Abstract
Escherichia coli possesses only one essential oligoribonuclease (Orn), an enzyme that can degrade oligoribonucleotides of five residues and shorter in length (nanoRNA). Firmicutes including Bacillus subtilis do not have an Orn homolog. We had previously identified YtqI (NrnA) as functional analog of Orn in B. subtilis. Screening a genomic library from B. subtilis for genes that can complement a conditional orn mutant, we identify here YngD (NrnB) as a second nanoRNase in B. subtilis. Like NrnA, NrnB is a member of the DHH/DHHA1 protein family of phosphoesterases. NrnB degrades nanoRNA 5-mers in vitro similarily to Orn. Low expression levels of NrnB are sufficient for orn complementation. YhaM, a known RNase present in B. subtilis, degrades nanoRNA efficiently in vitro but requires high levels of expression for only partial complementation of the orn– strain. A triple mutant (nrnA–, nrnB–, yhaM–) in B. subtilis is viable and shows almost no impairment in growth. Lastly, RNase J1 seems also to have some 5′-to-3′ exoribonuclease activity on nanoRNA and thus can potentially finish degradation of RNA. We conclude that, unlike in E. coli, degradation of nanoRNA is performed in a redundant fashion in B. subtilis.
INTRODUCTION
RNA turnover is an essential process associated with the regulation of gene expression in all organisms. Gram-positive and Gram-negative bacteria employ different principal mechanisms of RNA degradation and are equipped with different sets of ribonucleases. This can be exemplified by the fact that two essential RNases in the model bacterium E. coli, RNase E and oligoribonuclease (Orn), are absent in Bacillus subtilis. In E. coli, RNase E often initiates the degradation of mRNA endonucleolytically, which is followed by exonucleolytic degradation in a 3′ to 5′ direction catalyzed by several other enzymes (1). These exonucleases are unable to complete the degradation of RNA due to their inability to degrade RNA in the size range of 2–5 nt. For example, the end products of degradation catalyzed by RNase II and RNase R were found to be 3- to 5-mers (2) or 4- to 6-mers (3,4) and 2- to 3-mers (2,5) or 1- to 2-mers (4), respectively.
We had recently introduced the term ‘nanoRNA’ in order to distinguish these extremely short oligonucleotides from the longer microRNAs (6). We had chosen the term nano in reference to its roots: Nano originates from the Greek word nanos, which means dwarf. Micro on the other hand descends from the Greek word mikros, which means small. Nano is therefore used in this context simply to articulate ‘smaller than’ micro.
The fact that most exonucleases present in E. coli cannot complete the degradation of RNA gives importance to the only exoribonuclease capable of degrading nanoRNA: Orn (7–9).
In B. subtilis, the situation is less well understood. Here, RNase J1, an essential enzyme with no E. coli homolog, was originally identified as an endonuclease functionally analogous to E. coli RNase E, but was later shown to also have 5′–3′ exonuclease activity (10).
In a previous attempt to find a functional analog of Orn in B. subtilis, we had identified YtqI (NrnA) as an enzyme that has nanoRNase activity in vitro and can complement an E. coli orn mutant when expressed at low levels (6). This identification was done through the binding of NrnA to 3′-phosphoadenosine 5′-phosphate (pAp), exploiting the conserved strong interaction between pAp and oligoribonucleases, which was shown for Orn (E. coli) and the human homolog Sfn (11). In addition to its nanoRNase activity, NrnA has pAp-phosphatase activity, as demonstrated by the ability to complement a mutation in the E. coli gene coding for pAp-phosphatase, cysQ and to degrade pAp to AMP in vitro. Consistent with this second activity of NrnA, the phenotype of a ytqI (nrnA) mutant in B. subtilis resembles that of an E. coli cysQ mutant, namely its growth in the absence of cysteine is impaired. The non-essentiality of nrnA implied the existence of at least one more enzyme with an ability to degrade nanoRNA. Here we demonstrate that, unlike in E. coli, enzymes with nanoRNase activity are redundantly present in B. subtilis; in particular, we identify a protein of previously unknown function, YngD (now named NrnB), as a nanoRNase and we test the activity of YhaM and RNase J1 on nanoRNA.
MATERIALS AND METHODS
Strains, plasmids and growth conditions
Strains, plasmid and primers used in this study are listed in Table 1.
Table 1.
Bacterial strains, plasmids and primers
| Description | Reference | |
|---|---|---|
| Strains | ||
| MG1655 | E. coli K12 wild type | Ref. |
| B. subtilis 168 | B. subtilis 168 wild type | Ref. |
| CF10230 | nic+ derivative of DY329 (29) | M. Cashel, unpublished results |
| UM285 | As CF10230 but ΔcysQ, KmR | (11) |
| UM341 | As CF10230 but orn under control of P LtetO-1, TetR, KmR | (6) |
| UM517 | B. subtilis 168, yhaM::PhleoR | This study |
| UM545 | B. subtilis 168, nrnB::CmR | This study |
| UM579 | B. subtilis 168, PSpac::nrnA | This study |
| UM599 | B. subtilis 168, nrnA::KmR | This study |
| UM612 | B. subtilis 168, PSpac::nrnA, nrnB::CmR | This study |
| UM623 | B. subtilis 168, PSpac::nrnA, nrnB::CmR, yhaM::phleoR | This study |
| UM629 | UM623 carrying pMAP65 | This study |
| UM638 | B. subtilis, nrnB::CmR, nrnA::KmR | This study |
| UM645 | B. subtilis 168, nrnB::CmR, yhaM::PhleoR | This study |
| UM647 | B. subtilis 168, nrnB::CmR, yhaM::PhleoR, nrnA::KmR | This study |
| UM651 | B. subtilis 168, nrnB::CmR, yhaM::PhleoR, nrnA::EmR | This study |
| BFS66 | B. subtilis 168, nrnA::pMUTIN2MCS, EmR | (13) |
| Plasmids | ||
| pBAD18 | Vector, Para, pBR replicon, ApR | (30) |
| pBSL10 | Genomic library clone containing nt 1 068 397–1 070 326 of the B. subtilis genome in pCDNA 2.1 | This study |
| pGEM-T Easy | Vector for TA cloning | Promega |
| pMAP65 | Vector containing lacI | (31) |
| pMutin4 | Vector for gene inactivation in B. subtilis | (32) |
| pEC23 | pMutin derivative for replacement of EmR by KmR | M. Simon and P. Stragier, unpublished results |
| pUM404 | As pBAD18, CysQ with C-terminal his-tag | (11) |
| pUM408 | As pBAD18, Orn with C-terminal his-tag | (11) |
| pUM413 | As pBAD18, YhaM with C-terminal his-tag | This study |
| pUM414 | As pBAD18, NrnB with C-terminal his-tag | This study |
| pUM416 | As pUC18, YhaM with C-terminal his-tag under control of PLac | This study |
