Abstract
The universally conserved eukaryotic initiation factor (eIF), eIF1A, plays multiple roles throughout initiation: it stimulates eIF2/GTP/Met-tRNAiMet attachment to 40S ribosomal subunits, scanning, start codon selection and subunit joining. Its bacterial ortholog IF1 consists of an oligonucleotide/oligosaccharide-binding (OB) domain, whereas eIF1A additionally contains a helical subdomain, N-terminal tail (NTT) and C-terminal tail (CTT). The NTT and CTT both enhance ribosomal recruitment of eIF2/GTP/Met-tRNAiMet, but have opposite effects on the stringency of start codon selection: the CTT increases, whereas the NTT decreases it. Here, we determined the position of eIF1A on the 40S subunit by directed hydroxyl radical cleavage. eIF1A's OB domain binds in the A site, similar to IF1, whereas the helical subdomain contacts the head, forming a bridge over the mRNA channel. The NTT and CTT both thread under Met-tRNAiMet reaching into the P-site. The NTT threads closer to the mRNA channel. In the proposed model, the NTT does not clash with either mRNA or Met-tRNAiMet, consistent with its suggested role in promoting the ‘closed’ conformation of ribosomal complexes upon start codon recognition. In contrast, eIF1A-CTT appears to interfere with the P-site tRNA-head interaction in the ‘closed’ complex and is likely ejected from the P-site upon start codon recognition.
INTRODUCTION
Eukaryotic translation initiation requires at least nine initiation factors (eIFs) and occurs in two stages: assembly of 48S initiation complexes and their joining with 60S subunits (1,2). 48S complexes form by the scanning mechanism. First, eIF2, GTP and initiator tRNA form a ternary complex, which in cooperation with eIFs 3, 1 and 1A, binds to a 40S subunit yielding a 43S pre-initiation complex. Next, the 43S complex is loaded onto mRNA in a process that requires eIFs 4A, 4B and 4F. eIFs 4A/4B/4F bind to the cap-proximal region of mRNA and unwind its secondary structure, allowing 43S complexes to attach. The 43S complex then scans the 5′-untranslated region until it encounters an AUG triplet in a favorable context, stops and forms a 48S initiation complex with an established P-site codon–anticodon interaction. Codon–anticodon base-pairing induces eIF5-mediated hydrolysis of eIF2-bound GTP and release of Pi (3,4), which leads to partial dissociation of eIF2-GDP (5). eIF5B mediates subsequent dissociation of eIFs 1, 1A and 3 and residual eIF2-GDP from the 40S subunit and its joining with a 60S subunit. Hydrolysis of eIF5B-bound GTP is required for eIF5B's own release from the assembled 80S ribosome.
eIF1A plays pleiotropic roles throughout the initiation process. First, it stimulates attachment of eIF2-ternary complexes to 40S subunits (2). Second, together with eIF1, which enables scanning 43S complexes to discriminate against codon–anticodon mismatches and prevents premature eIF5-induced hydrolysis of eIF2-bound GTP and Pi release (3,4,6), eIF1A is also involved in scanning and initiation codon selection. According to a current model, eIF1 together with eIF1A promotes a scanning-competent ‘open’ conformation of the 43S complex that is characterized by opening of the entry ‘latch’ between h18 in the body and h34/rpS5 in the neck and establishment of a new head–body connection between h16 and rpS3 (7). The establishment of codon–anticodon base-pairing leads to tightening of the eIF1A-40S interaction (8) and dissociation of eIF1 (9), which switches the complex to a ‘closed’ conformation and relieves repression of hydrolysis of eIF2-bound GTP. Finally, eIF1A also acts during subunit joining: it binds directly to eIF5B (10,11), and this interaction is required for efficient subunit joining and GTP hydrolysis by eIF5B (12,13).
eIF1A is a universally conserved initiation factor. Its prokaryotic ortholog IF1 consists of a single oligonucleotide/oligosaccharide-binding (OB) domain (14). The central region of eIF1A forms an OB domain with a structure similar to that of IF1, but eIF1A also has a small folded C-terminal subdomain with two α-helices that constitutes a rigid body with the OB domain, and long unstructured N- and C-terminal tails (15; Figure 1A). The ∼25 amino acid-long N-terminal tail (NTT) is conserved and positively charged, whereas the ∼20-35 amino acid-long C-terminal tail (CTT) is negatively charged and less conserved.
Figure 1.
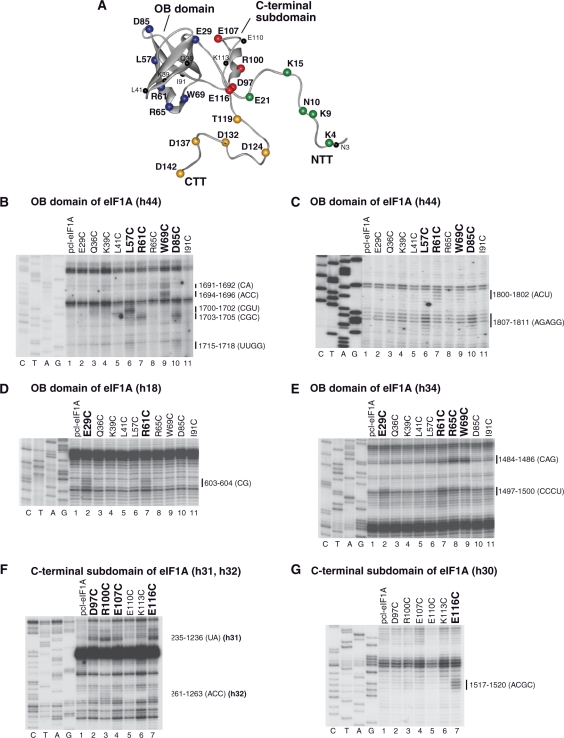
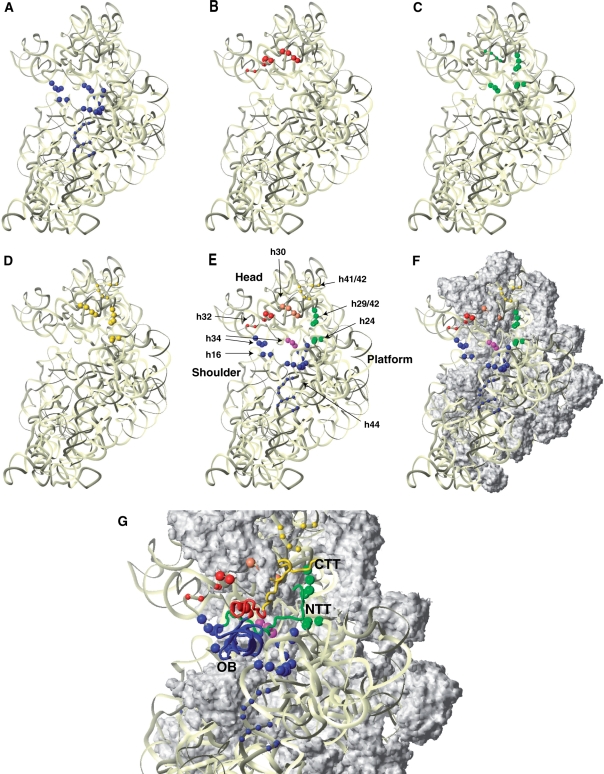
Directed hydroxyl radical cleavage of 18S rRNA in 40S/eIF1A binary complexes from Fe(II) tethered to cysteines in the OB domain and C-terminal subdomain of eIF1A. (A) Ribbon diagram of human eIF1A (15). Spheres indicate positions of cysteines introduced onto its surface. Cysteines from which hydroxyl radicals cleaved 18S rRNA are shown in colors. (B–G) Analysis of directed hydroxyl radical cleavage of 18S rRNA in 40S/eIF1A binary complexes. Sites of cleavage were mapped by primer extension inhibition. Positions, from which hydroxyl radicals cleaved 18S rRNA, are shown in bold. Positions of cleaved nucleotides are shown on the right. Lanes G, A, T, C depict 18S rRNA sequence generated from the same primer.
