Abstract
Every cell has to duplicate its entire genome during S-phase of the cell cycle. After replication, the newly synthesized DNA is rapidly assembled into chromatin. The newly assembled chromatin ‘matures’ and adopts a variety of different conformations. This differential packaging of DNA plays an important role for the maintenance of gene expression patterns and has to be reliably copied in each cell division. Posttranslational histone modifications are prime candidates for the regulation of the chromatin structure. In order to understand the maintenance of chromatin structures, it is crucial to understand the replication of histone modification patterns. To study the kinetics of histone modifications in vivo, we have pulse-labeled synchronized cells with an isotopically labeled arginine (15N4) that is 4 Da heavier than the naturally occurring 14N4 isoform. As most of the histone synthesis is coupled with replication, the cells were arrested at the G1/S boundary, released into S-phase and simultaneously incubated in the medium containing heavy arginine, thus labeling all newly synthesized proteins. This method allows a comparison of modification patterns on parental versus newly deposited histones. Experiments using various pulse/chase times show that particular modifications have considerably different kinetics until they have acquired a modification pattern indistinguishable from the parental histones.
INTRODUCTION
The packaging of DNA into chromatin plays a crucial role in regulating its accessibility for RNA polymerases and transcription factors. The level of chromatin condensation is dependent on the differentiation state of a cell, ranging from a hyper-dynamic, highly accessible structure in embryonic stem cells (1,2) to a less accessible more condensed form in senescent cells (3). The posttranslational modification of histones can alter the properties of chromatin fibers and is therefore a prime candidate to mediate the stable inheritance of chromatin structures. The histone code hypothesis (4,5) is further supported by genome-wide mapping studies of histone modifications (6,7), which show a high degree of correlation between particular histone modifications and RNA–Pol II occupancy. Based on these data, a complex set of modifications has been postulated to stably mark specific regions of the genome with regard to their further activity. Experiments in yeast (8), however, suggest that a putative histone code may reflect a relatively simple binary signal (9). An alternative model to explain the generation of complex histone modification patterns has been put forward by Schreiber and Bernstein who suggested that histone modifications function similar to modification networks involving receptor tyrosine kinases (10).
In order to distinguish between an inheritable histone code and the generation of modification patterns in response to external signals, it is crucial to investigate the histone modifications that occur during and immediately after histone deposition. During S-phase, newly synthesized and parental histones are distributed randomly on the two daughter strands (11,12) resulting in a dispersive replication of histone modification patterns (13). Newly synthesized histones have a distinct modification pattern (14,15) that matures after assembly into a pattern similar to the one observed on parental histones (16–18). As the histone modifications are considered to be causal for the maintenance of particular chromatin structures (5), mechanisms have to exist that allow a faithful copying of modification patterns from ‘old’ paternal histones to new ones. It is striking that many enzymes that catalyze a posttranslational modification of histones also carry domains known to bind to modified histone molecules or interact with factors containing such a domain (19–24). These domains could target histone-modifying or demodifying enzymes to particular regions within the genome by recognizing a modification pattern on the ‘old’ histones and copying it to the new ones.
The kinetics with which the modifications are copied from old histones to the new ones may define the time frame during which a given cell is susceptible for incoming signals and therefore its epigenetic plasticity. We investigated the kinetics of how fast modification patterns on new histones resemble the one on old ones by pulsed stable isotope labeling with amino acids in cell culture (pSILAC) and mass spectrometric analysis of histones. In accordance with previously published work, we observed a rapid adjustment of lysine acetylation patterns within the first 2 h after deposition (14,25,26). In contrast to this rapid acetylation and deacetylation, the methylation of lysine residues requires much more time to become indistinguishable from the old histones. In fact some of the modification patterns on newly incorporated histones remain different through most of the next cell cycle after deposition indicating that the histone code needs one full cell cycle in order to become fully re-established. Our findings have a major implication in our view for how histone modification marks may mediate epigenetic inheritance.
EXPERIMENTAL PROCEDURES
Synchronization of HeLa cells
For G1/S-phase synchronization, a double thymidine block was used. Therefore, HeLa cells were seeded and cultured for 24 h at 37°C in the R0 SILAC medium (Invitrogen). Thymidine (Sigma) was added to a final concentration of 2 mM and incubation was maintained for 16 h. The block was released by exchanging the thymidine-containing medium with the R0 culture medium. The cells were grown for 9 h before adding thymidine again to 2 mM final concentration for further 16 h to synchronize the HeLa cells at the G1/S border. The arrest was finally released by refeeding the cells with the thymidine-free R4 SILAC medium (Invitrogen) to allow cell cycle progression. Whenever indicated, 10 mM NaBu was added directly into the medium and the cells were washed twice with PBS before transferring them into the medium without NaBu. The cells were maintained in a 37°C incubator with a humidified atmosphere of 5% CO2.