| pUM420 | pGEMT-Easy carrying a fragment allowing construction of the nrnB::CmR replacement allele | This study |
| pUM432 | nrnA (nt 1–327) under control of PSpac in pMUTIN4 | This study |
| pUM443 | As pBAD18, NrnB (D87A, H88A, H89A) with C-terminal his-tag | This study |
| pFM1 | YhaM in pGEMT-Easy under control of its native promoter and in opposite direction to PLac | This study |
| pMK4 | shuttle vector, source for cat-cassette | (33) and Genbank EU549778 |
| pDG148-rnjA | pDG148 carrying rnjA coding for RNase J1 under control of PSpac | This study |
| Primers | ||
| FM1 | 5′GGGGAATTCTAAAAAGAGGTTCTATAGCTGAAAATCGC3′ | |
| IV211bis | 5′GTACAGTCGGCATTATCTCAT3′ | |
| MF80 | 5′CGGCAATAGTTACCCTTATTAT3′ | |
| UM172 | 5′GTGCTGCAAGGCGATTAAGT3′ | |
| UM173 | 5′CGCCAAGCTATTTAGGTGAC3′ | |
| UM179 | 5′GGGGAATTCACCATGTATCATTTATATTCACATAACGACTTGGA3′ | |
| UM180 | 5′GGGGCTCGAGCTTGCGATGTTGATTTGCCAGCTTCAGATTCTCCGCTAAAAATGCGACAAACACTTCATCCC3′ | |
| UM183 | 5′AGGAGGCGTCTTATTCCACGATTC3′ | |
| UM184 | 5′CTTATCTTGATAATAAGGGTAACTATTGCCGATGTAGGCCCCTTTTTATGATAAACTGAC3′ | |
| UM185 | 5′GGCTTTTATAATATGAGATAATGCCGACTGTACAAAAATATGCATAGGGGAGCGTCAG3′ | |
| UM186 | 5′CTTATGGCGGCTATATGGTATCGG3′ | |
| UM187 | 5′ACAGCCTGTCGGCATTGTTG3′ | |
| UM188 | 5′CTAACTCTCCGTCGCTATTG3′ | |
| UM189 | 5′CAATAGCGACGGAGAGTTAGG3′ | |
| UM190 | 5′TTGCCGTAAGGAGCCGATCCA3′ | |
| UM196 | 5′GCCGAAGCTTTAAAAACAATAAAGGAGTATCAA3′ | |
| UM197 | 5′CGCGGATCCTGGATCTTCGTTCGGATGGT3′ | |
| UM198 | 5′GGGGCATGCTTATTTATGAAATGTCGGTTTATAAAAGG3′ | |
| UM209 | 5′CAGCTTCCAGCCGTGCTCTT3′ | |
| UM210 | 5′TGCCTACCTAGCTTCCAAGA3′ | |
| UM211 | 5′GTGCTGCAAGGCGATTAAGT3′ | |
| UM238 | 5′GTGAAGCTCATTGCGGCCGCGAAAACAGCCCTTCATTTGAATG3′ | |
| UM239 | 5′GAAGGGCTGTTTTCGCGGCCGCAATGAGCTTCACTTTGCCCCCTGCA3′ | |
| UM240 | 5′GGGGAGCTCTGTGAATTGATCTAAGGCGTTTG3′ | |
| RD1 | 5′TATTAAGCTTGTATTGGAGTTATGAGCGGTATGAAATTTG3′ | |
| RD2 | 5′TATATCTAGAGTAAAATCATTTCAACACATATCACTGC3′ |
E. coli and B. subtilis strains were grown in LB. Arabinose was present for induction of the Para promoter as indicated. Ampicillin (100 μg/ml), kanamycin (5–10 μg/ml), erythromycin (1 μg/ml) together with lincomycin (25 μg/ml), phleomycin (1 μg/ml), chloramphenicol (5 μg/ml) was added for plasmid maintenance or to select for chromosomal markers. Anhydrotetracycline (Atc) was added at 250 ng/ml for induction of PLtetO-1.
The plasmids for the expression of his-tagged YhaM and NrnB (pUM413 and pUM414, respectively) were constructed as follows: primer UM177 plus UM178 and UM179 plus UM180 were used to amplify yhaM or yngD (nrnB), respectively from B. subtilis 168 chromosomal DNA. The EcoRI, XhoI digested fragments were used to replace the CDS from pUM407 leaving the region coding for the C-terminal his-tag and the ribosomal binding site intact. pUM416 expressing yhaM under control of PLac in pUC18 was constructed by PCR-amplifying yhaM including the region coding for the his-tag from pUM413 using primer UM191 and UM193 and inserting the EcoRI/XbaI digested fragment into pUC18 digested with EcoRI and XbaI.
pFM1 was generated by PCR amplifying yhaM including its own promoter from the chromosome of B. subtilis 168 and direct TA-cloning. This method exploits the terminal transferase activity of Taq polymerases that adds a 3′-A overhang to each end of the PCR products for cloning into a vector with 3′-T overhangs [pGEMT-Easy (Promega)]. Clones carrying an insert were sequenced using primers UM172 and UM173.
For the construction of pUM443 expressing his-tagged NrnB-DHH mutant protein with alanine replacements of amino acids D86, H87 and H88, two PCR fragments were amplified using pUM414 as template: PCR1 was performed with primers UM238 and UM240, and PCR2 was performed with primers UM179 and UM239. The outside primers UM179 and UM240 and equimolar amounts of PCR fragments 1 and 2 were used to perform overlapping PCR. The obtained PCR fragment was digested with EcoRI and SacI, and was used to replace the EcoRI/SacI fragment of pUM414. The obtained clone was verified by sequence analysis.
pDG148-rnjA was constructed by PCR-amplifying the rnjA gene from the chromosome of B. subtilis 168 using oligos RD1 and RD2. The obtained PCR fragment was cleaved with HindIII and XbaI and cloned into pDG148 (12) digested with HindIII and XbaI.
The B. subtilis nrnB mutant (UM545) was created by replacing nrnB with a cassette coding for chloramphenicol acetyl transferase (cat). To this end, three PCR fragments were generated: PCR1 and 2 amplified the 730-nt upstream and 539-nt downstream regions of nrnB using primers UM183 plus UM184 and UM185 plus UM186, respectively from B. subtilis 168 chromosomal DNA. PCR3 amplified the cat-cassette from pMK4 using primers IV211bis and MF80. Equimolar amounts of PCR1, 2 and 3 and the outside primers UM183 and UM186 were used for overlapping PCR. The obtained PCR fragment was cloned by TA-cloning into pGEMT-Easy. Plasmid DNA of a sequence-verified clone (pUM420) was linearized by ScaI and used to transform B. subtilis. Correct integration of the cat-cassette was verified on chromosomal DNA of chloramphenicol-resistant clones using primers UM187 plus UM188 and UM189 plus UM190 amplifying a 1286 or 1358 nt long PCR fragment for the 5′ or 3′ site of integration, respectively.
The nrnA::EmR mutant (BFS66) was part of the European/Japanese effort to inactivate the complete gene set of B. subtilis 168 and has an insertion after amino acid 108 (13). In order to create an nrnA::KmR allele (UM599), pEC23 was linearized by ScaI and transformed into the nrnA::EmR mutant. Erythromycin-sensitive/kanamycin-resistant clones were verified by PCR using primers UM196 plus UM211 and UM209 plus UM210. The yhaM::PhleoR allele (14) was obtained from David Bechhofer [strain BG395 (15)] and transformed into B. subtilis 168 selecting for phleomycin resistant/chloramphenicol sensitive clones to create UM517. For the construction of UM579 carrying nrnA under control of PSpac, pUM432 was transformed into B. subtilis 168. Erythromycin-resistant clones were verified by PCR using primers UM196 plus UM211 and UM209 plus UM210. pUM432 was generated as follows: a PCR fragment containing nt 1–328 plus 23-nt upstream of the ATG start codon was amplified from chromosomal DNA of B. subtilis 168 using primers UM196 and UM197, digested with HindIII and BamHI and cloned into pMUTIN4 cut with HindIII and BamHI.
