Abstract
Non-coding RNAs are emerging as key players in many fundamental biological processes, including specification of higher-order chromatin structure. We examined the implication of RNA transcribed from mouse centromeric minor satellite repeats in the formation and function of centromere-associated complexes. Here we show that the levels of minor satellite RNA vary during cell-cycle progression, peaking in G2/M phase, concomitant with accumulation of proteins of the chromosomal passenger complex near the centromere. Consistent with this, we describe that murine minor satellite RNA are components of CENP-A-associated centromeric fractions and associate with proteins of the chromosomal passenger complex Aurora B and Survivin at the onset of mitosis. Interactions of endogenous Aurora B with CENP-A and Survivin are sensitive to RNaseA. Likewise, the kinase activity of Aurora B requires an RNA component. More importantly, Aurora B kinase activity can be potentiated by minor satellite RNA. In addition, decreased Aurora B activity after RNA depletion can be specifically rescued by restitution of these transcripts. Together, our data provide new functional evidence for minor satellite transcripts as key partners and regulators of the mitotic kinase Aurora B.
INTRODUCTION
The centromeres of eukaryotic chromosomes are genomic regions featuring a unique and specific chromatin architecture, necessary for proper chromosome segregation during mitosis. The common trait of centromeres in all species is the presence of nucleosomes containing a specific variant of histone H3, the centromere protein A (CENP-A) (1). Flanking pericentromeric regions are devoid of CENP-A, but exhibit a high density of histone H3 tri-methylated on its lysine 9 (H3K9Me3), consistent with the heterochromatin nature of these domains (2). In addition to CENP-A, numerous proteins identified as essential for centromere assembly and function occupy centromeric regions in a constitutive manner (3). In contrast, the Chromosomal Passenger Complex (CPC), composed of Aurora B kinase and its regulatory subunits inner centromere protein (INCENP), Survivin and Borealin, shows dynamic changes in its subcellular localization in a cell-cycle-dependent manner (4).
Sequences of centromeric DNA repeats are not conserved among species, but transcripts originating from them have been described in a broad range of organisms (5–12). It remains unclear how these RNA participate in the formation and/or stabilization of large-scale centromeric chromatin structures. However, in many chromatin complexes, including pericentromeric heterochromatin, RNA is an integral component (13,14).
The repeats of minor satellite DNA on mouse chromosomes are found at the primary constriction, adjacent to repeats of major satellite that define pericentromeric regions. We previously described new RNA transcribed from minor satellite repeats and their accumulation on chromocenters, which are clusters of several centromeres, and proposed that they might participate in the formation of specific centromeric complexes (15). We now report that minor satellite transcripts are integral components of the CENP-A chromatin fraction and associate with endogenous CPC proteins Aurora B, Survivin and INCENP, at the onset of mitosis. Moreover, these transcripts potentiate Aurora B kinase activity on its mitotic substrate, the histone H3. Together with the cell-cycle regulated accumulation of minor satellite RNA during G2/M transition, our data provide new insights into the functional implication of non-coding minor satellite RNA in favouring Aurora B specific interaction with CENP-A-associated chromatin domains and enzymatic function at the onset of mitosis.
MATERIALS AND METHODS
Cell culture and synchronization
Murine Erythroleukemic (MEL) cells were grown in RPMI-1640 medium (Gibco) supplemented with 10% Fetal Calf Serum (FCS, Invitrogen). Exponentially growing MEL cells were stained with 5 μg/ml of Hoechst 33342 (Sigma) and sorted in different phases of the cell cycle by Fluorescence Activated Cell Sorting (FACS) (Epics ALTRA, Beckman Coulter) on the basis of their DNA content. Exponentially growing MEL cells were arrested in mitosis by 12 h treatment with 100 ng/ml of nocozadole (Sigma).
Murine NIH/3T3 fibroblasts were grown in DMEM (Gibco) supplemented with 10% FCS and were synchronized in mitosis by mechanical shake off.
Cell-cycle stage was determined by flow cytometry following cell fixation with 70% cold ethanol and propidium iodide-staining PI (Sigma) 20 μg/ml, 0.1% Triton X-100, 0.2 mg/ml RNaseA (Sigma).