SILAC labeling
We used three different SILAC DMEM media: R0 SILAC (l-arginine), R4 (L-12C6 15N4-arginine) and R10 (l-13C6 15N4-arginine). HeLa cells were synchronized at the G1/S border in the R0 SILAC medium and released into the cell cycle in the R4 SILAC medium in order to label all newly synthesized histones. For all pulse-chase experiments, we fed the synchronized cells with the R4 SILAC medium for 6 h and chased with the R10 SILAC medium. All materials for SILAC labeling were purchased from Invitrogen and prepared according to the manufacturer's instructions.
Flow cytometric analysis of the cell cycle
For fluorescence activated cell sorting (FACS) analysis, the cells (1 × 106) were harvested, washed twice in PBS followed by fixation in 70% ethanol at −20°C for a minimum of 1 h. Fixed cells were washed in PBS and incubated with 100 µg/ml of RNase A in PBS for 30 min at 37°C. Afterward, propidium iodide (Sigma) to a final concentration of 50 µg/ml was added and the cells were incubated at 37°C for 30 min. The samples were stored at 4°C in the dark until analysis on BD Biosciences FACSCanto. A minimum of 10 000 cells were counted, and the raw data were analyzed and histograms plotted using the FlowJo software.
RT–PCR
RNA was extracted from snap-frozen HeLa cell pellets using the RNeasy kit (Qiagen) according to the manufacturer's manual and dissolved in RNase-free water. Total RNA concentration was quantified using a spectrophotometer (Peqlab, Nanodrop). Reverse transcription was primed with 250 µg of random primers (Promega) with 1 μg of total RNA per sample at 70°C for 5 min. The samples were then incubated with 20 U MuLV in 20 µl of buffer containing 1000 µM dNTP and manufacturer's RT buffer for 10 min at 25°C, then heated up to 37°C for 1 h, 70°C for 10 min, chilled on ice and frozen at −20°C. PCR reaction was conducted with the following primers: H3.2 (5′-GCTACCAGAAGTCCACGGAG, 5′-GATGTCCTTGGGCATAATGG) and 18S (5′-TTGTTGGTTTTCGGAACTGAGG, 5′-CATCGTTTATGGTCGGAACTACG).
Histone extraction
The cell pellets were redissolved in 0.4 N HCl in a total volume of 0.5 ml per 1 × 106 cells and centrifuged for a minimum of 1 h at 4°C. After centrifugation at 13 000 rpm for 30 min, the supernatant was dialyzed at 4°C against 100 mM ice-cold acetic acid for three times for 1 h using 6–8000 MWCO. The sample was concentrated using a speed vac and redissolved in SDS-loading buffer and applied to a SDS–PAGE gel for further analysis.
MALDI-TOF analysis
Histones were separated by 18% SDS–PAGE. G-250 Coomassie blue stained bands were excised and destained with 50 mM ammonium bicarbonate in 50% ACN (Sigma) for 30 min at 37°C. After washing the gel pieces with HPLC grade water, histones were chemically modified by treating with 2 µl propionic anhydride (Sigma) and 48 µl of ammonium bicarbonate (1M) at 37°C. After 1 h, the modified histones were washed with HPLC grade water and digestions were carried out at 37°C overnight with 200 ng of sequencing grade trypsin (Promega) according to the manufacturer's manual. Digestion products were collected and the gel pieces were acid extracted in addition with 25 mM ammonium bicarbonate and afterward with 5% formic acid. The pooled digestions were concentrated using a speed vac and redissolved in 0.1% TFA. Afterward, the samples were desalted with ziptip µc18 (Millipore) according to the manufacturer's instructions and directly eluted onto the target plate with a saturated solution of α-cinnamic acid in 0.3% TFA and 50% ACN. The target plate was loaded into the Voyager DE STR spectrometer (Applied Biosystems) and analyzed. Peptide mass fingerprint covered the mass range from 500–2000 amu.
Quantification of MALDI signals
To determine all modifications occurring on histones, the Manuelito software was used. Spectra were processed and analyzed with the Data Explorer software (Applied Biosystems). For quantification, the integrated area of the peaks was used with a signal-to-noise ratio of 2. The sum of the area from all peaks derived from a single peptide was defined as 100%. Charts were drawn in Excel.