The B. subtilis mutants containing multiple mutant alleles were created by transforming chromosomal DNA of the donor to the recipient strains. Mutant alleles in the recipient strains were verified by PCR analysis confirming both the 5′ and 3′ site of integration using the above-mentioned primers that were used to confirm correct integration of the original nrnA- or nrnB-mutant alleles. The yhaM mutant allele was confirmed by PCR using primers UM177 and UM178. The order of combining the mutant alleles to create the triple mutant was as follows: 1. nrnB::CmR, 2. yhaM::PhleoR, 3. nrnA::KmR or nrnA::EmR.
The genomic library of B. subtilis 168 was kindly supplied by Florence Hommais and was prepared as described hereafter: genomic DNA was fragmented by nebulization. Fragments of ∼2-kb length were purified from an agarose gel and integrated into the BstXI site of pCDNA 2.1 using linkers.
Purification of his-tagged proteins and activity assays
Purification of C-terminally his-tagged NrnB protein from E. coli carrying pUM414 seemed to be difficult due to problems of solubility. We therefore tested several buffers and salts and found that surprisingly, NrnB seemed to be most soluble in water. As nickel-agarose purification cannot be performed in water, we decided to take advantage of this unique property to purify NrnB. Resuspension of the in 50 mM KPi pH 8.0, 300 mM NaCl insoluble fraction in water solubilized mainly NrnB. The two major contaminants were identified by mass-spectrometry to be membrane proteins OmpA and OmpC, and could therefore be removed by high-speed centrifugation (45 000 rpm, 30 min). The purified enzyme was estimated to be >95% pure and was stable for several months when kept on ice. The NrnB-DHH mutant protein was purified according to an identical protocol from E. coli carrying pUM443.
C-terminally his-tagged YhaM was purified from 1 l cultures of MG1655 carrying pUM413 grown at 30°C. Expression was induced at an OD600 of 0.5 by 0.2% arabinose for 3.5 h. Subsequent purification was according to the protocol described before (11). Purified YhaM was estimated to be ≥90% pure. YhaM was less stable than other purified enzymes and lost activity over time. Experiments comparing activity on RNA and DNA substrates were therefore always performed simultaneously.
C-terminally his-tagged wild-type RNAse J1 and RNase J1 (H46A) mutant protein were purified as described before (10).
Activity assays on nanoRNA were performed on custom-made RNA oligos essentially as described before (6), but taking into account the buffer conditions that were found to be optimal for NrnB activity: 50 mM Tris pH 8.0, 5 mM MnCl2. YhaM activity assays were performed in 5 mM MnCl2, 50 mM Tris pH 8.0, 100 mM KCl. RNase J1 activity was tested in 50 mM Tris pH 8.0, 8 mM MgCl2, 100 mM NH4Cl, and 0.1 mM DTT.
Assays determining degradation activity of NrnB on longer substrate were done using a custom made RNA 24-mer (5′CACACACACACACACACACACACA3′) 5′-end labeled with [γ-33P]ATP. This oligonucleotide was labeled using the MirVana Probe and Marker Kit (Ambion) in a 90 μl reaction containing 450 pmol oligo, 20 pmol [γ-33P]ATP (35 μCi), 180 pmol ATP and 4.5 μl T4 Polynucleotide Kinase, incubated for one hour at 37°C. The reaction was stopped by the addition of 10 μl of 10 mM EDTA and incubation at 95°C for two minutes. The reaction mixture was purified from the unincorporated nucleotides using NucAway spin columns (Ambion) according to the supplier's instructions. Concentration of labeled substrate was estimated to be 1.76 pmol/μl by measuring A260. Quantities of 5, 10 or 20 μl of substrate were used in 40 μl reactions containing 0.5 or 1 μg NrnB or 5 μg NrnB-DHH under the same buffer and salt conditions used in activity assays on nanoRNA. Reactions were started by adding enzyme after 5 min preincubation at 37°C. Six-microliter aliquots were taken at times indicated and added to an equal volume of loading buffer and set on ice in order to stop the reaction. After 3 min at 95°C, 7 μl of the samples were resolved on a 20% PAA, 7 M Urea gel containing 2XTBE that was run in 2XTBE. Labeling of the Decade-marker was performed as described previously (6).
Expression of his-tagged proteins was monitored by PAA gel electrophoresis, followed by staining with BioSafe Coomassie stain (BIO-RAD) or by western blot using Anti-His6 peroxidase antibodies (Roche) as described before (6).
Phylogenetic analysis
NrnB orthologs were defined by limiting the results of an NCBI Blast search according to the following criteria: ≤20% difference in protein length, alignment extends over ≥80% of the protein, and ≥40% amino acid similarity. Phylogenetic trees were constructed on http://www.phylogeny.fr.
RESULTS
Identification of NrnB as potential nanoRNase
The viability of an nrnA mutant in B. subtilis 168 pointed to the possible existence of a second enzyme that is capable of degrading nanoRNA in this organism. To search for another potential nanoRNase we used a genomic library of B. subtilis to screen for genes that complement a conditional promoter mutant of E. coli oligoribonuclease (orn) (strain UM341). In this strain, orn is under control of the anhydrotetracycline (Atc)-inducible promoter PLtetO-1. Transformation of this strain with a vector control (pBAD18) in the absence of Atc produced pinpoint-sized colonies that stopped growing, whereas transformants with a plasmid expressing orn (pUM408) continued to grow and formed normal size colonies compared to growth in the presence of Atc (Figure 1). Among the clones allowing complementation of the orn– strain was a plasmid with a fragment spanning nt 1 947 002–1 948 086 of the B. subtilis genome, containing one complete CDS (from nt 1 947 525–1 948 721) namely yngD (now nrnB). The function of YngD was unknown, but like NrnA, it belongs to the DHH/DHHA1 protein family (16) and was therefore a good candidate for a nanoRNase. Homology between NrnA and NrnB does not extend over the full-length protein and is restricted instead to small areas containing the conserved motifs characteristic for the protein family.
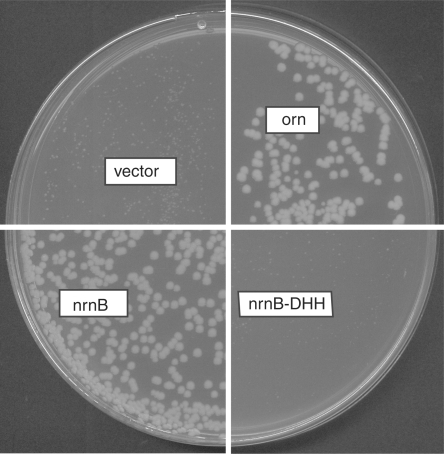
Figure 1.
Complementation of the conditional orn mutant by expression of NrnB but not the NrnB-DHH mutant. Transformants of strain UM341 with pBAD18 (vector control), pUM408 (arabinose-inducible orn), pUM414 (arabinose-inducible nrnB) or pUM443 (arabinose-inducible NrnB-DHH mutant) were spread on LB plates containing 0.2% arabinose in the absence of anhydrotetracycline (Atc).