MEL cells have a near-diploid karyotype whereas NIH/3T3 cells contain hyper-tetraploid cells. Since transcription of satellite sequences can be induced in stress conditions (15–17), culture conditions were controlled to ensure these cells have not undergone any stresses in culture (lack of nutriments, heat shock, etc.).
RNA extraction and semi-quantitative RT-PCR
For cell-cycle analysis, total RNA was isolated with Trizol reagent (Invitrogen) according to the manufacturer's instructions. Genomic DNA was removed by digestion with 2 U of DNaseI (Ambion). cDNA were synthesized from an equivalent of 500 000 cells (Figure 1A) or 100 000 cells (Figure 1B and C), using random hexamers and Superscript II reverse transcriptase (Invitrogen), and amplified by PCR. The amount of input cDNA was normalized to the signal obtained using primers that amplify the mouse β-actin transcripts. All PCR assays were calibrated to 23 cycles to stay in a range of linear amplification for minor satellite RNA. When using lower number of cells (100 000 cells) an additional hybridization step was performed rather than increasing the number of cycles.
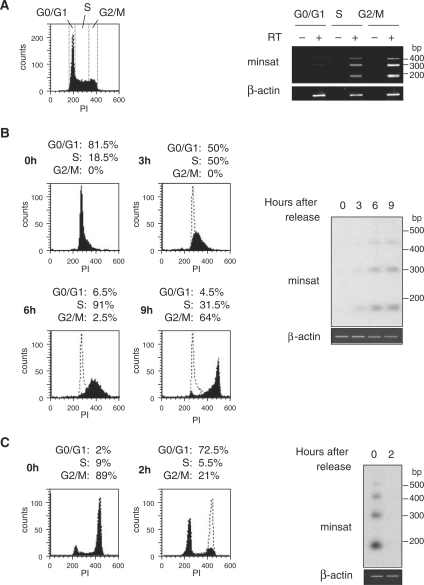
Figure 1.
Minor satellite transcripts accumulate in G2/M phase of the cell cycle. (A) FACS plot showing the gates used to sort MEL cells at different stages (G0/G1, S, G2/M) of the cell cycle (left panel). Levels of minor satellite RNA (minsat) from each fraction were evaluated by RT–PCR, in reactions containing (+) or not (−) RT (right panel), and normalized to the signal of a RT–PCR reaction amplifying β-actin transcripts, which levels do not change during cell cycle. (B) Cell-cycle distribution of PI-labelled MEL cells after FACS-sorting in G0/G1 phase and release in culture for 9 h (left panel). Total RNA at time 0, 3, 6 and 9 h was analysed by RT–PCR followed by hybridization with a minor satellite probe (right panel). Control RT-PCR reaction amplifying β-actin transcripts are shown below. (C) FACS analysis of PI-labelled NIH/3T3 cells after mitotic shake off and release into culture (left panel). Total RNA at time 0 and 2 h was analysed by RT–PCR followed by hybridization with a minor satellite probe (right panel). Control RT–PCR reactions amplifying β-actin transcripts are shown below.
The primers used in PCR or RT-PCR analysis were:
Mouse minor satellite:
forward 5′-GAACATATTAGATGAGTGAGTTAC-3′ and
reverse 5′-GTTCTACAAATCCCGTTTCCAAC-3′
Mouse β-actin:
forward 5′-AAGAGCTATGAGCTGCC-3′ and
reverse 5′-ACTCCTGCTTGCTGATCC-3′
Primers used to amplify mouse major satellite and ribosomal DNA are described in ref. (18).
For Southern blot hybridization, the minor satellite fragment used as a probe corresponds to region 20–100 of the consensus sequence (19) shown in Supplementary Figure S4.
Antibodies
The primary antibodies used in the present study were directed against CENP-A (ab33565, Abcam), H3K9Me3 (Upstate), H3Ac (Upstate), HP1γ (Euromedex), HDAC1 (2E10; Upstate), Aurora B (ab2254, Abcam), Survivin (ab469, Abcam), INCENP (ab36453, Abcam), H3K9Me3-S10ph (ab5819, Abcam) and Sir2 (Upstate).