The NTT and CTT of eIF1A are important for its activity, and have been implicated in stimulating ribosomal recruitment of eIF2-ternary complexes and in initiation codon selection (16,17). Mutations in both the NTT and CTT impair eIF1A's ability to promote ribosomal loading of eIF2-ternary complexes, although the defect caused by N-terminal mutations could be rescued by eIF1. However, the NTT and CTT have opposite effects on initiation codon selection: whereas mutations in the CTT enhance initiation on non-AUG triplets, mutations in the NTT result in leaky scanning and hyperaccuracy. It has therefore been suggested that mutations in the NTT promote the ‘open’ conformation of the scanning ribosomal complex, whereas mutations in the CTT have the opposite effect (17). The CTT is also responsible for eIF1A's interaction with eIF5B, and NMR spectrometry revealed that the last six residues of the CTT lie along the narrow groove on the surface of the helical subdomain of the C-terminal domain (CTD) of eIF5B (10,11). In addition, the NTT was reported to interact weakly with eIF2 and eIF3 (10), and a potential interaction between eIF1A and eIF5 has also been suggested (9).
The bacterial eIF1A homolog IF1 binds to the A site of the 30S ribosomal subunit in a cleft between ribosomal protein (rp) S12, helix (h) 44 and loop 530 in such a way that it sterically blocks binding of tRNA to the A site but does not interfere with binding of mRNA (18). Binding of IF1 induces local and long-range conformational changes in 30S subunits, which include flipping out of A1492/A1493 in h44 of 16S rRNA and tilting of the head and the platform of the 30S subunit towards the A site (18). Mutations of the putative ribosome-binding surface of eIF1A impair its function (15), and it is tempting to speculate that the position of eIF1A's OB domain on the 40S subunit could be similar to that of IF1 on the 30S subunit. However, this hypothesis has not yet received experimental support. Although a cryo-EM study of complexes of yeast 40S subunits with eIF1 and eIF1A reported conformational changes induced by these factors, the factors themselves could not be seen (7).
To establish a foundation for understanding the molecular mechanism of eIF1A's action and the functions of its NTT and CTT, we determined the position of all of eIF1A's segments on the 40S subunit by directed hydroxyl radical probing, using 27 Fe(II)-BABE-derivatized eIF1A mutants. The resulting model for the eIF1A/40S interaction indicates that eIF1A binds to the A site and its helical subdomain contacts the head, forming a bridge over the mRNA channel. The NTT and CTT extend into the P-site. Importantly, we also found that in contrast to yeast, mammalian truncated eIF1A mutants lacking 16 to 33 C-terminal amino acids not only lost the ability to stimulate ribosomal recruitment of eIF2-ternary complexes, but unexpectedly, also efficiently dissociated 43S complexes preassembled without eIF1A. Establishment of codon–anticodon base-pairing protected ribosomal complexes from dissociation by C-terminally truncated eIF1A mutants.
MATERIALS AND METHODS
Construction and purification of eIF1A mutants
To generate expression vectors for eIF1A containing C-terminal or N-terminal His6-Tags (N-Tag-eIF1A and C-Tag-eIF1A), human eIF1A coding regions appropriately flanked by His6 sequences were subcloned between NcoI/HindIII restriction sites of pET28a (Novagen). The resulting vectors were employed to generate plasmids for expression of eIF1A cysteine mutants using a ‘QuikChange’ site-directed mutagenesis kit (Stratagene). First, Cys50Ser substitutions were introduced to obtain the partially cysteine-less mutants, N-Tag-eIF1A-pcl and C-Tag-eIF1A-pcl. Cysteine residues were then introduced individually at residues 3, 4, 9, 10, 15, 21 (the NTT) and 29, 36, 39, 41, 57, 61, 65, 69, 85, 91 (the OB domain) in C-Tag-eIF1A-pcl, and at residues 97, 100, 107, 110, 113, 116 (the C-terminal subdomain) and 119, 124, 132, 137, 142 (the CTT) in N-Tag-eIF1A-pcl to create eIF1A mutants with surface-exposed cysteines. Vectors for expression of N-terminally truncated ΔeIF1A21–143 and ΔeIF1A26–143 containing C-terminal His6-Tags and C-terminally truncated ΔeIF1A1–134 and ΔeIF1A1–110 containing N-terminal His6-Tags were also generated by subcloning the corresponding human eIF1A-coding regions flanked by His6 sequences between NcoI/HindIII restriction sites of pET28a. To obtain expression vectors for ΔeIF1A1–117 and ΔeIF1A1–127, the sequence of full-length eIF1A flanked by an upstream His7-Tag, followed by a TEV protease cleavage site, was first inserted into a pENTR-D entry vector, and then transferred into a Gateway destination vector in frame with an N-terminal GB1 tag, under the control of a T7 promoter. The vectors for expression of ΔeIF1A1–117 and ΔeIF1A1–127 were obtained by introducing in-frame stop codons. All recombinant proteins were expressed in Escherichia coli BL21(DE3). All mutant proteins except ΔeIF1A1–117 and ΔeIF1A1–127 were purified on Ni2+-NTA and MonoQ columns as described (19). ΔeIF1A1–117 and ΔeIF1A1–127 were first purified on TALON CellThru His-Tag affinity resin (Clontech) and then loaded onto IgG sepharose (GE Bioscience). The GB1 tag and His7-Tag were cleaved off with TEV protease on the resin. After elution, proteins were >95% pure. Purified eIF1A cysteine mutants were concentrated on Microcon YM-10 filter units (Millipore), transferred to H300 buffer (80 mM HEPES (pH 7.5), 300 mM KCl, 2.5 mM MgCl2, 10% glycerol) and stored at −80°C.
Purification of initiation factors, ribosomal subunits and aminoacylation of initiator tRNA
Native eIFs 2, 3, 4F and 40S ribosomal subunits were purified from rabbit reticulocyte lysate, and recombinant eIFs 1, 4A and 4B were expressed and purified from E. coli as described (19). For hydroxyl radical cleavage experiments, ribosomal subunits were transferred into H100 buffer (80 mM HEPES, pH 7.5, 100 mM KCl, 5% Glycerol, 2.5 mM MgCl2) on Microcon YM-10 filter units. In vitro transcribed tRNAiMet (20) was aminoacylated as described (21). For analysis of 43S complex formation, tRNAiMet was aminoacylated in the presence of [35S]Met to a specific activity of 200 000 c.p.m./pmol.
Fe(II)-BABE modification of eIF1A mutants
eIF1A mutant proteins were derivatized with Fe(II)-BABE (22,23) by incubating 30 μg eIF1A with 1-mM Fe(II)-BABE in 100 μl H300 buffer for 30 min at 37°C. Derivatized proteins were separated from unincorporated reagent by buffer exchange on Microcon YM-10 filter units and stored at –80°C.
Directed hydroxyl radical cleavage
Binary 40S/[Fe(II)-BABE]-eIF1A complexes were formed by incubating 25 pmol 40S subunits and 50 pmol [Fe(II)-BABE]-eIF1A in 70 μl buffer A (40 mM HEPES, pH 7.5, 100 mM KCl, 2% glycerol and 2.5 mM MgCl2) for 10 min at 37°C. 43S complexes containing [Fe(II)-BABE]-eIF1A were formed by incubating 10 pmol 40S subunits, 20 pmol eIF2, 15 pmol eIF3, 10 pmol eIF1, 10 pmol Met-tRNAiMet and 50 pmol [Fe(II)-BABE]-ΔeIF1A in 50 μl buffer A supplemented with 1 mM ATP and 0.2 mM GMPPNP for 10 min at 37°C. To generate hydroxyl radicals, reaction mixtures were supplemented with 0.025% H2O2 and 5 mM ascorbic acid (22) and incubated on ice for 10 min. Reactions were quenched by adding 20 mM thiourea. Ribosomal RNA and initiator tRNA were phenol-extracted, ethanol-precipitated and analyzed by primer extension using AMV reverse transcriptase (RT) and [32P]-labeled primers complementary to different regions of 18S rRNA. cDNA products were resolved in 6% or 10% polyacrylamide sequencing gels.
Analysis of 43S complex formation
In experiments shown in Figure 7C and D, ribosomal complexes were assembled by incubating 4 pmol 40S subunits with 8 pmol eIF2, 6 pmol [35S]Met-tRNAiMet (200 000 c.p.m./pmol) and 20 pmol wt or mutant eIF1A in 100 μl buffer B (20 mM Tris pH 7.5, 100 mM KAc, 1 mM DTT, 2.5 mM MgCl2, 0.25 mM spermidine) supplemented with 0.2 mM GTP for 10 min at 37°C. In experiments shown in Figure 7E, ribosomal complexes were assembled by incubating 4 pmol 40S subunits with 8 pmol eIF2, 8 pmol eIF3, 20 pmol eIF1, 6 pmol [35S]Met-tRNAiMet (200 000 c.p.m./pmol) and 20 pmol wt or mutant eIF1A in 100 μl buffer B (20 mM Tris pH 7.5, 100 mM KAc, 1 mM DTT, 2.5 mM MgCl2, 0.25 mM spermidine) supplemented with 0.2 mM GTP for 10 min at 37°C. Assembled complexes were analyzed by centrifugation through 10–30% sucrose density gradients prepared in buffer B in a Beckman SW55 rotor at 53 000 rpm for 1 h 15 min. The presence of [35S]Met-tRNAiMet in ribosomal fractions was monitored by scintillation counting.