NrnB has nanoRNase activity in vivo
To confirm that it was the expression of NrnB that allowed complementation of E. coli orn– from the genomic library clone, we subcloned nrnB under the control of the arabinose-inducible Para promoter in pBAD18. As seen in Figure 1, expression of nrnB from pUM414 completely rescued the growth defect of the E. coli orn mutant. Here, expression was induced by the addition of 0.2% arabinose. Figure 1 shows that the colony sizes of NrnB-expressing transformants in the absence of Atc (orn– background) are similar to those expressing Orn. Complementation could be seen even in the absence of arabinose (data not shown). Under these conditions we could not detect NrnB either on a Coomassie-stained protein gel or by western blotting using Anti-His6 antibodies (data not shown) suggesting that low levels of NrnB are sufficient for orn– complementation. The NrnB protein expressed from pUM414 was C-terminally his-tagged. As this protein allowed orn– complementation, we conclude that the C-terminal his-tag does not interfere with nanoRNase activity of NrnB.
NrnB activity on nanoRNA in vitro
Recombinant NrnB was purified to near homogeneity by taking advantage of its unique solubility in water (see ‘Materials and Methods’ section).
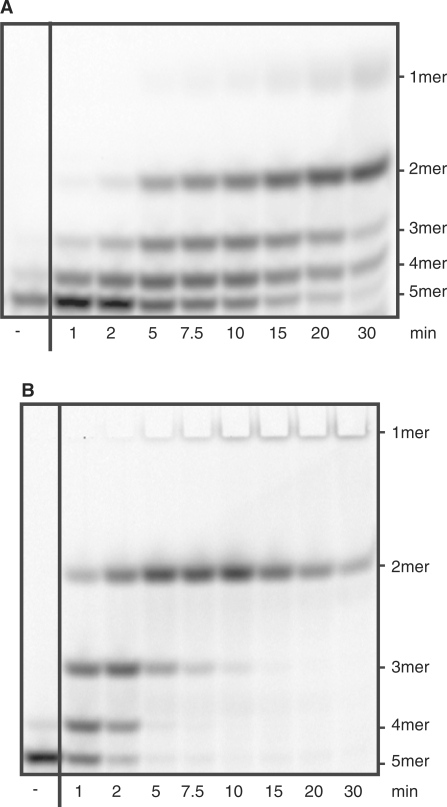
NanoRNase activity was tested on nanoRNA 5-mers (5′Cy5-CCCCC3′) and was easily detected in the presence of manganese (Figure 2A and C). Manganese could be replaced by cobalt with almost similar activity and with magnesium with considerably lower, but still appreciable activity (data not shown). NrnB was more active at pH 8.0 and pH 7.5 than at pH 7.0, and in Tris compared to HEPES (data not shown). Therefore, subsequent experiments were performed in Tris pH 8.0 in the presence of manganese. The sequential appearance of reaction products (Figure 2A) resembled Orn-catalyzed reactions (6) and pointed to a similar reaction mechanism. Under the conditions tested, the activity of NrnB measured as the disappearance of substrate 5-mer can be estimated to be 1 nmol/min/μg.
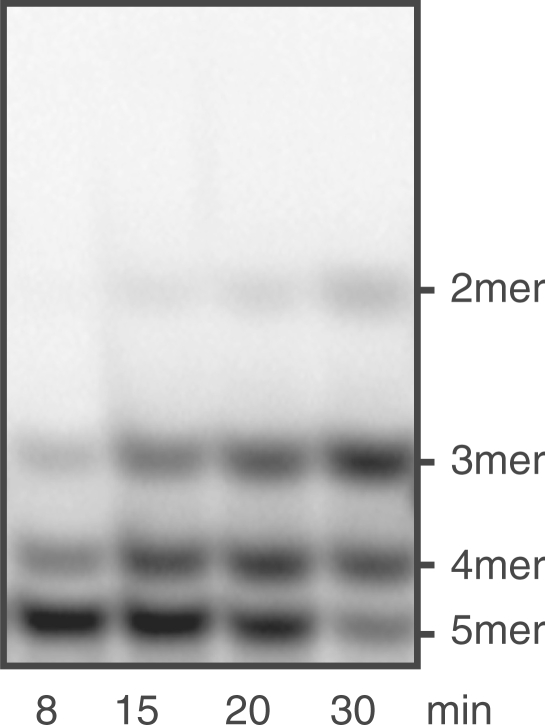
Figure 2.
NrnB-catalyzed degradation of nanoRNA. Shown is the separation of reaction products on 22% PAA gels (A, B, E, F). The reverse migration can be accounted for by the fact that cyanine dyes have a lower net negative charge than nucleic acids: thus, removing nucleotides will reduce the charge relative to the mass of the oligonucleotide and cause it to shift up instead of down. Panels (C) and (D) show quantifications of reaction products and intermediates of degradation of 5′Cy5-CCCCC3′ and 5′Cy5-AAAAA3′, respectively. 50 μl reactions contained 0.25 μg NrnB and 6.0 μM RNA (5′Cy5-CCCCC3′ for A and C, 5′Cy5-AAAAA3′ for B and D, 5′Cy5-CCC3′ for E, and 5′Cy5-AAA3′ for F). The minus indicates controls lacking enzyme. (C and D) Closed circles: 5-mers, open circles: 4-mers, closed triangles: 3-mers, open triangles: 2-mers, squares: 1-mers. (G) Comparison of initial rates of degradation of 5′Cy5-AAAAA3′, 5A; 5′Cy5-CCCCC3′, 5C; 5′Cy5-AAA3′, 3A; and 5′Cy5-CCC3′, 3C. Specific activities measured as disappearance of substrate were normalized according to the activity on 5′Cy5-AAAAA3′, which was set to be 1.
We also tested the activity of NrnB on RNA oligos different from 5′Cy5-CCCCC3′, namely 5′Cy5-AAAAA3′ (Figure 2B and D), 5′Cy5-CCC3′ (Figure 2E) and 5′Cy5-AAA3′ (Figure 2F). The degradation patterns observed in NrnB-catalyzed degradations of 5-mers 5′Cy5-CCCCC3′ and 5′Cy5-AAAAA3′ were slightly different but did not suggest any strong preference for substrates shorter than 5 nt, as we had seen in the case of NrnA (6). In agreement with this, the levels of NrnB activity on 5-mers and 3-mers varied only within one order of magnitude (Figure 2G). NrnB was somewhat (1.4-fold) more active on 5-mer A's than on 5-mer C's. In contrast to NrnA, NrnB was less active on 3-mers than on 5-mers with a larger difference for the oligos containing A's as compared to the oligos containing C's (0.5-fold versus 0.2-fold, respectively). This might explain why 3-mer intermediates accumulate to a larger extent in reactions containing 5-mer C's as compared to 5-mer A's (Figure 2C and D).
In addition to its activity on nanoRNA, NrnA is able to degrade pAp (6). We therefore asked whether NrnB could also degrade pAp. NrnB was unable to complement an E. coli cysQ mutant (UM285), however, when expressed from pUM414 in the presence of 0.2% arabinose (data not shown), precluding this possibility. NrnB activity was also tested on DNA 5-mers (5′Cy5-CCCCC3′) under identical conditions. NrnB could degrade DNA 5-mers with an activity of a similar order of magnitude (0.7 nmol/min/μg) (data not shown).