Nuclear extracts and immunoprecipitation
Cells were resuspended in hypotonic buffer [10 mM Tris pH 7.6, 10 mM KCl, 1.5 mM MgCl2, complete protease inhibitors (Roche) in presence of 2 mM Ribonucleoside Vanadyl Complex-RNase Inhibitor (VRC, New England BioLabs]. After disruption of cytoplasmic membranes with a dounce, nuclei were extracted in ice-cold lysis buffer (10 mM Tris–HCl pH 7.5, 5 mM EDTA, 150 mM NaCl, 30 mM sodium pyrophosphate, 50 mM NaF, 1 mM NaVO4, 10% glycerol, 1% NP40, 1 mM DTT and complete protease inhibitors) in presence of 1 U/µl RNaseOut (Invitrogen) and sonicated (three pulses of 20 s, 5 W, Ultrasonic processor, Bioblock Scientific). Magnetic protein-G beads (Dynabeads) were pre-incubated with lysis buffer containing 200 μg/ml bovine serum albumin (Pierce) and 0.2 mg/ml yeast tRNA (Ambion). Nuclear extracts were incubated for 2 h at 4°C with antibody bound to the beads, in lysis buffer containing 15 μg/ml yeast tRNA, 1 U/µl RNaseOut. Beads were washed twice with lysis buffer adjusted to 300 mM NaCl and once with lysis buffer in presence of VRC.
For RNA NChIP, precipitated RNA was extracted with Trizol reagent, digested with DNaseI, and the totality of precipitated RNA was reverse transcribed. To assess presence of minor satellite RNA in the precipitate, 1 µl of the resulting cDNA was used for PCR analysis and 30 cycles of PCR were performed.
For co-IP analysis, recovered proteins were analysed by western blot using the indicated antibodies. RNaseA treatment was performed with 100 μg/ml RNaseA (Ambion) on nuclear extracts or after IP.
RNA pull-down
A scheme of the procedure is shown in Supplementary Figure S4. One hundred and twenty base pair repeat unit of minor satellite cDNA was generated by PCR amplification and cloned into EcoRI-BamHI sites of pcDNA3 vector (Invitrogen). Minor satellite RNA was in vitro transcribed in both orientations using T7 or SP6 RiboMax large-scale production system (Promega). Four hundred picomoles of RNA probe was annealed with 500 pmol of biotinylated oligonucleotide in 50 mM KCl, 1 U/µl RNaseOUT (Invitrogen), for 1 h at RT. Different types of biotinylated oligonucleotides were tested for their ability to precipitate minor satellite RNA (Supplementary Figure S4): a 2′-O-methyl RNA oligonucleotide complementary to regions 1–27 of minor satellite consensus sequence and biotinylated in 5′ (lane 1), a DNA oligonucleotide complementary to regions 1–27 of consensus sequence and biotinylated in 5′ (lane 2), a DNA/locked nucleic acids (LNA) mixmer oligonucleotide complementary to regions 4–27 of consensus sequence and biotinylated in 3′ (lane 3), a DNA/LNA mixmer oligonucleotide complementary to regions 89–115 of consensus sequence and biotinylated in 3′ (lane 4) and a DNA/LNA mixmer oligonucleotide complementary to regions 4–27 of consensus sequence and biotinylated in 5′ (lane 5). As shown in Supplementary Figure S4, the DNA/LNA mixmer oligonucleotide complementary to regions 4–27 of consensus sequence and biotinylated in 3′ (lane 3) retained more efficiently minor satellite RNA and was further used in all experiments. Streptavidin magnetic beads (Dynabeads) were washed according to the manufacturer's instructions twice in 0.1 M NaOH, 0.05 M NaCl and once in 0.1 M NaCl and then incubated with the RNA/oligonucleotide complex in binding/washing buffer (10 mM Tris–HCl pH 7.5, 1 mM EDTA, 1 M NaCl) for 30 min at RT. Pre-incubated beads were washed twice in binding/washing buffer and added to 4 mg of nuclear extract in RNA binding buffer (10% glycerol, 10 mM HEPES pH 7, 150 mM KCl, 1 mM EDTA, 0.5% TritonX-100) containing 15 μg/ml yeast tRNA, 1 mM DTT, 240 U RNaseOut for 2 h at 4°C. RNA-bound proteins were washed twice with RNA binding buffer adjusted to 300 mM KCl and once with RNA binding buffer before protein denaturation. Precipitated proteins were then resolved on a gradient-like SDS–PAGE (Prosieve, Cambrex), transferred on a Hybond C-extra membrane (Amersham) and blotted with the indicated antibodies.