RESULTS
Pulse labeling can be used to mark newly synthesized histones
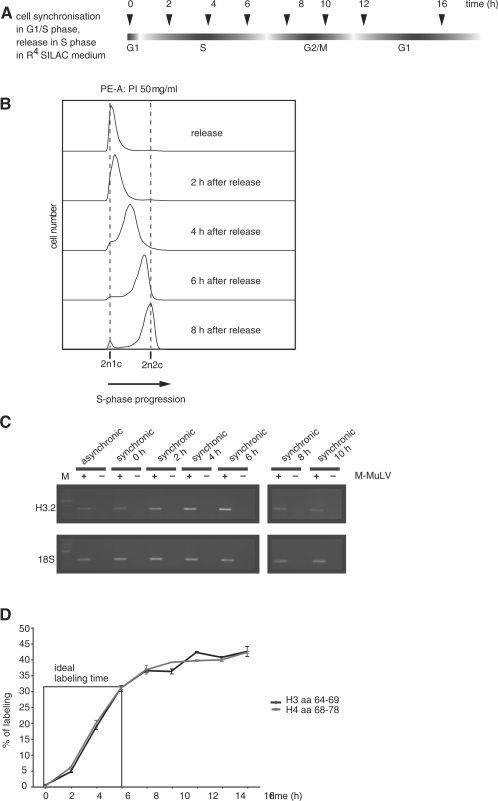
Most of the new histones that are incorporated during replication are synthesized during S-phase [(27) and Figure 1B and C]. To study the dynamics of histone modifications on the newly synthesized histones in comparison to the parental histones, we designed a method to selectively label new histones by isotopic labeling (Figure 1A). To do this, we arrested HeLa cells at the G1/S boundary using a double thymidine block (28) and verified the cell cycle arrest using fluorescence-activated cytometry (Figure 1B). After removal of the block, the cells were placed into the labeling medium containing isotopically labeled arginine (15N4). To control the correct replication-dependent synthesis of histone mRNA, we analyzed RNA at different times after removal of the cell cycle block (Figure 1C). As cells went into S-phase, the amount of histone H3.2 mRNA (29) increased substantially indicating that the histone mRNA synthesis is not affected by the heavy medium. Isotopically labeled histones also accumulate with a kinetic similar to the mRNA (Figure 1D). We measured the total level of incorporation by comparing the integrated peak areas of H3 and H4 peptides that were not modified in our samples [H3 64–69, m/z = 844.5 (light) or 848.5 (heavy) and H4 68–78, m/z = 1346.7 (light) or 1350.7 (heavy)]. A continuous labeling during the first 8-h postrelease resulted in a continuous increase in the histone synthesis. Theoretically, one would expect the level of incorporation to be 50% assuming that the total amount of histones is doubled and that all newly synthesized histones incorporate exclusively labeled arginine. In our experiments, the labeling efficiency is ∼40% of all H3 and H4 molecules (Figure 1D). This discrepancy may either be due to the fact that (i) not all cells enter S-phase after synchronization (Figure 1B), (ii) not all histones are incorporated into chromatin or that (iii) residual pools of light arginine are used in HeLa cells to synthesize new histone molecules. To quantify the latter, we measured the incorporation of heavy arginine in the H3 peptide containing amino acids 41–49. This peptide has the sequence YRPGTVALR, which is not cleaved by trypsin after R42 and therefore contains two arginines. It can get singly or doubly labeled depending on the amount of light arginine still present in the cell (Supplemental Figure S1). We do get a low level of singly labeled peptide of ∼5%, suggesting that ∼5% of all arginines used for the histone synthesis are in fact derived from endogenous amino-acid pools.
Figure 1.
pSILAC can be used to distinguish old from newly synthesized histones. (A) Schematic experimental overview. HeLa cells were synchronized using a double thymidine block. Cells were released into S-phase and simultaneously grown in the R4 SILAC medium in order to label all newly synthesized proteins. The cells were harvested at indicated time points. (B) DNA content of synchronized HeLa cells after different time points of release into S-phase using FACS analysis. The fluorescence intensity (DNA content) is depicted on the abscissa. (C) mRNA amount of H3.2 for asynchronic and synchronic cells detected by reverse transcriptase PCR. 18S serves as a loading control. M, marker. (D) Labeling efficiency when using the SILAC medium (R4). Two peptides H3 aa 64–69 and H4 aa 68–78 are analyzed by MALDI-TOF from 0 h to 16 h after release into S-phase. Error bars indicate the standard error of the mean (SEM) from three independent biological replicates.