Activity of NrnB on a RNA 24-mer
We next asked whether NrnB specifically degraded nanoRNA or whether it could also degrade longer substrates as well, by testing its activity on a 24-mer (5′CACACACACACACACACACACACA3′) that was 5′-labeled with 33P. Initial experiments showed some activity of NrnB on the RNA 24-mer. As this activity could potentially arise from impurities in the protein preparation, we constructed a mutant enzyme to determine whether or not activity on the 24-mer was inherent to NrnB. We chose to mutagenise one of the motifs characteristic of the DHH protein family, namely motif III including Asp86, His87, His88. According to structural and mutational analyses of other members of the DHH protein family [RecJ from E. coli (17,18), PPX1 from Saccharomyces cerevisiae (19), and human Prune (20)], this motif is important for activity, as it contributes to the catalytic center. Amino acids Asp86, His87 and His88 were replaced by alanine. We will refer to this mutant from here on as the DHH mutant or NrnB-DHH. To confirm that this replacement affected NrnB activity, we first tested its ability to complement E. coli orn–. As expected, this mutant was unable to rescue the effects of Orn deficiency (Figure 1). Next we purified both wild-type NrnB and NrnB-DHH according to an identical protocol. An activity of the DHH mutant protein on nanoRNA 5-mers (5′Cy5-CCCCC3′) was still detectable (Figure 3), but was dramatically reduced with an estimated activity of less than 1% of that of wild-type NrnB.
Figure 3.
Activity of NrnB-DHH on nanoRNA (5′Cy5-CCCCC3′). Fifty-microliter reactions contained 5 μg NrnB-DHH and 6 μM RNA 5-mer (5′Cy5-CCCCC3′).
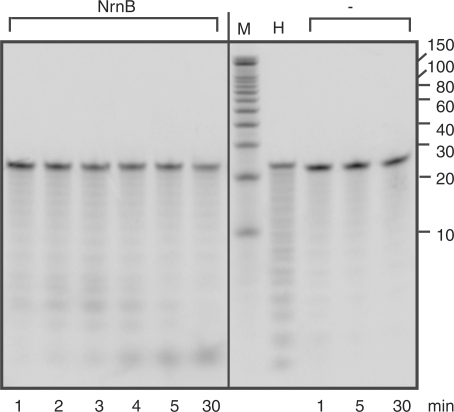
The activity of the NrnB-DHH mutant on the 24-mer was not visible by eye and amounted to less than 10% of that of NrnB (data not shown), which clearly showed activity on the 24-mer (Figure 4). However, the activity of NrnB on the 24-mer was considerably less than that on nanoRNA. The NrnB-catalyzed degradation of 24-mers, measured as disappearance of substrate, was roughly estimated at 2 pmol/μg/min (1–4). This amounts to three orders of magnitude lower than its activity on nanoRNA 5-mers, but is 200-fold greater than that of NrnA on 24-mers.
Figure 4.
Activity of NrnB on RNA 24-mers. Reactions containing 5′ 33P-labeled RNA 24-mers (5′CACACACACACACACACACACACA3′) and 0.5 μg NrnB or no enzyme (minus) were incubated at 37°C. M, decade marker; H, alkaline hydrolysis control.
Phylogenetic distribution of NrnB
We searched the databases for homologs of NrnB. Table S1 lists NrnB orthologs that were defined by fulfilling the following criteria: ≤20% difference in protein length, alignment extends over ≥80% of the protein, and ≥40% amino-acid similarity. The 41 sequences fulfilling these criteria (sequences from one particular strain of one species are counted for one) originated mainly from species closely related to B. subtilis. However, orthologs were also found in ε-proteobacteria like Helicobacter sp. and Campylobacter sp. and in some archaea. We constructed a phylogenetic tree on http://www.phylogeny.fr using the 41 sequences defined in Table S1. Figure S1 shows a simplified version of this tree that was obtained by omitting multiple sequences that took an identical position in the tree and belonged to the same species.
YhaM activity on nanoRNA and DNA
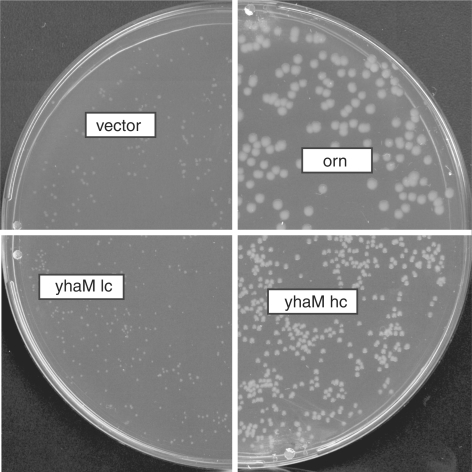
In our screen for B. subtilis genes that were able to complement E. coli orn–, we also identified a clone containing nt 1 068 397–1 070 326 of the B. subtilis genome (pBSL10). This region contains partial CDSs coding for YhaM, PrsA and a complete CDS coding for YhaL. YhaL is a gene of unknown function, implicated in sporulation (21). prsA codes for a molecular chaperone that is involved in protein secretion, this CDS was largely deleted in pBSL10. This plasmid coded only for the C-terminal part of PrsA protein (one-third of the full length). The CDS coding for YhaM was incomplete and allowed only for the expression of an N-terminally modified protein with the first 16 amino-acid residues replaced by a 24 amino-acid peptide encoded by the vector sequence. Despite that fact, we considered YhaM as the most likely candidate to be responsible for the complementation of E. coli orn–, as it is a previously characterized RNase (14,22). We therefore subcloned yhaM under the control of the arabinose-inducible promoter Para in pBAD18. This plasmid (pUM413), however, did not allow complementation of orn–. We subsequently constructed two additional high copy number plasmids expressing YhaM. pUM416 expressed C-terminally his-tagged YhaM under control of the PLac promoter in pUC18. pFM1 expressed native yhaM under control of its own promoter in pGEMT-Easy. pFM1 was the only construct allowing partial complementation of orn– (Figure 5). This could suggest that either high expression levels were necessary for complementation, or that the his-tag interfered with the activity of YhaM. Comparing expression levels, we found that indeed the original library clone that complementated orn– allowed very high expression of YhaM, which was easily detectable on a colloidal coomassie-stained gel. On the contrary, none of the other YhaM-expressing constructs allowed protein detection, partially due to presence of other proteins in this gel region. However, using western blot analysis with Anti-His6 the expression of all his-tagged proteins was detected (data not shown).
Figure 5.
Lacking or partial complementation of the conditional orn mutant by expression of YhaM from different constructs differing in copy number. Transformants of strain UM341 with pBAD18 (vector control), vector; pUM408 (arabinose-inducible orn), orn; pUM413 (arabinose-inducible yhaM in pBAD18, a low-copy (lc) number vector), yhaMlc; or pFM1 (yhaM in pGEMT-Easy, a high-copy (hc) number vector), yhaMhc were spread on LB plates containing 0.2% arabinose in the absence of anhydrotetracycline (Atc).
His-tagged YhaM purified from E. coli carrying pUM413 was able to degrade nanoRNA in vitro (Figure 6A). This suggests that the presence of his-tag does not account for the inability of pUM413 and pUM416 to complement the orn– mutation, although we cannot exclude partial inhibition of YhaM nanoRNase activity by the his-tag. The pattern of degradation of the nanoRNA 5-mer (5′Cy5-CCCCC3′) was similar to that produced by Orn- or NrnB-catalyzed degradation. The high expression levels required for complementation of E. coli orn–, however, made us consider that nanoRNA might not be a physiological substrate for YhaM. We therefore decided to compare the activity of YhaM on RNA and DNA substrates 5 nt in length.
Figure 6.
Comparison of YhaM activity on RNA and DNA substrates. Shown is the separation of reaction products on 22% PAA gels. 50 μl reactions contained (A) 20 μg YhaM and 5.4 μM RNA oligo (5′Cy5-CCCCC3′) or (B) 4 μg YhaM and 5.4 μM DNA oligo (5′Cy5-CCCCC3′).