Figure 3.
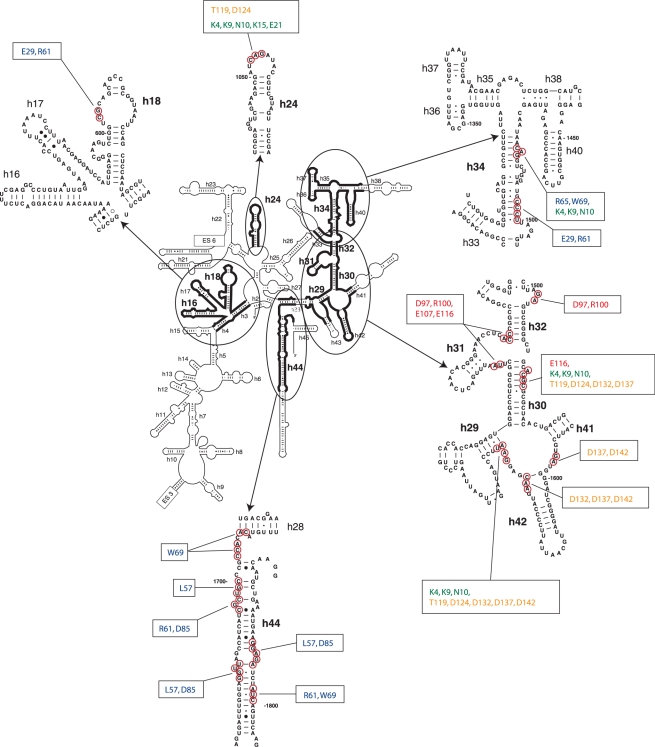
Sites of directed hydroxyl radical cleavage from positions on the surface of eIF1A mapped onto the secondary structure of 18S rRNA. Sites of directed hydroxyl radical cleavage are circled on close-up views of named 18S rRNA elements. The corresponding positions on eIF1A that mediated cleavage are indicated in boxes.
Figure 7.
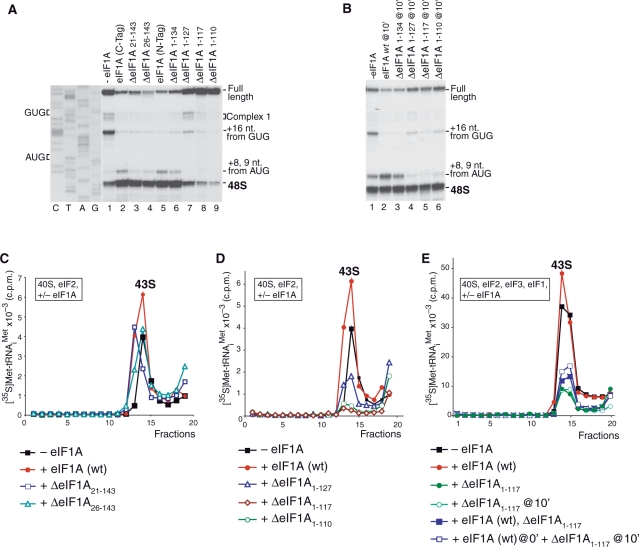
Activities of eIF1A truncation mutants in 43S and 48S complex formation. (A and B) Toe-printing analysis of initiation complexes assembled on β-globin mRNA in the presence of 40S subunits, Met-tRNAiMet, eIFs 2/3/4A/4B/4F/1 and eIF1A mutants as indicated. Positions of assembled ribosomal complexes are marked on each panel. (C–E) Influence of N-terminally truncated (C) and C-terminally truncated (D and E) eIF1A mutants on binding of eIF2·GTP·Met-tRNAiMet ternary complexes to 40S subunits in the absence (C and D) and in the presence (E) of eIF3 and eIF1. Ribosomal complexes were fractionated by centrifugation in 10–30% sucrose gradients and analyzed by scintillation counting of [35S]Met-tRNAiMet. Sedimentation was from right to left. Upper fractions have been omitted for clarity.
Analysis of 48S complex formation on a native β-globin mRNA
48S complexes were assembled on native capped β-globin mRNA (Invitrogen). Reaction mixtures (40 μl) containing 1 pmol mRNA, 2 pmol 40S subunits, 4 pmol eIF2, 3 pmol eIF3, 10 pmol eIF4A, 3 pmol eIF4B, 2.5 pmol eIF4F, 2 pmol (Figure 7A) or 20 pmol (Figure 7B) of eIF1, 4 pmol Met-tRNAiMet and 10 pmol eIF1A mutants were incubated for 10 min at 37°C in buffer B supplemented with 1 mM ATP and 0.2 mM GTP. The concentration of eIF1 in experiments shown in Figure 7A was reduced so that the effect of eIF1A would be more pronounced, whereas in experiments shown in Figure 7B, the concentration of eIF1 was substantially increased to obtain a higher yield of 48S complexes even in the absence of eIF1A. Assembled initiation complexes were analyzed by primer extension using AMV RT and [32P]-labeled primers complementary to coding regions of mRNAs (19). cDNA products were resolved in 6% polyacrylamide sequencing gels.
Modeling
The eIF1A/40S ribosomal subunit interaction was modeled using the crystal structures of the T. thermophilus 30S subunit in complex with IF1 (18: PDB code 1HR0) and of the 70S ribosome in complex with mRNA and tRNAs (24: PDB code 1JGO; 25: PDB code 1JGP), and the NMR structure of human eIF1A (15: PDB code 1D7Q). Docking was done interactively in MOLMOL (26). The distances between Cβ atoms of eIF1A residues and RNA backbone phosphates of their corresponding targets were interpreted as <15 Å for strong cleavages, <20 Å for medium cleavages and <25 Å for weak cleavages. Cleavages in the distal portion of h44 were not used in the docking. Docking was first done for the folded domain of eIF1A as a rigid body, using only cleavages in the body of the 40S subunit. The resulting docking model, obtained independently of the cleavages in the head, provided information concerning the mutual orientation of the head and the body in the eIF1A/40S subunit complex. Docking of the folded domain of eIF1A done using cleavages both in the body and the head of the 40S subunit yielded similar results (data not shown). The NTT and CTT were docked using the structure of the 30S subunit with P-site tRNA. Since the tails are unfolded in free eIF1A, it was not possible to dock them as rigid bodies, or to determine their bound conformation on the 40S subunit. The docking was therefore aimed to minimize the distances between the positions of cysteines and the corresponding cleavage sites. Thus, the resulting backbone conformations are essentially random.
RESULTS
Construction and activity of eIF1A mutants
To determine the orientation of eIF1A on the 40S subunit by directed hydroxyl radical probing, 27 eIF1A mutants containing unique surface-exposed cysteines were generated (Figure 1A). Wild-type (wt) eIF1A contains cysteines at positions 50 and 58, neither of which is surface-exposed. Whereas substitution of C50 by serine in eIF1A tagged with His6 at either N- or C-terminus (partially cysteine-less eIF1A mutants, pcl-eIF1A) did not influence eIF1A's activity in 43S complex formation (data not shown) or in assembly of 48S complexes on native β-globin mRNA (Figure S1), substitution of C58 reduced eIF1A's activity in both assays (data not shown). pcl-eIF1A mutants with a C50S substitution and the non-exposed C58 were therefore used to construct eIF1A mutants containing additional surface-exposed cysteines. Cysteines were introduced individually at positions 3, 4, 9, 10, 15, 21 (in the NTT) and 29, 36, 39, 41, 57, 61, 65, 69, 85, 91 (in the OB domain) of C-terminally His6-tagged pcl-eIF1A, and at positions 97, 100, 107, 110, 113, 116 (in the C-terminal subdomain) and 119, 124, 132, 137, 142 (in the CTT) of N-terminally His6-tagged pcl-eIF1A to create mutants with surface-exposed cysteines (Figure 1A). Although residue 29 is not part of the OB fold, it is on its immediate periphery, and the C29 mutant will therefore be discussed with other OB domain mutants. All eIF1A cysteine mutants were active in 48S complex formation on β-globin mRNA (Figure S1).