Histone acetylation
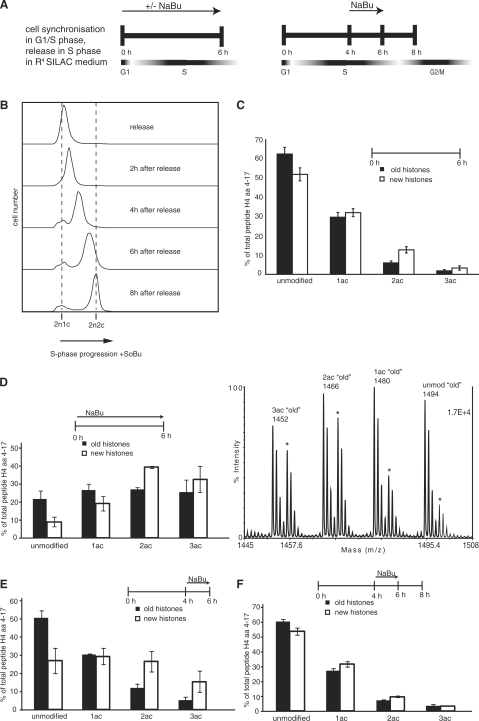
One of the best-known modifications on histones is the acetylation of lysines, which is considered to lead to a more open and hence to a more active chromatin structure (30). Histones are acetylated at specific sites before deposition (14,15), which get deacetylated after histone assembly (16,18,25). We wanted to determine, whether our analysis would allow us to dissect the process of acetylation and deacetylation coupled with histone deposition in vivo. Therefore, we compared the levels of lysine acetylation on newly synthesized histones with that on old histones. When histones were analyzed 6 h after the release into S-phase, we observed a small but reproducibly an increased proportion of the diacetylated isoform in the new histones (Figure 2C). As free H4 is exclusively diacetylated before deposition (14,15,25), this increased level of diacetylated H4 reflects the higher percentage of pre-deposition forms with the pool of new histones. In order to get an estimate of how quickly the pre-deposition markers disappear, we used the broad-range deacetylase inhibitor sodium butyrate (NaBu) (Figure 2A). When treating the cells with NaBu throughout the labeling period, we observed an increase in acetylation on the old as well as on the new histones, suggesting that histone acetyltransferases as well as histone deacetylases do not distinguish between old and new histones. However, whereas NaBu treatment leads to similar amounts of all acetylated isoforms among old histones [mono-, di- and triacetylated; tetraacetylated H4 could not be analyzed by MALDI-TOF due to an overlap of the heavy peptide H4ac4 (4–17) with an unmodified H4 peptide], in the case of the new histones, H4ac2 is the most abundant isoform (Figure 2D). This was not due to a NaBu-induced delay of the cell cycle, as treated cells entered S-phase approximately at the same rate as untreated ones (compare Figures 1B and 2B). Besides the enrichment of the diacetylated H4, we also saw a strong increase in the triacetylated form of the new H4 molecules, suggesting that a subsequent acetylation that follows incorporation is independent of a deacetylation event at the other residues (Figure 2D). This acetylation occurs very likely at K16, as this is the major acetylation site in human cells (31), and the diacetylated deposition form of H4 is acetylated at K5 and K12 (14–17). When NaBu was added 4 h after the start of the histone synthesis, we observed a substantial increase in the monoacetylated and the unmodified forms of new H4 suggesting that the pre-deposition modifications are removed within <2 h after assembly (Figure 2E). The higher percentage of diacetylated H4 on the new histones compared to the old ones is very likely due to the ongoing assembly at 4–6 h, after the beginning of S-phase. If, after treatment, the histones were incubated for another 2 h in the absence of NaBu, the acetylation patterns become indistinguishable between the old and new histones (Figure 2F). In summary, we conclude from the analysis of acetylation kinetics of new and old histones that the acetylation is rapidly adjusted to equalize the modification patterns between old and new histones. Due to the high turnover of acetyl groups (32), the pattern of acetylation does not seem to be well suited to confer stable epigenetic memory. Moreover, as acetylation events occur independent from each other, we have no evidence for a regulated establishment of a specific modification pattern that is based on distinct pre-existing modifications. This is in agreement with what has been previously described in yeast (33) and human cells (8).
Figure 2.
Deacetylation of H4 peptide 4–17 happens fast. (A) Scheme of pulse-chase experiments including 10 mM sodium butyrate treatment. Sodium butyrate (NaBu) was added either at the time of release for 6 h or for a shorter period of time (2 h) during S-phase. (B) FACS analysis of synchronized HeLa cells treated with sodium butyrate harvested at 0, 2, 4, 6 and 8 h after release. (C) Acetylation patterns of H4 peptide 4–17 (GKGGKGLGKGGAKR) of ‘old’ (R0) and ‘new’ (R4) histones after 6 h after a release into G1/S-phase. Error bars indicate the SEM from three independent biological experiments. 1ac, monoacetylation; 2ac, diacetylation; 3ac, triacetylation. (D) Comparison of acetylation patterns of ‘old’ and ‘new’ histones after 6 h of NaBu treatment. Left: quantification; right: MALDI-TOF spectrum; asterisk indicates peaks of ‘new’ histones. (E) Comparing acetylation patterns of ‘old’ and ‘new’ histones when treating for 2 h with NaBu without additional chase or with an additional 2 h chase (F).