Kinase assay
Immunoprecipitated Aurora B kinase was included into a 20 μl reaction containing 1 μg of canonical core histones (Upstate), 50 mM HEPES pH 7.5, 10 mM MgCl2, 1 mM DTT, 0.1 mM NaVO4, 1 mM NaF, 0.5 μl of [γ-32P]ATP (PerkinElmer Life Sciences) and increasing amounts of in vitro transcribed minor satellite RNA or β-tubulin RNA. Protein samples were separated by 12% SDS–PAGE and phosphate incorporation determined by phosphorimager (Typhoon 8600, Amersham Pharmacia Biotech). For rescue experiment, RNaseA treatment was performed after IP and kinase reactions were carried out in presence of 1 U/µl RNaseOut and in vitro transcribed minor satellite or β-tubulin RNA.
RESULTS
Levels of minor satellite transcripts vary with cell cycle
We previously reported that minor satellite transcripts accumulate on chromocenters in a subset of cycling cells, suggesting that levels of these RNA may vary during cell cycle (15). We therefore analysed the levels of these RNA in MEL cells sorted in G0/G1, S or G2/M phase (Figure 1A). Semi-quantitative RT–PCR using primers amplifying minor satellite transcripts was normalized to the signal of a RT–PCR reaction amplifying mouse β-actin transcripts, whose levels do not change during the cell cycle. RT–PCR analysis of minor satellite RNA showed a long ladder of discrete bands differing by about 120 bp consistent with transcription through multiple tandem repeats of the 120-nt minor satellite unit, and revealed that the levels of minor satellite transcripts were barely detectable in G0/G1 phase, increased in S phase and peaked in G2/M phase (Figure 1A).
MEL cells were then synchronized in G0/G1 phase using FACS sorting and released in culture for 9 h. Flow cytometry analysis confirmed the synchronous progression of the sorted population through early and late S phase, then G2 and mitosis, at 3, 6 and 9 h, respectively after release in culture (Figure 1B). In agreement with the results described above, RT–PCR analysis on total RNA isolated at various time points after release in culture showed that the levels of minor satellite RNA were below detection levels in G0/G1 sorted cells, started to increase 3 h after release in culture, concomitantly with entry of MEL cells into S phase, accumulated gradually during the course of S phase and reached a maximum in G2/M phase (Figure 1B). We then performed similar experiments in a different murine cell line. NIH/3T3 cells were synchronized in mitosis by mechanical shake-off and cultured over a 2 h time course that was sufficient for the majority of the cells to complete mitosis and enter the subsequent G1 phase of the cell cycle (Figure 1C). Minor satellite transcripts strongly accumulated in mitotic NIH/3T3 cells and became undetectable as early as 2 h after mitosis, when the cells re-entered the next G1 phase (Figure 1C).
Taken together, these results indicated that the pool of minor satellite RNA varies significantly during cell cycle, from barely detectable in G1 phase to a distinct peak in G2/M.