Directed hydroxyl radical probing of the 18S rRNA regions surrounding eIF1A
To investigate the ribosomal position of eIF1A, the locations of all eIF1A's segments were first determined in 40S/eIF1A binary complexes, followed by verification of whether the CTT and NTT retain their positions in 43S complexes. The position of eIF1A in 48S complexes was not studied because of the relatively low efficiency of 48S complex formation under conditions of hydroxyl radical cleavage resulting in a high background of 43S complexes.
Probing the rRNA region surrounding the OB domain of eIF1A in 40S/eIF1A binary complexes
Binary 40S/[Fe(II)-BABE]-eIFA complexes were assembled from 40S subunits and eIF1A mutants, in which Fe(II) was tethered to engineered cysteines via the 1-(p-bromoacetamidobenzyl)-EDTA (BABE) linker. Hydroxyl radicals were induced by Fenton chemistry, and the cleavage sites in 18S rRNA were mapped by primer extension inhibition. Hydroxyl radicals generated from 6 positions in the OB domain (Figure 1A, blue spheres) cleaved 18S rRNA in h44, h18 and h34 (Figures 1B–E, 3; Table 1). The most intense cleavage in h44 was observed in its basal region when Fe(II) was tethered to C57, C61, C69 and C85 (Figure 1B). Weak cleavage also occurred in the more central region of h44 from C57, C61, C69 and C85 (Figure 1B and C). Hydroxyl radicals generated from C29 and C61 yielded strong cleavage in the loop of h18 (Figure 1D). Strong cleavage was also induced in h34 in the head of the 40S subunit by C29, C61, C65 and C69 (Figure 1E). The cleavage pattern observed for the OB domain of eIF1A indicates that the factor occupies the area of the ribosomal A-site, which is consistent with the position of IF1 on the 30S subunit (18).
Table 1.
[Fe(II)-BABE]-eIF1A cleavage sites in 18S rRNA
| Domain | Residue | rRNA helices | rRNA cleavage sites (nt) |
|---|---|---|---|
| OB domain | E29C | h18 | 603–604 (CG) strong |
| h34 | 1497–1500 (CCCU) strong | ||
| L57C | h44 | 1700–1702 (CGU) strong; 1807–1811 (AGAGG) weak; 1715–1718 (UUGG) weak | |
| R61C | h44 | 1703–1705 (CGC) strong; 1800–1802 (ACU) weak | |
| h18 | 603–604 (CG) strong | ||
| h34 | 1497–1500 (CCCU) strong | ||
| R65C | h34 | 1484–1486 (CAG) strong | |
| W69C | h44 | 1694–1696 (ACC) strong; 1691–1692 (CA) medium; 1800–1802 (ACU) weak | |
| h34 | 1484–1486 (CAG) strong | ||
| D85C | h44 | 1703–1705 (CGC) strong; 1715–1718 (UUGG) weak; 1807–1811 (AGAGG) weak | |
| C-terminal helical domain | D97C | h31 | 1235–1236 (UA) strong |
| h32 | 1261–1263 (ACC) weak | ||
| h32a (43S) | 1503–1504 (GA) weak | ||
| R100C | h31 | 1235–1236 (UA) strong | |
| h32 | 1261–1263 (ACC) weak | ||
| h32a (43S) | 1503–1504 (GA) weak | ||
| E107C | h31 | 1235–1236 (UA) strong | |
| h32 | 1261–1263 (ACC) weak | ||
| E116C | h31 | 1235–1236 (UA) strong | |
| h32 | 1261–1263 (ACC) weak | ||
| h30 | 1517–1520 (ACGC) strong | ||
| N-terminus | K4C, K9C, N10C | h30 | 1517–1520 (ACGC) weak |
| h29/h42 | 1634–1637 (GAAU) strong | ||
| h34 | 1484–1486 (CAG) medium | ||
| h24 | 1053–1055 (CAG) medium | ||
| K15C | h24 | 1053–1055 (CAG) weak | |
| E21C | h24 | 1053–1055 (CAG) very weak | |
| C-terminus | T119C | h29/42 | 1634–1637 (GAAU) weak |
| h30a (43S) | 1517–1520 (ACGC) strong | ||
| h24 | 1053–1055 (CAG) medium | ||
| D124C | h29/42 | 1634–1637 (GAAU) strong | |
| h30* (43S) | 1517–1520 (ACGC) strong | ||
| h24 | 1053–1055 (CAG) weak | ||
| D132C | h29/42 | 1634–1637 (GAAU) strong | |
| h42 | 1628–1630 (AAC) weak | ||
| h30a (43S) | 1517–1520 (ACGC) strong | ||
| h24 | 1053–1055 (CAG) very weak | ||
| D137C | h29/42 | 1634–1637 (GAAU) strong | |
| h42 | 1628–1630 (AAC) weak | ||
| h41/h42 | 1595–1596 (GA) weak | ||
| h30a (43S) | 1517–1520 (ACGC) medium | ||
| h24 | 1053–1055 (CAG) very weak | ||
| D142C | h29/42 | 1634–1637 (GAAU) weak | |
| h42 | 1628–1630 (AAC) weak | ||
| h41/h42 | 1595–1596 (GA) weak | ||
| h30a (43S) | 1517–1520 (ACGC) very weak |
aSites of cleavage observed in 43S complexes but not in binary 40S/eIF1A complexes.
Probing the rRNA region surrounding the C-terminal subdomain of eIF1A in 40S/eIF1A binary complexes
Hydroxyl radicals generated from 4 positions in the C-terminal subdomain (Figure 1A, red spheres) cleaved 18S rRNA in h30, h31 and h32 in the head of the 40S subunit (Figures 1F–G, 3; Table 1). Strong cleavage was observed in h31 from C97, C100, C107 and C116 (Figure 1F), and in h30 from C116 (Figure 1G). Hydroxyl radicals generated from C97, C100, C107 and C116 also cleaved weakly in h32 at its junction with h31 (Figure 1F).
Probing the rRNA region surrounding the NTT of eIF1A in 40S/eIF1A binary and 43S complexes
In 40S/eIF1A binary complexes, hydroxyl radicals generated from five positions in the NTT (Figure 1A, green spheres) cleaved 18S rRNA in h30 and h34 in the head, in the loop connecting h29 and h42 at the head/neck junction, and in the 1060 loop of h24 on the platform (Figures 2A, C, E, 3, 4, and 5A; Table 1). Hydroxyl radicals generated from C4, C9 and C10 induced strong cleavage of comparable intensity in the loop connecting h29 and h42 (Figure 2A), the strong to medium intensity cleavage in h34 from C4, C9 and C10 (Figure 2C), the weak cleavage in h30 from C4, C9 and C10 (Figure 2C), and the medium to weak intensity cleavage in h24 from C4, C9, C10, C15 and C21 (Figure 2E). These data indicate that upon binding of eIF1A to the 40S subunit, the NTT undergoes some ordering and extends into the P-site.
Figure 2.
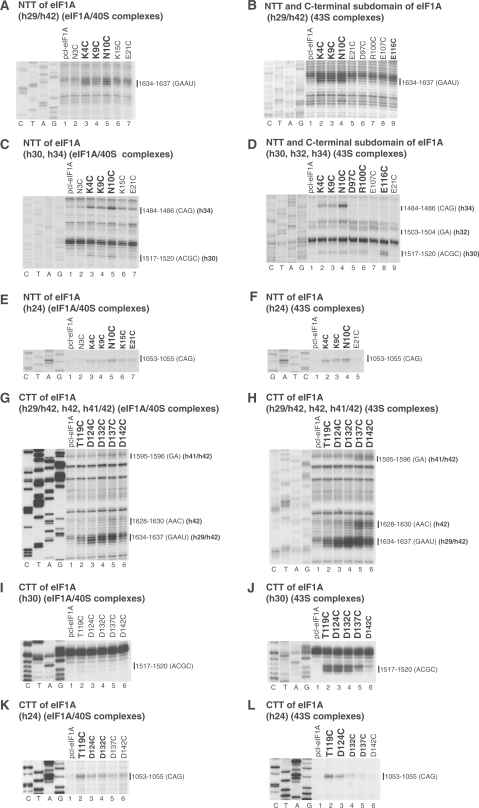
Directed hydroxyl radical cleavage of 18S rRNA in 40S/eIF1A binary and 43S complexes from Fe(II) tethered to cysteines in the NTT, CTT and C-terminal subdomain of eIF1A. Primer extension analysis of directed hydroxyl radical cleavage of 18S rRNA in 40S/eIF1A binary complexes (A, C, E, G, I, K) and 43S complexes (B, D, F, H, J, L) assembled from 40S subunits, Met-tRNAiMet, eIF2, eIF3, eIF1 and Fe(II)-tethered eIF1A mutants. Positions, from which hydroxyl radicals cleaved 18S rRNA, are shown in bold. Positions of cleaved nucleotides are shown on the right. Lanes G, A, T, C depict 18S rRNA sequence generated from the same primer.