Histone methylation
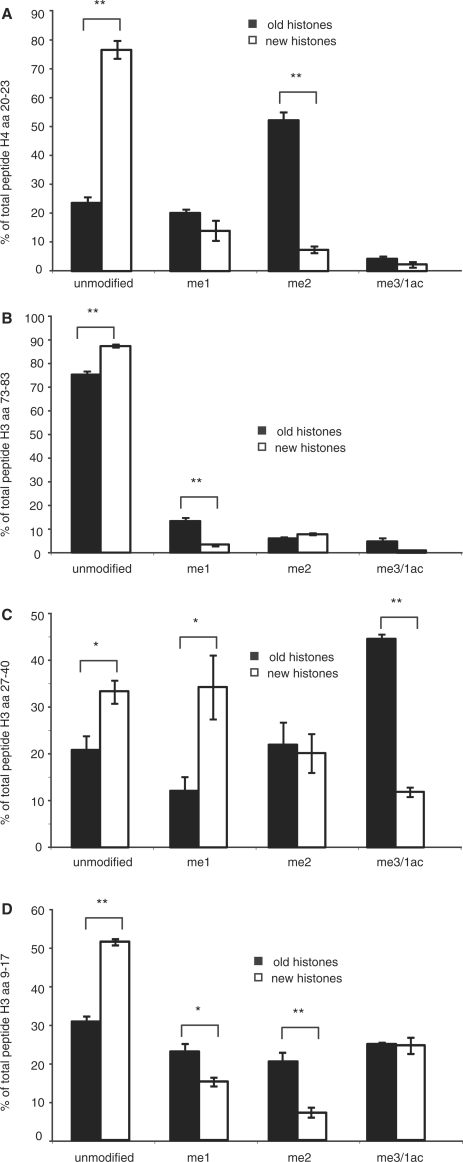
In contrast to acetylation, the methylation of lysine residues on histone tails has been suggested to constitute a major factor in the establishment and inheritance of stable chromatin states (4,5,34). We therefore wondered whether the methylation of histones might show a different behavior than acetylation. We focused our studies mainly on the most prominent methylations on H3 and H4 (K4, 9, 27, 36 and 79 on H3 and K20 on H4). In contrast to the highly dynamic lysine acetylation, lysines on the histone tails of newly synthesized histones are methylated with a much slower kinetic. When we compared the methylation levels of H3 and H4 after 6 h of pSILAC labeling, we observed a striking difference in modification patterns between old and new histones (Figure 3). As the cells progress through the cell cycle, methylation patterns on new histones gradually adopt those on the old histones. This is most obvious for the methylation of lysine 20 within histone H4 (Figure 3A). On newly synthesized H4 molecules, K20 is usually not modified (15,16) and becomes monomethylated after incorporation into chromatin (16,26). This is in sharp contrast to the methylation pattern on the pre-existing H4 molecules that are mainly dimethylated at K20 (Figure 3A).
Figure 3.
Differences in methylation patterns of ‘old’ and ‘new’ histones 6 h after release in G1/S-phase. (A) Methylation patterns of H4 peptide 20–23 of ‘old’ and ‘new’ histones analyzed by MALDI-TOF. 1ac, monoacetylation; me1, monomethylation; me2, dimethylation; me3, trimethylation. (B) H3 73–83 (C) H3 27–40 (D) H3 9–17. Error bars indicate the SEM of three independent biological replicates. The P-values are calculated using Student's unpaired t-test. The significance of the differences in methylation levels were assessed by unpaired Student's t-tests and indicated on the top of the bars **P < 0.01: a highly significant difference; *P < 0.05: a significant difference; and no bracket means no significant difference.
We observed a similar difference for the methylation on the H3 tail. Like in the H4 molecule, the replication-dependent H3 variants are not methylated to a measurable extent before incorporation (15,16). We concentrated our analysis on K9, K27, K36 and K79, as these have been suggested to play important roles during the inheritance of epigenetic states (5,34). In contrast to yeast cells, most of the H3 molecules remain unmodified at K79 in human cells (35,36). This makes it difficult to quantitatively compare the methylation levels of K79 on old and new histones. However, we observed a small but discernible difference between old and new histones with regard to K79 methylation. Interestingly, we saw more dimethylation of K79 on the new histones at 6 h after synthesis, whereas the old histones contain a higher percentage of monomethylated K79 (Figure 3B). The other lysines that carry methyl groups within H3 are K4, K9, K27 and K36. We limited our analysis on K9, K27 and K36, as the peptide containing K4 (3–8) gave only a very weak signal and appeared to be mostly unmodified in new as well as in old histones. Like H4K20me, the differences in methylation levels between old and new histones 6 h after the release into S-phase were clearly visible (Figure 3C and D). In the case of H3K9, the degree of mono- or dimethylation was considerably lower in the new histones when compared to the old ones and a large percentage of the new H3 molecules were still unmodified at that time point (Figure 3D). As we used MALDI-TOF for quantification, we could not distinguish between trimethylation and acetylation of the peptide, but based on earlier experiments (15) we suggest that most of the peptides contributing to this mass carry an acetyl group at K14. The methylation profiles of the peptides carrying K27 and K36 that were derived from the old and new histones are shown in Figure 3D and also show a marked difference between the two histone types. In contrast to the methylation at K9, we observed a higher percentage of monomethyation of this peptide, which was predominantly due to monomethylation at K27 (data not shown). Interestingly, within the first 6 h after release, this monomethylation does not seem to get further methylated to the trimethyl state on the new histones, and we do not observe a strong trimethylated peptide, which is the predominant one in the pre-existing histones (Figure 3D). In light of recent reports that show a localization of the main H3K27-specific histone methyltransferase EZH2 to sites of replication (37), the rapid monomethylation of H3K27 is very likely a product of the enzyme bound to replication foci.