CENP-A chromatin fraction contains minor satellite RNA
Our initial characterization of minor satellite transcripts subcellular location by RNA-FISH revealed that these RNA localize to chromocenters, suggesting that they belong to specific ribonucleoprotein complexes located at centromeric or pericentromeric regions (15). To further characterize centromeric RNA-containing complexes we isolated subnuclear fractions enriched in chromocenters by discontinuous sucrose gradient (Supplementary Figure S1), as described in the Supplementary Data. Chromocenters were separated from dispersed fragments of low density chromatin in a fraction characterized by enrichment in DNA sequences and proteins specifically found at centromeric regions, i.e. CENP-A (Supplementary Figure S1B) and centromeric minor satellite DNA sequences (Supplementary Figure S1C and D). In addition, the CPC proteins Aurora B and Survivin were predominantly recovered in the chromocenter fraction. In contrast, histone deacetylase HDAC1 was recovered in both low density and chromocenter fractions, consistent with its subnuclear localization in MEL cells (20), whereas the repressor Sir2 was recovered only in low density chromatin fractions, also consistent with its previously reported subnuclear location (21) (Supplementary Figure S1B). In those conditions, RT-PCR analysis on total RNA isolated after nuclear fractionation showed that minor satellite RNA were specifically enriched in the chromocenter fraction (Supplementary Figure S1E).
To further characterize minor satellite RNA-containing ribonucleo-complexes located at centromeric regions, we performed Native Chromatin Immunoprecipitation (NChIP) using antibodies directed against CENP-A. As controls, we used antibodies directed against H3K9Me3, a heterochromatin mark found at pericentromeric regions, or against acetylated H3 (H3Ac), an activating mark mainly associated with euchromatic regions. DNA or RNA were then isolated from the precipitated material and analysed by PCR or RT-PCR. The immuno-precipitated material was first tested for the presence of centromeric minor and pericentromeric major satellite DNA sequences. To assess the NChIP background, we used primers specific for ribosomal DNA. Under these conditions, CENP-A indeed exclusively co-precipitated with minor satellite DNA, whereas both murine minor and major satellite DNA sequences associated with H3K9Me3 chromatin fractions (Supplementary Figure S2), in agreement with previous reports (2,18). In addition, association between these repetitive sequences and H3Ac was significantly lower consistent with the hypoacetylated status of these regions (22). In contrast, control ribosomal DNA (rDNA) was only detected in the H3Ac fraction (Supplementary Figure S2). To further assess the association of minor satellite transcripts with murine centromeric fractions, NChIP was performed from asynchronous and nocodazole-arrested MEL cells, using the same antibodies, in the presence of RNase inhibitor, and followed by a DNaseI treatment of the collected material. Following nocodazole treatment 86% of MEL cells accumulated in G2/M phase (Supplementary Figure S3, left and middle panel), a stage at which minor satellite RNA strongly accumulated in the cells (Supplementary Figure S3, right panel). RT-PCR analysis of precipitated RNA revealed that minor satellite RNA were specifically associated with the CENP-A immune complex in both asynchronous and nocodazole-arrested MEL cells (Figure 2A). In contrast, minor satellite RNA were not associated with H3K9Me3 or H3Ac chromatin fractions (Figure 2A), even in G2/M arrested cells, a stage at which minor satellite RNA strongly accumulated in the cells (Supplementary Figure S3, right panel). As a negative control, abundant control rDNA transcripts were not precipitated with any of the antibodies used, whereas RT-PCR performed on supernatants from NChIP reactions showed correct amplification of minor satellite RNA and rDNA transcripts in all samples, indicating that RNA was not degraded (Figure 2A).
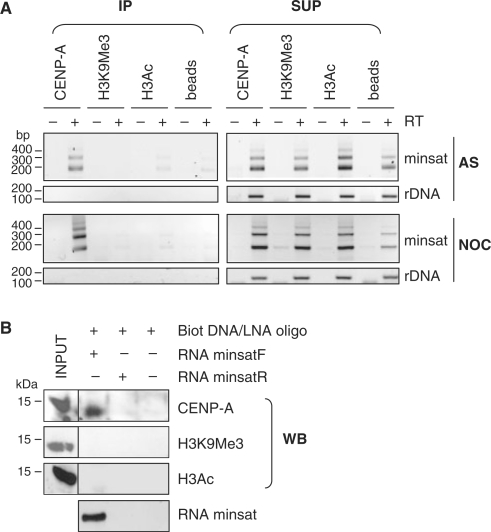
Figure 2.