We then determined whether the NTT retains the same position in 43S complexes as in 40S/eIF1A complexes. 43S complexes were assembled from 40S subunits, eIF2, eIF3, eIF1, Met-tRNAiMet and Fe(II)-derivatized eIF1A mutants. As in 40S/eIF1A complexes, hydroxyl radicals generated from C4, C9 and C10 of eIF1A induced strong cleavage in 43S complexes at the h29/h42 junction (Figure 2B), strong to medium intensity cleavage in h34 (Figure 2D), weak cleavage in h30 (Figure 2D) and medium cleavage in h24 (Figure 2F). These data indicate that the position of the NTT is similar in both complexes.
Comparable cleavage in 40S/eIF1A binary and 43S complexes also occurred from the surface of the C-terminal subdomain. Thus, as in 40S/eIF1A binary complexes (Figure 1G, lane 7), strong cleavage from C116 in the C-terminal subdomain of eIF1A was observed in 43S complexes in h30 (Figure 2D, lane 8). Weak cleavage at this site also occurred from C97 and C100 (Figure 2D, lanes 5, 6). In 43S complexes, hydroxyl radicals generated from C97 and C100 also weakly cleaved h32 (Figure 2D, lanes 5, 6).
Probing the rRNA region surrounding the CTT of eIF1A in 40S/eIF1A binary and 43S complexes
In 40S/eIF1A complexes, hydroxyl radicals generated from five positions (Figure 1A, yellow spheres) cleaved in the loop connecting h29 and h42 at the head/neck junction, at the base of h42 and in the loop between h42 and h41 in the head, and in the 1060 loop at the top of h24 on the platform (Figures 2G, K, 3, 4, 5C and D; Table 1). The strongest cleavage occurred in the loop between h29 and h42 from C124, C132 and C137 (Figure 2G, lanes 3–5). Cleavage of this region from C119 and C142 was weak (Figure 2G, lanes 2 and 6). C132, C137 and C142 also weakly cleaved h42, and even weaker cleavage occurred from C137 and C142 in the loop between h41 and h42 (Figure 2G, lanes 4–6). Cleavage in h24 was quite diffuse, occurring with medium intensity from C119 and with similarly low intensities from C124, C132, C137 and C142 (Figure 2K). Importantly, cleavage in the h29/h42 loop and in h24 from the CTT coincided with cleavage from the NTT (Figure 2A and E).
In 43S complexes, the cleavage pattern in the loop connecting h29 and h42, in h42, in the loop between h42 and h41, and in h24 (Figure 2H and L) resembled that of 40S/eIF1A complexes, but the intensity of all cleavages was substantially higher. Cleavage in h24 also became less diffuse with distinctly preferential cuts from C119. Importantly, whereas no cleavage in h30 was observed in 40S/eIF1A binary complexes (Figure 2I), in 43S complexes, strong cleavage in h30 occurred from C119, C124 and C132, medium from C137 and weak from C142 (Figure 2J).
These data indicate that, like the NTT, the CTT extends into the P-site. While the CTT appears to remain quite flexible in 40S/eIF1A binary complexes, at least part of the time it resides near the head of the 40S subunit, possibly also contacting the NTT. In 43S complexes, the CTT remains in the P-site, but its mobility is significantly restricted. However, although modification of eIF1A, NTT and CTT mutants with Fe(II)-BABE did not affect their activity in 48S complex formation on β-globin mRNA (data not shown), it is nevertheless important to note that the possibility that such modification might affect the position of the NTT and CTT in ribosomal complexes cannot formally be excluded.
Modeling the eIF1A/40S subunit interaction
The structure of human eIF1A (15) was docked onto the T. thermophilus 30S subunit structure in complex with IF1 (18) and the 70S ribosome structures (24,25), using directed radical cleavage data (Table 1) as distance constraints. While the structure of the free 30S subunit in complex with IF1 and as part of the 70S ribosome are very similar, using the 70S ribosome structures additionally allowed us to infer the positions of mRNA and P-site tRNA. Since the NTT and the CTT of eIF1A are unfolded and flexible in the free protein (15, see also Figure 1A), they were docked separately from the folded domain comprising the OB-fold and helical subdomain.
The folded domain of eIF1A binds in the ribosomal A site
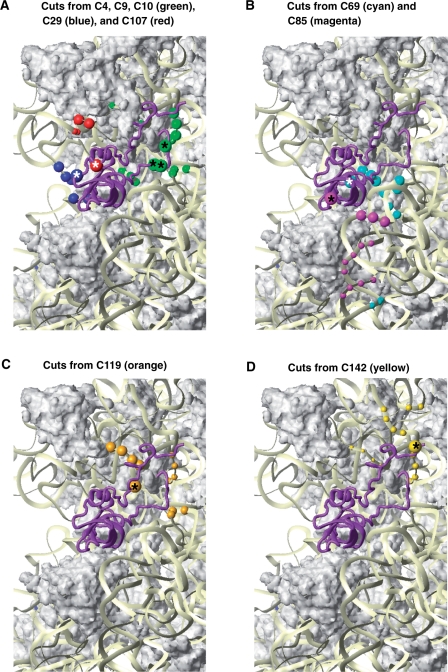
The positions of directed hydroxyl radical cleavages (Figure 4) indicates that the folded domain of eIF1A binds in the A site of the 40S subunit, with the OB-fold subdomain facing the body and the helical subdomain oriented toward the head. The resulting docking model (Figure 4G) places eIF1A in contact with h18, h44 and rpS23, similar to its bacterial homolog IF1 (18). G530, A1492 and A1493 in the E. coli 30S ribosomal subunit are protected by IF1 binding (27) and contact IF1 or are implicated in the 30S/IF1 interaction (18). No cleavage was observed at or around the corresponding positions in the 40S subunit (G616, A1818 and A1819), even though in the docking model, these nucleotides are all within 20 Å of the positions of cysteines used for directed radical cleavage, and the latter two nucleotides directly contact eIF1A. Therefore, the region of h44 around A1818/A1819 appears shielded from cleavage by eIF1A itself. The same could also be the case for the segment around G616 in h18, although that would require conformational changes in h18, as suggested for the bacterial ribosome (18).
Figure 4.
Positions of directed hydroxyl radical cleavage in 18S rRNA from Fe(II) tethered to cysteines on the surface of eIF1A mapped onto corresponding regions of 16S rRNA. (A–D) Cleavages in 18S rRNA mapped onto 16S rRNA in the T. thermophilus 30S subunit crystal structure (18: PDB code 1HR0) from the OB subdomain (A), the C-terminal helical domain (B), the NTT (C), or the CTT (D). 16S rRNA is shown as light yellow ribbon. The radius of the spheres is proportional to the strongest cleavage observed: weak or medium/strong. (E and F) Cleavages in 18S rRNA from all parts of eIF1A, showing rRNA alone (E) or with ribosomal proteins (F). Ribosomal proteins, in surface representation, are colored grey. Positions of cleavage are shown as colored spheres: blue (from the OB-fold), red (from the helical subdomain), yellow (from the CTT only), magenta (from both the NTT and the OB-fold), green (from both the NTT and the CTT) and orange (from the NTT, the CTT and the helical subdomain). The radius of the spheres is proportional to the strongest cleavage observed: weak or medium/strong. (G) The modeled position of eIF1A (ribbon) on the small ribosomal subunit. The OB-fold is blue; the helical subdomain is red; the NTT is green, and the CTT is yellow. The ribosome orientation in panels A–G is identical.
Whereas most cleavages were consistent with the docking model (Figure 4G), cleavages in the distal portion of h44 (nts. 1715–1718, 1800–1802) were clear outliers and were not used in the docking. The basal and distal segments on h44 are cleaved from the same cysteines (e.g. C69), whereas the middle segments are cleaved from other positions (e.g. C85) (Figure 5B). Such a cleavage pattern could be explained by bending of h44, bringing its distal segment closer to its basal segment. However, the available structural and biochemical data provide no indication that such a major rearrangement in h44 can occur. It is therefore more likely that the weak cleavage in the distal segments of h44 resulted from eIF1A attached to a second ribosomal binding site with a significantly lower affinity. We note that two binding sites for the IF3-CTD were reported on the 30S subunit (28,29).
Figure 5.