Figure 6 compares YhaM activity on oligo RNA and on oligo DNA. YhaM degraded the RNA 5-mer at an initial rate of 0.76 pmol/min/μg, while it degraded 5-mer DNA at a rate of 10.14 pmol DNA 5-mer/min/μg enzyme during the first minute of the reaction. Since the DNA monomer did not enter the gels, it could not be included in the quantification. The values of all other reaction intermediates, as well as the value for the substrate 5-mer were therefore overrestimated, as soon as monomer appeared in the reaction. The turnover rate of the 5-mer DNA after 1 min might therefore be slightly underestimated. The appearance of 2-mer was much more rapid for the DNA than for RNA substrate, and the degradation of 270 pmol nanoRNA was less complete after 30 min by 20 μg YhaM, compared to the degradation of the same quantity of nanoDNA by 4 μg YhaM.
A triple mutant lacking nrnA, nrnB and yhaM is viable
As Orn is essential in E. coli, we wanted to ask if deleting all sources of nanoRNase activity known to us, i.e. nrnA, nrnB and yhaM would be lethal in B. subtilis. For this purpose we constructed an nrnB::CmR replacement allele and an allele carrying nrnA under control of the IPTG-inducible promoter PSpac. We combined these two alleles with the yhaM::PhleoR allele (14). Table 2 shows the results of growth experiments performed with mutants containing one or multiple mutant alleles. Growth of the single mutants in nrnA or nrnB as well as the double mutant was not affected. Growth of the triple mutant carrying nrnA under control of PSpac and replacement alleles for nrnB and yhaM (strain UM629) was only slightly affected in the absence of IPTG.
Table 2.
Doubling times of mutant strains in nrnA, nrnB and yhaM
| Strain | Genotype | Doubling time (min) |
|
|---|---|---|---|
| –IPTG | +IPTG | ||
| B. subtilis 168 | Wild type | 24 | |
| UM516 | yhaM::PhleoR | 24 | |
| UM545 | nrnB::CmR | 23 | |
| UM599 | nrnA::KmR | 24 | |
| UM638 | nrnB::CmR, nrnA::KmR | 25 | |
| UM629 | PSpac::nrnA, nrnB::CmR, yhaM::PhleoR (pMAP65 i.e. lac-IQ) | 29 | 25 |
| UM647 | nrnA::KmR, nrnB::CmR, yhaM::PhleoR | 26 | |
| UM651 | nrnA::EmR, nrnB::CmR, yhaM::PhleoR | 27 | |
We also created triple mutants with three replacement/insertional alleles. These strains (UM647 and UM651) were also only marginally affected in growth like the triple mutant carrying nrnA under the control of PSpac in the absence of IPTG (Table 2).
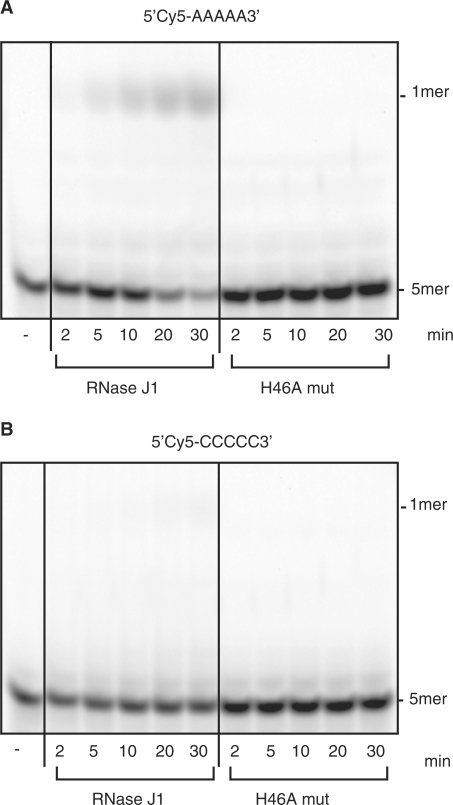
RNase J1 is able to degrade nanoRNA
According to in vitro data, the essential enzyme RNase J1 does not produce large quantities of 2- to 5-mers as end products of RNA 5′-to-3′ degradation (10). This encouraged us to test activity of RNase J1 on nanoRNA. Despite the fact that our in vitro conditions were not ideal (our substrate was labeled at the 5′ end with Cy-5, which could potentially interfere with 5′–3′ directed degradation), the release of mononucleotides (Figure 7) indicated activity of RNase J1 on nanoRNA 5-mers (5′Cy5-AAAAA3′). Activity on 5′Cy5-CCCCC-3′ was barely detectable (Figure 7). Estimations of specific activities of RNase J1 on 5′Cy5-AAAAA3′ and 5′Cy5-CCCCC-3′ revealed a roughly 10-fold difference in favor of 5′Cy5-AAAAA3′ with 0.5 pmol/min/μg as compared to 0.03 pmol/min/μg, respectively. An inactive mutant RNAse J1 (H46A) was included in order to exclude the possibility of contaminating activity in the RNAse J1 preparation and showed no detectable activity on either substrate.
Figure 7.
Degradation of nanoRNA 5-mers by RNase J1. Thirty-microliter reactions contained 3 μg RNase J1 or 5 μg RNase J1 mutant protein (RNase J1 H46A, H46A mut) and 5 μM RNA oligo (5′Cy5-AAAAA3′, Figure 7A; or 5′Cy5-CCCCC3′, Figure 7B). The minus indicates controls lacking enzyme.
To test whether expression of RNase J1 could complement E. coli orn–, we transformed pDG148-rnjA into UM341. This plasmid carries rnjA coding for RNase J1 under the control of the IPTG-inducible PSpac promoter. When transformed into the conditional orn mutant, colony sizes depended strongly on the concentration of IPTG added to the plates (Table 3). Complementation seemed to be partial in the absence of IPTG or at very low concentrations of the inducer (10 μM). At higher concentrations of IPTG, however, no colonies could be detected. The absence of growth at high IPTG concentrations may be due to the detrimental effect of RNase J1 overexpression even in Orn+ conditions, indicated by the fact that the colony size of transformants spread under permissive conditions in the presence of ≥50 μM IPTG was adversely affected (Table 3).
Table 3.
Growth of UM341 (E. coli orn–) carrying a vector control or expressing RNase J1 in the presence of varying concentrations of IPTG
| Plasmid | +Atc IPTG in μM |
−Atc IPTG in μM |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 10 | 50 | 100 | 200 | 0 | 10 | 50 | 100 | 200 | |
| pBAD18 (vector) | ++++ | ++++ | ++++ | ++++ | ++++ | + | + | + | + | + |
| pDG148-rnjA (RNase J1) | ++++ | ++++ | +++ | ++ | – | ++ | ++ | – | – | – |
| pUM408 (Orn) | ++++a | ++++a | ||||||||
Colony sizes: ++++, wild type; +++, smaller; ++, very small; +, tiny; –, no growth, Atc: anhydrotetracycline.
aIn the presence of 0.02% arabinose.
DISCUSSION
The purpose of this study was to search for enzymes that allow B. subtilis to degrade nanoRNA. We identify here one new nanoRNase, NrnB (previously YngD) and we tested nanoRNase activity of two known RNases, YhaM and RNase J1. All three enzymes are able to degrade nanoRNA in vitro to various extents. NrnB, however, is the only one of these three enzymes that allows full complementation of an E. coli orn mutant when expressed at low levels. This suggests that nanoRNA might not be the preferred or physiological substrate for YhaM or RNase J1.