Minor satellite RNA associate with CENP-A-containing chromatin. (A) RNA NChIP from nuclear extracts of asynchronously growing (AS) or nocodazole-arrested (NOC) MEL cells using antibodies against CENP-A, H3K9Me3 or H3Ac. Following DNaseI digestion, minor satellite RNA (minsat) and rDNA transcripts (rDNA) were analysed by RT-PCR, from the immunoprecipitated (IP) or supernatant fractions (SUP), in reactions containing (+) or not (−) RT. Control reactions in the absence of immunoprecipitating antibody (beads) were used as a negative control. (B) RNA pull-down experiments using in vitro transcribed minor satellite RNA (minsatF), followed by western blot (WB) analysis with the indicated antibodies. As a control, a similar experiment was performed with in vitro transcribed minor satellite RNA from the reverse strand (minsatR), therefore not complementary to the biotinylated DNA/LNA oligonucleotide used, or in the absence of RNA.
These results provide evidence that minor satellite transcripts preferentially associate with the CENP-A chromatin fraction ex vivo.
To confirm association of minor satellite RNA with CENP-A chromatin, RNA pull-down assays were performed, using minor satellite RNA transcribed in vitro from a 120-nt repeat unit (minsatF), bound to a complementary biotinylated oligonucleotide, and incubated with MEL cells nuclear extracts. We first tested several types of oligonucleotides for their efficiency to pull down minor satellite transcripts in vitro, and chose a 3′ biotinylated compound DNA/LNA oligonucleotide (Supplementary Figure S4). After pull-down using streptavidin-coated magnetic beads, and verification that the oligonucleotide efficiently retained minor satellite RNA (Figure 2B, bottom line), the associated proteins were analysed by western blot. Immunoblot analysis of minor satellite RNA-associated histone H3 species revealed that these RNA preferentially precipitated CENP-A, but not H3K9Me3 nor H3Ac (Figure 2B). CENP-A was not precipitated in various control experiments in which centromeric RNA was omitted (Figure 2B, far right column) or reactions treated with RNaseA prior or after precipitation (not shown). As an additional control for the specificity of the DNA/LNA oligonucleotide, we used in vitro transcribed RNA from the reverse strand (minsatR), therefore not complementary to the oligonucleotide used, which did not precipitate CENP-A (Figure 2B). Together, our experiments showed that CENP-A was detected in the precipitated material only when minor satellite RNA were efficiently precipitated.
Minor satellite RNA associate with proteins of the CPC in G2/M phase
The association of minor satellite transcripts with CENP-A-associated chromatin together with their steady-state levels peaking in G2/M phase suggests that they could play a role in the assembly of centromeric complexes during G2/M transition. The CPC exhibits a highly dynamic localization throughout the cell cycle. During G2 phase and metaphase, the complex localizes to centromeres, transfers to the central spindle at the onset of anaphase, and finally flanks the midbody during telophase and cytokinesis (4). To determine whether proteins of the CPC associate with minor satellite transcripts, we performed RNA pull-down assays using in vitro transcribed 120-nt minor satellite RNA, and analysed the presence of the kinase Aurora B, its obligate co-activator Survivin and the inner centromere protein INCENP, in the precipitated material. RNA pull-down assays revealed that Aurora B, Survivin and, to a lesser extent, INCENP, were efficiently precipitated when the oligonucleotide was complementary to the minor satellite RNA (Figure 3A, minsatF). No signal was observed when the reaction was carried out in presence of RNaseA or when the RNA was omitted from the reaction (Figure 3A, two left panels). As described above, an additional control consisted in using in vitro transcribed RNA not complementary to the oligonucleotide used in the pull-down assay (minsatR). Interestingly, although heterochromatin protein HP1γ and the histone deacetylase HDAC1 associated with chromocenters in MEL cells (20), they were not retained on minor satellite transcripts (Figure 3A).