Cleavage positions in 18S rRNA from individual positions on the surface of eIF1A. Close-up views of cleavage positions in 18S rRNA (colored spheres) from different positions on the surface of eIF1A (colored spheres with asterisks). Colors of cleavage sites correspond to colors of cysteines on the surface of eIF1A. (A) Cleavages from C4, C9 and C10 in the NTT (green), C29 in the OB-fold (blue) and C107 in the helical subdomain (red). (B) Cleavages from C69 (cyan) and C85 (magenta) in the OB fold. (C) Cleavages from C119 in the beginning of the CTT (orange). (D) Cleavages from C142 near the C-terminus (yellow). The radius of the spheres is proportional to the efficiency of cleavage: very weak, weak, medium and strong. Ribosomal protein S13/rpS18 in the head is not shown in panels C and D as it blocks the view of some of the cleavages in h31. The ribosome orientation in panels A, B, C and D is identical to that in Figure 4.
eIF1A contacts the head of the 40S subunit
Remarkably, the docking model (Figure 4G) places the helical subdomain of eIF1A in direct contact with h31 in the head. No contacts have been observed between bacterial IF1 and the head of the small ribosomal subunit, likely because IF1 has no counterpart of the helical subdomain of eIF1A. In the eIF1A docking model (Figure 4G), the sites of weak cleavages in h32 are within 25–35 Å of this subdomain. This prompted us to consider possible head rotation in the 40S/eIF1A complex. The observed cleavage pattern fits qualitatively better to a moderate head rotation toward the E-site (6–12°) such as that found in the E. coli ribosome structures (30) than to ribosome structures showing no head rotation (24). Possible head rotation is also consistent with the strong cleavages in h34 from the OB-fold (Figures 4 and 5A) and could be more pronounced (or stabilized) in 43S complexes, where additional cleavages in h32 from the helical subdomain are observed (Figure 2D).
The NTT and the CTT of eIF1A extend into the ribosomal P-site
Hydroxyl radicals generated from C4, C9, C10, C15, and C21 in the NTT produced a distinct cleavage pattern predominantly in the P-site (Figure 4). Thus, whereas the NTT is flexible in free eIF1A (15), it is at least partially immobilized upon binding to the 40S subunit. On the other hand, since the cleavages from C4, C9 and C10 were identical and covered a rather large surface area, the NTT likely retained some degree of mobility.
Cleavages from several positions within the CTT in 40S/eIF1A binary complexes were also localized to the P-site, but mostly weak and diffuse, indicating that the CTT occupied the P-site but remained mobile. In 43S complexes, stronger cleavages were observed, with more distinct patterns changing gradually from one end of the CTT to the other. This indicated that the CTT's mobility was restricted, presumably by the presence of the Met-tRNAiMet.
Since the cleavages from the NTT (and presumably its location) are the same in 40S/eIF1A binary and 43S complexes, we used the position of the Met-tRNAiMet in the P-site (inferred from the 70S ribosome structure; 24) to restrict the space available for the NTT. The location of the CTT could only be modeled in 43S complexes, since it remains quite mobile in 40S/eIF1A binary complexes. As the NTT and the CTT have no intrinsic structure, they could not be docked as rigid bodies, and it was not possible to pinpoint their exact locations on the 40S subunit with the same certainty as for the folded domain of eIF1A. Similarly, the conformation of the tails (if any) is unknown, their locations in the docking model (Figure 4G) are therefore only approximate, and the backbone conformations shown are essentially random. Both eIF1A tails extend into the P-site and appear to contact the head at the loop between h29 and h42, as well as h30. The two tails could also directly contact each other. The NTT appears to be threading deeper, closer to the mRNA channel. The extreme C-terminus likely remains mobile and samples the vicinity of h42 on the periphery of the P-site.
The influence of N- and C-termini of eIF1A on 43S and 48S complex formation
The NTT of yeast eIF1A has been implicated in promoting ribosomal recruitment of eIF2/GTP/Met-tRNAMeti ternary complexes and in AUG selection (17). Like yeast eIF1A N-terminal deletion mutants, N-terminally truncated mammalian eIF1A mutants ΔeIF1A21–143 and ΔeIF1A26–143 had a strongly reduced ability to promote ribosomal recruitment of eIF2-ternary complexes in minimal reaction mixtures containing 40S subunits, eIF2 and Met-tRNAMeti (Figure 7C). However, these mutants were active in 48S complex formation on β-globin mRNA, promoting efficient formation of 48S complexes on the initiation codon and suppressing assembly of aberrant ribosomal complexes at the 5′-end of the mRNA (complex I) (31) and at the GUG triplet (6) (Figure 7A, lanes 2–4). Toe-prints +8–9 nt downstream of the AUG codon most likely represent initiation complexes, in which the 3′-portion of mRNA is not fixed in the mRNA-binding cleft of the 40S subunit (15,32). The low activity of N-terminally truncated eIF1A mutants in promoting ribosomal recruitment of eIF2-ternary complexes can therefore be compensated by the presence of eIF3 and eIF1 during 48S complex formation. eIF1 and eIF3 have been implicated in promoting ribosomal recruitment of eIF2-ternary complexes (for review see 2), and we previously reported that eIF1A had only a slight additional stimulatory effect on binding of eIF2-ternary complexes to 40S subunits in the presence of eIF3 and eIF1 (33).
The CTT of yeast eIF1A has also been implicated in promoting ribosomal recruitment of eIF2-ternary complexes, scanning and initiation codon selection (16,17). Surprisingly, mammalian C-terminally truncated eIF1A mutants ΔeIF1A1–127, ΔeIF1A1–117 and ΔeIF1A1–110 not only lost the ability to stimulate ribosomal recruitment of eIF2-ternary complexes, but also strongly inhibited this process in minimal reaction mixtures containing 40S subunits, eIF2 and Met-tRNAMeti. Their inhibitory effect increased in parallel with the extent of the deletion (Figure 7D). Moreover, these mutants also inhibited 43S complex formation in the presence of eIF3 and eIF1, and even dissociated preassembled 43S complexes upon their delayed addition (shown for ΔeIF1A1–117 in Figure 7E, filled and open green circles). The presence of wt eIF1A only slightly relieved the inhibition (Figure 7E, filled blue squares). C-terminally truncated eIF1A mutants also dissociated 43S complexes preassembled with wt eIF1A (Figure 7E, open blue squares), which indicates that in 43S complexes, the wt eIF1A can be displaced by the mutant proteins. Consistent with their inhibitory effect on 43S complex formation, C-terminally truncated eIF1A mutants also inhibited 48S complex assembly on β-globin mRNA when added simultaneously with other translation components, and again, the inhibitory effect increased with the extent of the deletion (Figure 7A, lanes 7–9). Importantly, in contrast to preassembled 43S complexes, C-terminally truncated eIF1A mutants could not dissociate 48S complexes preassembled on β-globin mRNA in the absence of eIF1A (Figure 7B, compare lanes 4–6 with lane 1), even after eIF5-induced hydrolysis of eIF2-bound GTP (data not shown). This indicates that establishment of codon–anticodon base-pairing protects ribosomal complexes from dissociation by these eIF1A mutants. However, C-terminally truncated eIF1A mutants dissociated preassembled 48S complexes containing codon–anticodon mismatches (complexes formed on a GUG codon) and initiation complexes with presumably incomplete accommodation of mRNA in the mRNA-binding cleft (complexes yielding +8–9 nt toe-prints) (Figure 7B, compare lanes 4–6 with lane 1). The ΔeIF1A1–134 mutant lacking 9 C-terminal amino acids had only low inhibitory effect on ribosomal recruitment of eIF2-ternary complexes (data not shown) and retained activity in 48S complex formation (Figure 7A, lane 6).
DISCUSSION
Whereas bacterial IF1 consists of only an OB-fold domain, its eukaryotic homologue eIF1A additionally contains a helical C-terminal subdomain, a positively charged N-terminal tail, and a negatively charged C-terminal tail (Figure 1). Many eukaryote-specific functions of eIF1A are clearly linked to these new segments (see Introduction). Here we demonstrate that the folded domain of eIF1A, comprising the OB domain and the C-terminal subdomain, binds in the A site of the 40S subunit, with its N- and C-terminal tails extending into the P-site (Figure 4G).
The position of the folded domain of eIF1A on the 40S subunit and implications thereof
The position of the OB domain of eIF1A in the A site is consistent with that of IF1 (18). IF1 binding to the bacterial 30S subunit causes conformational changes that involve movement of the head, shoulder and platform of the 30S subunit towards the A site (18). Since both the location and ribosomal contacts of eIF1A are similar to those of IF1, it is possible that binding of eIF1A induces similar conformational changes in the 40S subunit. However, since the changes induced in the 30S subunit by IF1 are rather modest, the resolution of directed radical cleavage is not sufficient to determine whether eIF1A also induces similar movements within the 40S subunit.