YhaM
Despite the fact that a genomic library clone (pBSL10) containing most of the CDS for YhaM was found among the clones complementing the E. coli orn mutant, when subcloned, yhaM could only partially complement this mutant, even when expressed from a high copy number plasmid. Possible explanations for this discrepancy that one could consider are: (i) the changed protein sequence of the protein expressed from pBSL10 (16 N-terminal amino acids replaced by 24 amino acids encoded by the vector) could affect substrate specificity of the protein. (ii) Expression of yhaL, another CDS present in pBSL10, might be necessary for YhaM in vivo in order to complement the orn mutant. This hypothesis could not be tested as we were unable to obtain any viable clones coding for YhaM and YhaL (data not shown). The few clones that we obtained carried mutations and formed very small colonies suggesting that expression of those two genes together might be toxic in E. coli. (iii) High expression levels of YhaM are necessary for complementation. We consider the latter explanation the most likely due to the fact that the library clone allowing complementation indeed showed very high expression levels of YhaM. All other YhaM constructs did not allow detection of the expressed protein on a coomassie-stained protein gel due to its overlap with a strong band on the gel. The expression of his-tagged YhaM constructs that did not allow complementation was however detectable using Anti-His6 antibodies. Importantly, the expression of other nanoRNases, NrnB and NrnA, in the absence of inducer was not detectable by Anti-His6 antibodies and yet allowed complementation. However, a note of caution is warranted due to the fact that direct comparison between protein levels of NrnB, NrnA and YhaM might not be possible as the his-tag of different proteins could react differently with the Anti-His6 antibody due to different accessibility of the epitope. Consequently we are left with some uncertainty concerning the amount of YhaM present in constructs that did not allow complementation and cannot completely exclude that nanoRNA is a physiological substrate for YhaM.
The strong preference of DNA over RNA 5-mers that we observed in vitro could be another argument against nanoRNase being the main activity of YhaM in bacterial cells. In addition, this preference for DNA could point to a potential role of YhaM in DNA metabolism. yhaM is located in an operon containing yhaO, yhaN and yhaM that was identified as a part of the SOS LexA regulon in B. subtilis (23). The LexA-binding site is located upstream of the promoter region of the operon preceding yhaO. The corresponding sequence (aGAACgTgcaTTCG) differs in five positions from the consensus sequence. Global transcriptome analysis by microarrays shows that YhaO and YhaM are induced in a RecA-dependent manner when the SOS response is induced by mitomycin C or UV radiation (23). Although the functions of YhaO and YhaN are unknown, YhaO is annotated as a member of the SbcD protein family containing metallophosphoesterases involved in DNA repair, recombination and replication (24) and YhaN contains a domain with homology to this protein family. Thus, both YhaO and YhaN likely play a role in DNA metabolism, and our enzymatic assays suggest that YhaM might as well. Furthermore, all three proteins were linked in an interaction network that connects DNA repair, recombination and replication. Direct interaction between YhaM and DnaC, as well as between DnaG and YhaN was shown by yeast two hybrids screens (25). YhaO was linked indirectly through interaction with YhaN (25). Altogether, this demonstrates that the role of YhaM in nucleic acid metabolism still needs to be clarified.
RNase J1
Despite the in vitro activity of RNase J1 on nanoRNA, it is difficult to ascertain whether or not nanoRNA is a physiological substrate of this enzyme. This is because expression of RNase J1 seems to be detrimental to growth in E. coli, at least in the orn mutant strain we used. According to in vitro evidence (10,26), it is likely that RNase J1 degrades RNA completely and does not leave 2- to 5-mers in vivo. As this essential enzyme does have many other substrates (27), nanoRNA may not be a preferred substrate and therefore despite the ability of RNase J1 to finish degradation once started on a longer substrate, it might not pick up 2- to 5-mers that arise from incomplete degradation catalyzed by other enzymes.
NrnB
NrnB degrades nanoRNA 5-mers in vitro in a manganese- or cobalt-dependent 3′–5′ directed reaction and can complement an E. coli orn– mutant when expressed at low levels. Both in vitro and in vivo evidence presented here allow annotation of NrnB as a nanoRNase. The large activity difference in favor of 5-mer as compared to 24-mer substrate implies that nanoRNA is a preferred substrate of NrnB. After NrnA, NrnB is the second protein in B. subtilis that we have identified as a nanoRNase. We therefore proposed to rename YtqI and YngD as NrnA and NrnB, respectively.
Both proteins are members of the DHH/DHHA1 protein family of phosphoesterases, it is therefore likely that one can find other nanoRNases among the DHH proteins of unknown function. As in the case of NrnA and NrnB, these proteins may differ in their substrate specificity: the substrate specificity of NrnA is not restricted to nanoRNA but extends to pAp. We show here that this is not the case for NrnB. Although NrnA has a strong preference for 3-mers in vitro, NrnB does not show such a preference and has in fact lower activity on 3-mers than on 5-mers. Thus, the two nanoRNases present in B. subtilis have different but overlapping substrate specificities, which could add some functional redundancy and/or specialization in this organism. Despite the fact that the specific activity of NrnA on 5-mer C's is considerably lower in vitro than that of NrnB (0.2 pmol/μg/min versus 1 nmol/μg/min, respectively) [data extracted from ref. (6)], both enzymes are able to complement the E. coli orn mutant when expressed at low levels. Due to the preference of NrnA for 3-mers, the difference between the specific activity of NrnA and NrnB is much smaller when considering activities on 3-mers (<6-fold). Nevertheless, even with 3-mers, NrnB is more active than NrnA. However, it is possible that conditions for activity assays of NrnA were not optimal and therefore, direct comparison of specific activities of the two enzymes should not be over interpreted. Also, the fact that many bacterial species that do not carry Orn, have only NrnA and not NrnB supports the importance of NrnA as a nanoRNase under physiological conditions.
NrnB is present in a smaller number of genomes as compared to NrnA, and several of those species are closely related to B. subtilis. However, we also found NrnB orthologs in ε-proteobacteria, namely Helicobacter sp. and Campylobacter sp. and in some Archaea. Intriguingly, ε-proteobacteria possesses neither Orn- nor NrnA-orthologs. NrnB might therefore play a more important role in the degradation of nanoRNA in these organisms, although the simultaneous presence of RNase J1 homologs in these species might contribute as well.
Simultaneous deletion of nrnA, nrnB and yhaM in B. subtilis has a minor effect, if any, on growth in complex medium. This result could be interpreted in two ways: (i) RNase J1/J2 can take over this essential function in the absence of these three genes. This hypothesis cannot be tested easily due to essentiality of RNase J1 itself. (ii) Yet another protein exists in B. subtilis that has nanoRNase activity. We cannot exclude this possibility, as our genomic library screen for the orn– complementing genes was not saturated. Candidates might be among the DHH proteins of unknown function in Bacillus: YorK and YybT, for example.
With the identification of NrnA and NrnB, however, our work demonstrates that the degradation of nanoRNA catalyzed by a single protein Orn in E. coli is performed by redundant proteins in B. subtilis. This is a remarkable example of xenologous gene replacement (28) in the case of coding for ubiquitous functions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union Network of Excellence BioSapiens [LSHG CT-2003-503265 to A.D.]; the French Research Agency (Agence nationale de la recherche) [BLAN06-3_135068 to C.C.]; La Ligue Contre le Cancer [9ADO1217/1B1-BIOCE to V.O.]; Institut national du Cancer [247343/1B1-BIOCE to V.O.]. Funding for open access charge: Institut Pasteur.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Emilie Cochet for assistance in mass spectrometry, Isabelle Martin-Verstraete, Anne-Marie Gilles, and Tingzhang Wang for helpful discussions, Florence Hommais for construction of the genomic library of B. subtilis, Saravuth Ngo for preliminary in vitro experiments, Andrew Martens and Nikolay Ogryzko for carefully reading the manuscript. We thank David Bechhofer and Patrick Stragier for the kind gift of yhaM::PhleoR and pEC23, respectively.