Pulse chase labeling to follow histone modifications over one cell cycle
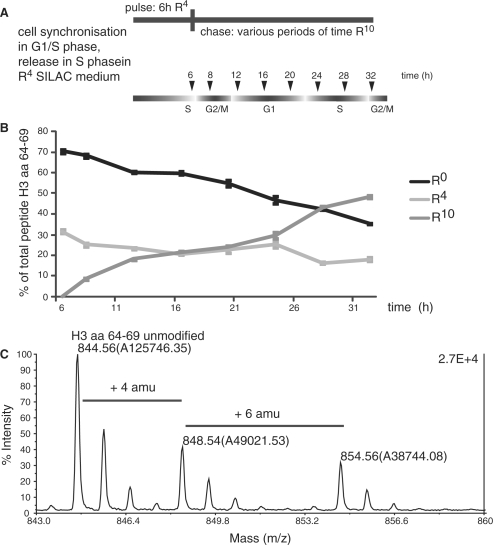
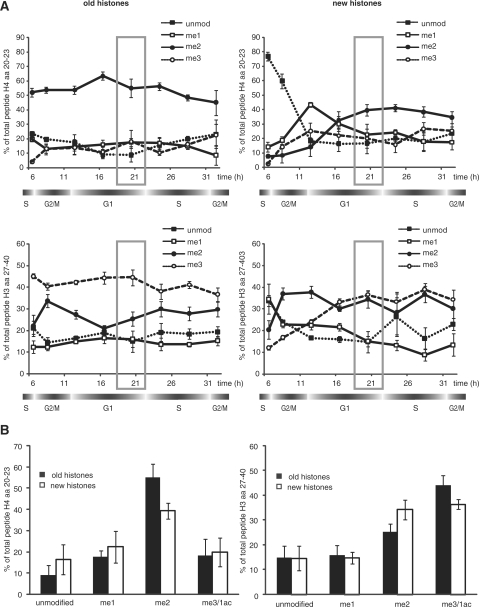
Our findings that the methylation patterns of lysine residues does not adopt the identical modification patterns immediately after deposition prompted us to ask how long it would take until the two modification patterns become indistinguishable. To do this, we pulse labeled the newly synthesized histones for 6 h using heavy arginine (R4) and subsequently chased the culture using super-heavy arginine (R10). This procedure is crucial for the analysis as it prevents the interference of histones incorporated at late S-phase, outside S-phase or during the next S-phase. As for R4 labeling, we measured incorporation by quantifying the peptides H364–69 and H468–78 (Figure 4 shows only the quantification of the H3 peptide; a representative spectrum is shown in Figure 4C). As the HeLa cells partly lost their synchrony during the second cell cycle, the incorporation of R10 into histones is not as clearly restricted to a defined time window but increases over the whole analysis time (Figure 4B). Therefore, we focused our analysis on the comparison between the old pre-existing histones and the ones incorporated during the first 6 h of S-phase. The samples were taken every 4 h after the end of the labeling period until the end of the following S-phase as judged by FACS analysis (∼30 h after release from the G1/S block). During this time, we only see minor changes in the global methylation patterns of the old histones arguing for a relatively stable marking system (Figure 5A, left panels). The methylation patterns of the newly synthesized histones on the contrary change slowly during the course of one cell cycle (Figure 5A, right panels). Similar to what had been reported before (26), H4K20 gets transiently monomethylated after deposition and subsequently dimethylated. At the beginning of the next S-phase, the H4K20 methylation levels of the new histones are very similar to the old ones (Figure 5B). This situation is slightly different in the case of the peptide containing H3K27 and K36, where a dimethylation transiently peaks during G2/M after which the amount of K27/36me2 declines and the trimethylated peak increases (Figure 5A, bottom right panel). As in the case of H4K20me, the methylation of K27/36 is adjusted to the pattern of the parental histones during the following G1, such that it is very similar to the parental one at the beginning of the next S-Phase (Figure 5B).
Figure 4.
Double labeling using different SILAC media allows to follow histones for a period longer than one cell cycle. (A) Experimental scheme. After synchronization, HeLa cells are released into S-phase under the R4-medium conditions. After 6 h, the cells were transferred into a medium containing R10 and then harvested at indicated time points. time (h); time after release. (B) Incorporation efficiency of double-labeled histones for R4 and R 10 showing H3 peptide aa 64–69. Error bars indicate the SEM of three independent biological replicates. (C) MALDI-TOF spectrum of H3 aa 64–69 from HeLa cells that were R4 labeled for 6 h and afterward R10 labeled for additional 6 h.
Figure 5.