The docking model (Figure 4G) places the C-terminal helical subdomain in direct contact with h31 in the head of the 40S subunit, consistent with the RNA-binding area on the surface of the subdomain (15). The interaction between h31 and eIF1A would create a bridge over the mRNA in the A site that could contribute to the stability of the scanning complex. However, it is necessary to emphasize that stable interaction of eIF1A with the head of the 40S subunit would impair mRNA binding to 43S pre-initiation complexes, which implies that the contacts between eIF1A and the head in RNA-free ribosomal complexes are most likely transient in order to allow mRNA loading to occur. Consistent with our findings, recent cryo-EM reconstructions of yeast 40S subunits in complex with eIF1A and/or eIF1 (7) showed more pronounced density for the entry channel latch between the head and body of the 40S subunit in the presence of eIF1A. Cleavages in h32 and h34 from the helical subdomain and the OB domain, respectively, suggest that the head of the 40S subunit might also be slightly rotated towards the E-site in 40S/eIF1A/40S binary complexes, and that such rotation may be more pronounced in 43S complexes.
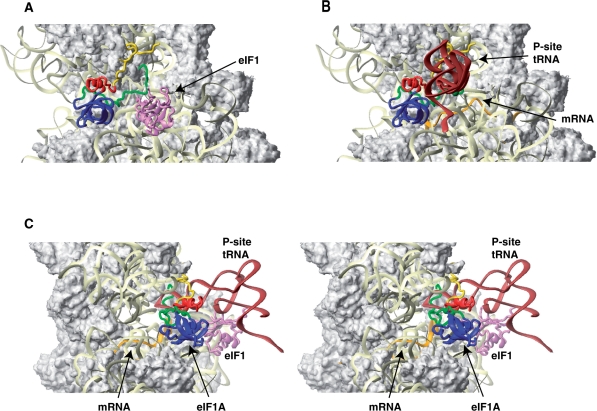
The folded domain of eIF1A does not come in proximity to either eIF1 or Met-tRNAiMet (Figure 6). It may, however, contact mRNA in the A site, especially during scanning, when mRNA mobility may be less restricted than after establishment of codon–anticodon base-pairing.
Figure 6.
Mutual orientations of eIF1A, eIF1, mRNA and Met-tRNAiMet on the 40S subunit. (A) Comparison of the positions of eIF1A (this study) and eIF1 (violet ribbon; 23) on the 40S subunit. Note that although the modeled position of eIF1A-NTT appears in proximity to eIF1 in this view, the two do not contact each other (see also panel C below). (B) Comparison of the positions of eIF1A with those of the mRNA (orange ribbon; 25: PDB code 1JGP) and the P-site tRNA in a P/P orientation (brown ribbon; 24: PDB code 1JGO). Note that the orientation of Met-tRNAiMet in the 43S complex is likely different from the P/P orientation: it may be rotated toward the E-site (23,37) and/or its anticodon loop may not be inserted as deep into the P-site (this study). The ribosome orientation in panels A and B is identical to that in Figures 4 and 5. (C) Stereo view of the mutual orientation of eIF1A, eIF1, mRNA, and Met-tRNAiMet on the 40S subunit, rotated 90° clockwise, compared to panels A and B. Ribosomal protein S13/rpS18 in the head is not shown as it blocks the view of a portion of eIF1A-CTT.
Positions of the NTT and CTT of eIF1A in the context of the 43S pre-initiation complex
The localization of both eIF1A-NTT and eIF1A-CTT in the P-site prompted us to examine more closely their orientation relative to eIF1, Met-tRNAiMet and mRNA in and around the P-site. To compare the locations of eIF1A and eIF1, we used the previously determined position of eIF1 on the mammalian 40S subunit (23). The positions of mRNA and Met-tRNAiMet were inferred from the structures of bacterial 70S ribosome complexes (24,25). However, it should be noted that, while the overall position of the mRNA in 48S complexes (34) is very similar to that in bacterial 70S ribosomes, the position of mRNA in scanning 43S complexes may be less restricted as the complexes are in an ‘open’ conformation, without codon–anticodon base-pairing. Moreover, taking into account that chemical probing analyses and crystallographic studies of prokaryotic ribosomal complexes show that base-pairing with mRNA causes tRNA to be inserted more deeply into the P-site (35,36), it is possible that in 43S pre-initiation complexes, the anticodon loop of Met-tRNAiMet might not be inserted as deeply into the P-site as in initiation complexes with established codon–anticodon base-pairing. In addition, its acceptor end might be rotated towards the E-site similarly to P/E-like or P/I conformations (23,37).
The NTT appears to thread under Met-tRNAiMet in proximity to the platform of the 40S subunit and to the mRNA-binding channel (Figure 6B and C). The NTT's position indicates that it could contact the anticodon loop of Met-tRNAiMet directly, and that in the context of mRNA-bound 43S complexes, could remain in the P-site and also contact mRNA. However, the NTT is unlikely to contact eIF1 (Figure 6A and C).
In 43S complexes, the N-terminal portion of eIF1A-CTT likely also threads under the Met-tRNAiMet, its middle portion contacts the head in the vicinity of h30 and the C-terminus extends past the P-site and likely remains mobile (Figure 6B and C). Since eIF1A-CTT remains more mobile in the absence of Met-tRNAiMet, theoretically it could come in proximity to eIF1 in eIF1A/40S/eIF1 complexes, but in 43S complexes, it would be sterically restricted by the Met-tRNAiMet. In 43S complexes, eIF1A-CTT could also at least transiently contact the Met-tRNAiMet, but such contacts may not be productive (stabilizing).
The results presented here also support the possibility that in 43S complexes, Met-tRNAiMet might not be inserted as deeply into the P-site as tRNA in the bacterial 70S complex. First, if Met-tRNAiMet were to occupy the same position as P-site tRNA in 70S ribosomal complexes (Figure 6B and C), the cleavages in h30 of the head and h24 of the platform from C18, C23, C24 and C119 would be on opposite sides of the Met-tRNAiMet anticodon loop, implying that hydroxyl radicals would have to diffuse through the anticodon loop to cleave rRNA on both sides of the Met-tRNAiMet. Second, in the 70S complex, the anticodon stem of the P-site tRNA is in close contact with h30 in the head, which would leave too little space for eIF1A-CTT to contact h30 in the same area (Figure 6B and C). This latter observation also indicates that, as has previously been suggested (17), eIF1A-CTT would likely be ejected from its P-site location upon start codon recognition and formation of the closed 48S complex (see below).
Functions of the N- and C-terminal tails of eIF1A
Although both eIF1A tails enhance 43S complex formation, they have opposite effects on start codon selection: whereas the CTT increases the stringency of start codon selection and was proposed to promote the ‘open’ conformation of scanning complexes, the NTT decreases the accuracy of initiation and likely promotes the ‘closed’ conformation (17). Our results offer mechanistic insights into the functions of eIF1A's tails.
How can the presence of eIF1A-NTT in the P-site stimulate 43S complex formation? The NTT stabilizes binding of eIF1A to the 40S subunit (17), which in turn might affect Met-tRNAiMet binding via induced conformational changes in the 40S subunit and/or interactions with other eIFs (e.g. eIF2, eIF3; 10). However, our data suggest that the NTT may additionally stabilize binding of Met-tRNAiMet directly, through interaction with its anticodon loop. Importantly, in the proposed 40S/eIF1A model, the NTT does not clash with either mRNA or Met-tRNAiMet, consistent with its suggested role in promoting the ‘closed’ conformation of ribosomal complexes upon start codon recognition (17).
In contrast to eIF1A-NTT, deletion of eIF1A-CTT enhances the 40S/eIF1A interaction (8,16), and eIF1A C-terminal deletion mutants effectively compete with wt eIF1A for 43S complexes (Figure 7E). The ability of such mutants to disrupt 43S complexes (Figure 7D and E) indicates that their binding to the 40S subunit induces conformational changes in it that result in release of Met-tRNAiMet. Since deletion of eIF1A-CTT decreases the stringency of start codon recognition, presumably by promoting the ‘closed’ conformation of ribosomal complexes (17), the eIF1A C-terminal deletion mutants could be causing Met-tRNAiMet release via premature ‘closing’ of 43S complexes in the absence of mRNA. Thus, just as in 48S complex formation, the role of eIF1A-CTT in 43S complex formation could also be to prevent premature ‘closing’. The docked position of eIF1A-CTT on the 40S subunit appears to interfere with the P-site tRNA-head interaction in the closed complex. Therefore, eIF1A-CTT is most likely ejected from the P-site upon start codon recognition.
