.24.6586-6596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter RJ, Dubchak I, Holbrook SR. A computational approach to identify genes for functional RNAs in genomic sequences. Nucleic Acids Res. 2001;29:3928–3938. doi: 10.1093/nar/29.19.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selinger DW, Cheung KJ, Mei R, Johansson EM, Richmond CS, Blattner FR, Lockhart DJ, Church GM. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 2000;18:1262–1268. doi: 10.1038/82367. [DOI] [PubMed] [Google Scholar]
- 31.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EGH. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawano M, Reynolds AA, Miranda-Rios J, Storz G. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 2005;33:1040–1050. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saetrom P, Sneve R, Kristiansen KI, Snove O, Jr, Grunfeld T, Rognes T, Seeberg E. Predicting non-coding RNA genes in Escherichia coli with boosted genetic programming. Nucleic Acids Res. 2005;33:3263–3270. doi: 10.1093/nar/gki644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Opdyke JA, Kang JG, Storz G. GadY, a small RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tutukina MN, Shavkunov KS, Masulis IS, Ozoline ON. Intragenic promoter-like sites in the genome of Escherichia coli. Discovery and functional implication. J. Bioinf. Comput. Biol. 2007;5:549–560. doi: 10.1142/s0219720007002801. [DOI] [PubMed] [Google Scholar]
- 36.Huerta A, Francino MP, Morett E, Collado-Vides J. Selection for unequal densities of σ70 promoter-like signals in different regions of large bacterial genomes. PLoS Genetics. 2006;2:e185. doi: 10.1371/journal.pgen.0020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isalan M, Lemerle C, Michalodimitrakis K, Horn C, Beltrao P, Raineri E, Garriga-Canut M, Serrano L. Evolvability and hierarchy in rewired bacterial gene networks. Nature. 2008;452:840–845. doi: 10.1038/nature06847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salgado H, Gama-Castro S, Peralta-Gil M, Diaz-Peredo E, Sanchez-Solano F, Santos-Zavaleta A, Martinez-Flores I, Jimenez-Jacinto V, Bonavides-Martinez C, Segura-Salazar J, et al. RegulonDB (version 5.0): Escherichia coli K12 transcriptional regulatory network, operon organization and growth conditions. Nucleic Acids Res. 2006;34:D394–D397. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S, Lesnik EA, Hall TA, Sampath R, Griffey RH, Ecker DJ, Blyn LB. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. BioSystems. 2002;65:157–177. doi: 10.1016/s0303-2647(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 40.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]