Footnotes
Present address: Ming Fang, Institut Curie, Centre de Recherche, UMR CNRS 7147 (Unité de Dynamique de l'information génétique: Bases fondamentales et Cancer), 75248 Paris Cedex 05, France.
REFERENCES
- 1.Condon C. Maturation and degradation of RNA in bacteria. Curr. Opin. Microbiol. 2007;10:271–278. doi: 10.1016/j.mib.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 2.Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 2002;277:21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- 3.Amblar M, Barbas A, Fialho AM, Arraiano CM. Characterization of the functional domains of Escherichia coli RNase II. J. Mol. Biol. 2006;360:921–933. doi: 10.1016/j.jmb.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 4.Amblar M, Barbas A, Gomez-Puertas P, Arraiano CM. The role of the S1 domain in exoribonucleolytic activity: substrate specificity and multimerization. RNA. 2007;13:317–327. doi: 10.1261/rna.220407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J. Biol. Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- 6.Mechold U, Fang G, Ngo S, Ogryzko V, Danchin A. YtqI from Bacillus subtilis has both oligoribonuclease and pAp-phosphatase activity. Nucleic Acids Res. 2007;35:4552–4561. doi: 10.1093/nar/gkm462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghosh S, Deutscher MP. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc. Natl Acad. Sci. USA. 1999;96:4372–4377. doi: 10.1073/pnas.96.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu D, Deutscher MP. Oligoribonuclease is distinct from the other known exoribonucleases of Escherichia coli. J. Bacteriol. 1995;177:4137–4139. doi: 10.1128/jb.177.14.4137-4139.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Zhu L, Deutscher MP. Oligoribonuclease is encoded by a highly conserved gene in the 3′-5′ exonuclease superfamily. J. Bacteriol. 1998;180:2779–2781. doi: 10.1128/jb.180.10.2779-2781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathy N, Benard L, Pellegrini O, Daou R, Wen T, Condon C. 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell. 2007;129:681–692. doi: 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- 11.Mechold U, Ogryzko V, Ngo S, Danchin A. Oligoribonuclease is a common downstream target of lithium-induced pAp accumulation in Escherichia coli and human cells. Nucleic Acids Res. 2006;34:2364–2373. doi: 10.1093/nar/gkl247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stragier P, Bonamy C, Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, et al. Essential Bacillus subtilis genes. Proc. Natl Acad. Sci. USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oussenko IA, Sanchez R, Bechhofer DH. Bacillus subtilis YhaM, a member of a new family of 3′-to-5′ exonucleases in gram-positive bacteria. J. Bacteriol. 2002;184:6250–6259. doi: 10.1128/JB.184.22.6250-6259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bechhofer DH, Oussenko IA, Deikus G, Yao S, Mathy N, Condon C. Analysis of mRNA decay in Bacillus subtilis. Methods Enzymol. 2008;447:259–276. doi: 10.1016/S0076-6879(08)02214-3. [DOI] [PubMed] [Google Scholar]
- 16.Aravind L, Koonin EV. A novel family of predicted phosphoesterases includes Drosophila prune protein and bacterial RecJ exonuclease. Trends Biochem. Sci. 1998;23:17–19. doi: 10.1016/s0968-0004(97)01162-6. [DOI] [PubMed] [Google Scholar]
- 17.Sutera V.A., Jr., Han ES, Rajman LA, Lovett ST. Mutational analysis of the RecJ exonuclease of Escherichia coli: identification of phosphoesterase motifs. J. Bacteriol. 1999;181:6098–6102. doi: 10.1128/jb.181.19.6098-6102.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamagata A, Kakuta Y, Masui R, Fukuyama K. The crystal structure of exonuclease RecJ bound to Mn2+ ion suggests how its characteristic motifs are involved in exonuclease activity. Proc. Natl Acad. Sci. USA. 2002;99:5908–5912. doi: 10.1073/pnas.092547099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tammenkoski M, Moiseev VM, Lahti M, Ugochukwu E, Brondijk TH, White SA, Lahti R, Baykov AA. Kinetic and mutational analyses of the major cytosolic exopolyphosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 2007;282:9302–9311. doi: 10.1074/jbc.M609423200. [DOI] [PubMed] [Google Scholar]
- 20.D'Angelo A, Garzia L, Andre A, Carotenuto P, Aglio V, Guardiola O, Arrigoni G, Cossu A, Palmieri G, Aravind L, et al. Prune cAMP phosphodiesterase binds nm23-H1 and promotes cancer metastasis. Cancer Cell. 2004;5:137–149. doi: 10.1016/s1535-6108(04)00021-2. [DOI] [PubMed] [Google Scholar]
- 21.Feucht A, Evans L, Errington J. Identification of sporulation genes by genome-wide analysis of the sigmaE regulon of Bacillus subtilis. Microbiology. 2003;149:3023–3034. doi: 10.1099/mic.0.26413-0. [DOI] [PubMed] [Google Scholar]
- 22.Oussenko IA, Abe T, Ujiie H, Muto A, Bechhofer DH. Participation of 3′-to-5′ exoribonucleases in the turnover of Bacillus subtilis mRNA. J. Bacteriol. 2005;187:2758–2767. doi: 10.1128/JB.187.8.2758-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Au N, Kuester-Schoeck E, Mandava V, Bothwell LE, Canny SP, Chachu K, Colavito SA, Fuller SN, Groban ES, Hensley LA, et al. Genetic composition of the Bacillus subtilis SOS system. J. Bacteriol. 2005;187:7655–7666. doi: 10.1128/JB.187.22.7655-7666.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noirot-Gros MF, Dervyn E, Wu LJ, Mervelet P, Errington J, Ehrlich SD, Noirot P. An expanded view of bacterial DNA replication. Proc. Natl Acad. Sci. USA. 2002;99:8342–8347. doi: 10.1073/pnas.122040799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deikus G, Condon C, Bechhofer DH. Role of Bacillus subtilis RNase J1 endonuclease and 5′-exonuclease activities in trp leader RNA turnover. J. Biol. Chem. 2008;283:17158–17167. doi: 10.1074/jbc.M801461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mader U, Zig L, Kretschmer J, Homuth G, Putzer H. mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol. Microbiol. 2008;70:183–196. doi: 10.1111/j.1365-2958.2008.06400.x. [DOI] [PubMed] [Google Scholar]
- 28.Pushker R, Mira A, Rodriguez-Valera F. Comparative genomics of gene-family size in closely related bacteria. Genome Biol. 2004;5:R27. doi: 10.1186/gb-2004-5-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petit MA, Dervyn E, Rose M, Entian KD, McGovern S, Ehrlich SD, Bruand C. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol. Microbiol. 1998;29:261–273. doi: 10.1046/j.1365-2958.1998.00927.x. [DOI] [PubMed] [Google Scholar]
- 32.Vagner V, Dervyn E, Ehrlich SD. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144(Pt 11):3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 33.Sullivan MA, Yasbin RE, Young FE. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene. 1984;29:21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.