Upon start codon recognition and GTP hydrolysis by eIF2, eIF5B-CTD presumably replaces eIF2 at the acceptor end of the Met-tRNAiMet, promoting displacement of eIF2-GDP (5) and the correct orientation of Met-tRNAiMet required for ribosomal subunit joining. eIF5B-CTD also binds to eIF1A-CTT, and this interaction is important for subunit joining (10–13). Efficient subunit joining therefore requires simultaneous interaction of eIF5B-CTD with the acceptor end of Met-tRNAiMet and eIF1A-CTT. Although in 43S complexes, the eIF5B-binding region of eIF1A-CTT protrudes out of the P-site and remains flexible and potentially accessible for interaction with eIF5B-CTD (Figures 4–6), its position would not permit simultaneous binding of eIF5B-CTD with eIF1A-CTT and Met-tRNAiMet. Ejection of eIF1A-CTT from the P-site upon start codon recognition, as discussed above, would restore its mobility allowing simultaneous interaction of eIF5B-CTD with eIF1A-CTT and Met-tRNAiMet in 48S complexes upon GTP hydrolysis by eIF2, as required for efficient ribosomal subunit joining.
The in vitro properties of the C-terminal deletion mutants of human eIF1A reported here indicate that such deletions would be lethal for the cell. However, an S. cerevisiae strain with a similarly truncated eIF1A exhibited a slow-growth phenotype but remained viable (16). This difference between yeast and human eIF1A occurs despite the high degree of sequence similarity between yeast and human eIF1A-CTTs (15) and generally analogous effects of C-terminal deletions on the properties of human and yeast eIF1A. The explanation for the viability of this mutant yeast strain may lie in the existence of a stable yeast multifactor complex (38). Thus, if similarly to human eIF1A, the yeast eIF1A-CTT deletion mutants eject Met-tRNAiMet from 43S complexes, eIF2-ternary complexes could remain associated with 40S subunits due to interactions with other eIFs, and Met-tRNAiMet might be eventually reinserted in the P site. However, the fact that the C-terminally truncated yeast eIF1A did not have a strong inhibitory effect on formation of mammalian 48S complexes even though yeast eIF1A was active in a mammalian reconstituted translation system (16) also suggests that the C-terminally truncated yeast eIF1A might have lower affinity to 40S subunits than human truncated eIF1A and might not cause equally dramatic conformational changes in pre-initiation complexes.
eIF1A has also been reported to interact with eIF2, eIF3 and, possibly, also eIF5 in solution, but the exact binding interfaces for these proteins are not yet known (9,10). If these interactions also occur on the 40S subunit, then the position of eIF1A determined here will allow the eIF1A-binding segments of these proteins within the translation initiation complex to be inferred.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (grant GM59660 to T.V.P., and grants GM47467 and CA68268 to G.W.); NCI (Howard Temin award K01 CA119107 to A.M.); Boston University (Peter Paul Career Development Professorship award to A.M). Funding for open access charge: National Institutes of Health (GM59660).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J.-P. Bachellerie for the 18S rRNA plasmid and V. Ramakrishnan and L. Passmore for their helpful comments on the manuscript.
REFERENCES
- 1.Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q. Rev. Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- 2.Pestova TV, Lorsch JR, Hellen C.UT. The mechanism of translation initiation in eukaryotes. In: Mathews MB, Sonenberg N, Hershey J.WB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 87–128. [Google Scholar]
- 3.Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV. Specific functional interactions of nucleotides at key -3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20:624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 8.Maag D, Algire MA, Lorsch JR. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J. Mol. Biol. 2006;356:724–737. doi: 10.1016/j.jmb.2005.11.083. [DOI] [PubMed] [Google Scholar]
- 9.Maag D, Fekete CA, Gryczynski Z, Lorsch JR. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol. Cell. 2005;17:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 10.Olsen DS, Savner EM, Mathew A, Zhang F, Krishnamoorthy T, Phan L, Hinnebusch AG. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J. 2003;22:193–204. doi: 10.1093/emboj/cdg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marintchev A, Kolupaeva VG, Pestova TV, Wagner G. Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: a new interaction between old partners. Proc. Natl Acad. Sci. USA. 2003;100:1535–1540. doi: 10.1073/pnas.0437845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Acker MG, Shin BS, Dever TE, Lorsch JR. Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J. Biol. Chem. 2006;281:8469–8475. doi: 10.1074/jbc.M600210200. [DOI] [PubMed] [Google Scholar]
- 13.Acker MG, Shin BS, Nanda JS, Saini AK, Dever TE, Lorsch JR. Kinetic analysis of late steps of eukaryotic translation initiation. J. Mol. Biol. 2008;385:491–506. doi: 10.1016/j.jmb.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sette M, van Tilborg P, Spurio R, Kaptein R, Paci M, Gualerzi CO, Boelens R. The structure of the translational initiation factor IF1 from E.coli contains an oligomer-binding motif. EMBO J. 1997;16:1436–1443. doi: 10.1093/emboj/16.6.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Battiste JL, Pestova TV, Hellen CU, Wagner G. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell. 2000;5:109–119. doi: 10.1016/s1097-2765(00)80407-4. [DOI] [PubMed] [Google Scholar]
- 16.Fekete CA, Applefield DJ, Blakely SA, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG. The eIF1A C-terminal domain promotes initiation complex assembly, scanning and AUG selection in vivo. EMBO J. 2005;24:3588–3601. doi: 10.1038/sj.emboj.7600821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fekete C, Mitchell S, Cherkasova V, Applefield D, Algire MA, Maag D, Saini A, Lorsch JR, Hinnebusch AG. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO J. 2007;26:1602–1614. doi: 10.1038/sj.emboj.7601613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carter AP, Clemons WM, Brodersen DE, Morgan-Warre RJ, Hartsch T, Wimberley BT, Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 19.Pisarev AV, Unbehaun A, Hellen C.UT, Pestova TV. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007;430:147–177. doi: 10.1016/S0076-6879(07)30007-4. [DOI] [PubMed] [Google Scholar]
- 20.Pestova TV, Hellen C.UT. Preparation and activity of synthetic unmodified mammalian tRNAi(Met) in initiation of translation in vitro. RNA. 2001;7:1496–1505. doi: 10.1017/s135583820101038x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Culver GM, Noller HF. Directed hydroxyl radical probing of RNA from iron(II) tethered to proteins in ribonucleoprotein complexes. Methods Enzymol. 2000;318:461–475. doi: 10.1016/s0076-6879(00)18070-x. [DOI] [PubMed] [Google Scholar]
- 23.Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 25.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 26.Koradi R, Billeter M, Wuthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 27.Moazed D, Samaha RR, Gualerzi C, Noller HF. Specific protection of 16 S rRNA by translational initiation factors. J. Mol. Biol. 1995;248:207–210. doi: 10.1016/s0022-2836(95)80042-5. [DOI] [PubMed] [Google Scholar]
- 28.Dallas A, Noller HF. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell. 2001;8:855–864. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 29.Pioletti M, Schlünzen F, Harms J, Zarivach R, Glühmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, et al. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001;20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 31.Pestova TV, Borukhov SI, Hellen C.UT. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 32.Pisareva VP, Pisarev AV, Komar AA, Hellen C.UT, Pestova TV. Translation initiation on mammalian mRNAs with structured 5’-UTRs requires DExH-box protein DHX29. Cell. 2008;135:1237–1250. doi: 10.1016/j.cell.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 2005;11:470–486. doi: 10.1261/rna.7215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schäfer MA, Tastan AO, Patzke S, Blaha G, Spahn CM, Wilson DN, Nierhaus KH. Codon-anticodon interaction at the P site is a prerequisite for tRNA interaction with the small ribosomal subunit. J. Biol. Chem. 2002;277:19095–19105. doi: 10.1074/jbc.M108902200. [DOI] [PubMed] [Google Scholar]
- 36.Berk V, Zhang W, Pai RD, Cate JH. Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl Acad. Sci. USA. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simonetti A, Marzi S, Myasnikov AG, Fabbretti A, Yusupov M, Gualerzi CO, Klaholz BP. Structure of the 30S translation initiation complex. Nature. 2008;45:416–420. doi: 10.1038/nature07192. [DOI] [PubMed] [Google Scholar]
- 38.Asano K, Clayton J, Shalev A, Hinnebusch AG. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes Dev. 2000;14:2534–2546. doi: 10.1101/gad.831800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.