Establishment of posttranslational modification patterns of histones differ in their kinetics. Pulse-chase experiments as described in Figure 4A. Underneath the charts, a cell cycle scheme is depicted. time (h); time after release. Three independent biological experiments are shown and analyzed by MALDI-TOF. Error bars indicate standard deviation of a mean. Grey square indicates 20 h after release (A) top left: H4 peptide aa 20–23 is shown of the ‘old’ histones. Top right: H4 peptide aa 20–23 is shown of the ‘new’ histones. Bottom left: R0 (‘old’)-labeled histones showing H3 peptide 27–40. Bottom right: R4 (‘new’)-labeled histones showing H3 peptide 27–40. (B) Methylation patterns after 20-h release of H4 peptide 20–23 (left) and H3 peptide 27–40 (right) of ‘old’ and ‘new’ histones analyzed by MALDI-TOF. 1ac, monoacetylation; me1, monomethylation; me2, dimethylation; me3, trimethylation. Error bars indicate the SEM from three independent biological replicates.
DISCUSSION
Histone modifications are considered to constitute a second (epi)genetic code that operates to establish distinct chromatin structures and maintain them throughout several cell divisions. In order for this to happen, the histone modifications on the pre-existing old histones have to be copied on the newly deposited histones, which have a fundamentally different modification pattern when placed onto DNA (15,18). It has been shown that histone modifications indeed change during the cell cycle (38). However, no distinction has been made with regard to modifications present on old and new histones except for H4 (26). In order to investigate the mechanisms and the kinetics of modification pattern inheritance, we used a pulsed SILAC labeling technique that enabled us to selectively label the newly synthesized histones and subsequently compare modifications on old and new histones. In agreement with previous results (32), we found that the acetylation of histones is highly dynamic and regulated by a tight equilibrium of acetyltransferases and deacetylases. The situation is different in the case of the observed methylations. For all lysines investigated (except H3K79), we observed a transient peak in the monomethylated isoform, suggesting that the monomethylation is put onto the histones either at or immediately after histone deposition. Several histone methyltransferases have been shown to interact with the replication machinery (37,39,40) where they probably catalyze the monomethylation of histones (41,42). This early burst of monomethylation is followed by a relatively slow progression to higher levels of methylation. This is in accordance with earlier observations, which suggest that monomethylation occurs first and may indeed be a prerequisite for further methylations (16,43). This is especially interesting in the case of H3K27, where the methylase EZH2 is responsible for all degrees of methylation. The stepwise methylation suggests a highly regulated progression from lower to higher methylated forms. Such a regulation of the generation of higher methylation states has been reported for several HMT complexes, where histone-binding subunits are required to achieve these states (44,45). The trimethylated forms are much better binding partners for the structural proteins HP1 and Polycomb (46–48) that are thought to condense chromatin by preventing nucleosome remodelers from acting on them (49). It is therefore tempting to speculate that the slow trimethylation on newly deposited histones prevents a premature chromatin condensation and allows chromatin to adopt a structure that is more susceptible for external signals. This window of opportunity could allow cells to stably shift their gene expression profile when they are exposed to changing external signals, such as stem cells leaving their specific niche. On the other hand, the methylation state could also provide a means for the cell to detect the cellular age, as trimethylated isoforms will increase when senescent cells do not undergo continuous cell division (3,50). Especially, as no demethylase has been characterized so far that is able to remove methyl groups from H4, H4K20 may have an exquisite function in measuring cellular age (26).
We and others (26) have analyzed the modification kinetics in immortalized human cells (HeLa) of a tumorigenic origin. Many tumor cells have a markedly different histone modification pattern when compared to normal cells (51). This has to be taken into account when interpreting our data. It will be interesting to see whether primary cells have a similar kinetic or whether the fast replication of tumor cells may in fact prevent the cells from fully replicating the epigenome, which could in fact explain the epigenetic instability that is frequently observed in tumors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Boehringer predoctoral fellowship to A.N.D.S.; European Union LSHG-CT2006-037415 grants and the Deutsche Forschungsgemeinschaft (SFB/TR5, M4). Funding for open access charge: LSHG-CT2006-037415.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank S. Hake for the primer pair to study H3.2 mRNA synthesis. We are grateful to L. Israel and I. Forne-Ferrer for expert technical assistance and to A. Villar-Garea, P.B. Becker and Richard Page for critical reading of the manuscript and helpful comments. We also would like to thank the whole Imhof lab for constant support during the progress of the work.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 3.Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 4.Turner BM. Histone acetylation and an epigenetic code. BioEssays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 5.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Rando OJ. Global patterns of histone modifications. Curr. Opin. Genet. Dev. 2007;17:94–99. doi: 10.1016/j.gde.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl Acad. Sci. USA. 2005;102:5501–5506. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henikoff S. Histone modifications: combinatorial complexity or cumulative simplicity? Proc. Natl Acad. Sci. USA. 2005;102:5308–5309. doi: 10.1073/pnas.0501853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111:771–778. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]
- 11.Jackson V, Chalkley R. Histone segregation on replicating chromatin. Biochemistry. 1985;24:6930–6938. doi: 10.1021/bi00345a027. [DOI] [PubMed] [Google Scholar]
- 12.Sogo JM, Stahl H, Koller T, Knippers R. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J. Mol. Biol. 1986;189:189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- 13.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 14.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl Acad. Sci. USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 16.Scharf AN, Meier K, Seitz V, Kremmer E, Brehm A, Imhof A. Monomethylation of lysine 20 on histone H4 facilitates chromatin maturation. Mol. Cell Biol. 2009;29:57–67. doi: 10.1128/MCB.00989-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimamura A, Worcel A. The assembly of regularly spaced nucleosomes in the Xenopus oocyte S-150 extract is accompanied by deacetylation of histone H4. J. Biol. Chem. 1989;264:14524–14530. [PubMed] [Google Scholar]
- 18.Benson LJ, Gu Y, Yakovleva T, Tong K, Barrows C, Strack CL, Cook RG, Mizzen CA, Annunziato AT. Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J. Biol. Chem. 2006;281:9287–9296. doi: 10.1074/jbc.M512956200. [DOI] [PubMed] [Google Scholar]
- 19.Carre C, Szymczak D, Pidoux J, Antoniewski C. The histone H3 acetylase dGcn5 is a key player in Drosophila melanogaster metamorphosis. Mol. Cell Biol. 2005;25:8228–8238. doi: 10.1128/MCB.25.18.8228-8238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 21.Syntichaki P, Topalidou I, Thireos G. The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature. 2000;404:414–417. doi: 10.1038/35006136. [DOI] [PubMed] [Google Scholar]
- 22.Melcher M, Schmid M, Aagaard L, Selenko P, Laible G, Jenuwein T. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol. Cell Biol. 2000;20:3728–3741. doi: 10.1128/mcb.20.10.3728-3741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J, Thompson JR, Botuyan MV, Mer G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat. Struct. Mol. Biol. 2008;15:109–111. doi: 10.1038/nsmb1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 25.Annunziato AT, Seale RL. Histone deacetylation is required for the maturation of newly replicated chromatin. J. Biol. Chem. 1983;258:12675–12684. [PubMed] [Google Scholar]
- 26.Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol. Cell Biol. 2008;28:468–486. doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osley MA. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- 28.Xeros N. Deoxyriboside control and synchronization of mitosis. Nature. 1962;194:682–683. doi: 10.1038/194682a0. [DOI] [PubMed] [Google Scholar]
- 29.Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- 30.Wolffe AP. Chromatin Structure and Function. 3rd. San Diego, CA: Academic Press; 1998. [Google Scholar]
- 31.Taipale M, Rea S, Richter K, Vilar A, Lichter P, Imhof A, Akhtar A. hMOF Histone Acetyltransferase Is Required for Histone H4 Lysine 16 Acetylation in Mammalian Cells. Mol. Cell Biol. 2005;25:6798–6810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chestier A, Yaniv M. Rapid turnover of acetyl groups in the four core histones of simian virus 40 minichromosomes. Proc. Natl Acad. Sci. USA. 1979;76:46–50. doi: 10.1073/pnas.76.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pesavento JJ, Bullock CR, Leduc RD, Mizzen CA, Kelleher NL. Combinatorial modification of human histone H4 quantitated by two-dimensional liquid chromatography coupled with top down mass spectrometry. J. Biol. Chem. 2008;28:468–486. doi: 10.1074/jbc.M709796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sims RJ, III, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 35.Garcia BA, Barber CM, Hake SB, Ptak C, Turner FB, Busby SA, Shabanowitz J, Moran RG, Allis CD, Hunt DF. Modifications of human histone H3 variants during mitosis. Biochemistry. 2005;44:13202–13213. doi: 10.1021/bi050906n. [DOI] [PubMed] [Google Scholar]
- 36.Thomas CE, Kelleher NL, Mizzen CA. Mass spectrometric characterization of human histone H3: a bird's eye view. J. Proteome Res. 2006;5:240–247. doi: 10.1021/pr050266a. [DOI] [PubMed] [Google Scholar]
- 37.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 38.Bonenfant D, Towbin H, Coulot M, Schindler P, Mueller DR, van Oostrum J. Analysis of dynamic changes in post-translational modifications of human histones during cell cycle by mass spectrometry. Mol. Cell Proteomics. 2007;6:1917–1932. doi: 10.1074/mcp.M700070-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 40.Huen MS, Sy SM, van Deursen JM, Chen J. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J. Biol. Chem. 2008;283:11073–11077. doi: 10.1074/jbc.C700242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 42.Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 43.Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, Tempst P, Roeder RG, Zhang Y. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell. 2003;12:475–487. doi: 10.1016/j.molcel.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 45.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 46.Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 47.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 48.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl–lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 50.Sarg B, Koutzamani E, Helliger W, Rundquist I, Lindner HH. Postsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with aging. J. Biol. Chem. 2002;277:39195–39201. doi: 10.1074/jbc.M205166200. [DOI] [PubMed] [Google Scholar]
- 51.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.