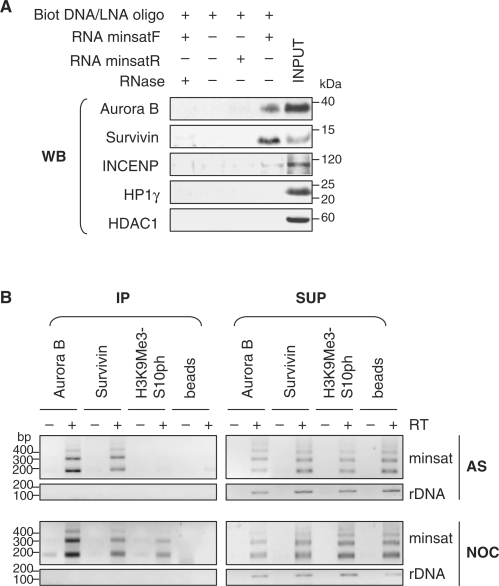
Figure 3.
Association of minor satellite RNA with proteins of the chromosomal passenger complex. (A) In vitro transcribed minor satellite RNA (minsatF) was used in RNA pull-down experiments, followed by western blot (WB) analysis with the indicated antibodies. As controls, similar experiments were performed in presence of RNaseA, in absence of minor satellite RNA, or using an RNA not complementary to the biotinylated oligonucleotide used (minsatR). (B) RNA NChIP from nuclear extracts of asynchronously growing (AS) and nocodazole-arrested (NOC) MEL cells using antibodies against Aurora B, Survivin and H3K9Me3-S10ph. As a negative control, a similar experiment was performed in the absence of immunoprecipitating antibody (beads). Following DNaseI treatment, immunoprecipitated (IP) RNA and RNA from supernatant fractions (SUP) were analysed by RT-PCR, in reactions containing (+) or not (−) RT, for the presence of minor satellite RNA (minsat) and rDNA transcripts (rDNA).
To validate these results ex vivo, we immunoprecipitated Aurora B and Survivin from asynchronous MEL cells extracts. RT-PCR analysis of the co-precipitated RNA resulted in specific amplification of minor satellite RNA from both Aurora B and Survivin immunoprecipitated material, whereas they were not detected in control reaction performed in absence of immuno-precipitating antibody (Figure 3B). In addition, no association was observed between these proteins and the abundant rDNA transcripts (Figure 3B). Similar experiments were conducted with nocodazole-arrested MEL cells. NChIP data revealed that minor satellite RNA were significantly associated with Aurora B and Survivin in nocodazole-arrested cells whereas rDNA transcripts were not precipitated (Figure 3B). Phosphorylation of H3 on its serine 10 by Aurora B kinase initiates at centromeric regions in late G2 interphase cells, and further spreads throughout the condensing chromatin. We found that minor satellite RNA co-precipitated with the H3K9Me3-S10ph in nocodazole-arrested cells (Figure 3B). In all experiments, RT-PCR from supernatant fractions showed correct amplification of both minor satellite RNA and rDNA transcripts, indicating that RNA was not degraded.
These data are consistent with a model in which minor satellite RNA could provide a scaffold to recruit and/or stabilize passenger proteins at centromeric regions in G2/M phase.
The assembly of Aurora B/Survivin complex and Aurora B kinase activity require an RNA component
The observation that the CPC and minor satellite RNA have the potential to form a protein–RNA complex, prompted us to test the functional relevance of this association. Assembly of the CPC is a prerequisite for its correct localization, kinase activity of Aurora B and substrate specificity during G2/M transition (23). We first examined the ability of Aurora B to form a complex in vivo with Survivin, in presence or absence of RNA. We tested the sensitivity of the interactions to RNaseA treatment, before immunoprecipitation of Aurora B complexes and omission of RNase inhibitors during the process, or after immunoprecipitation of Aurora B complexes. Under these conditions, the amount of recovered Aurora B was not affected by RNaseA treatment (Figure 4A). In contrast, analysis of co-immunoprecipitated material showed that the interaction of Aurora B with Survivin was significantly diminished after RNA depletion compared to control experiments where RNA was protected from degradation (Figure 4A). In addition, we tested whether association of Aurora B with CENP-A-associated chromatin was dependent on an RNA molecule. Interestingly, our results showed that RNaseA treatment completely abolished the interaction between Aurora B and CENP-A (Figure 4A).
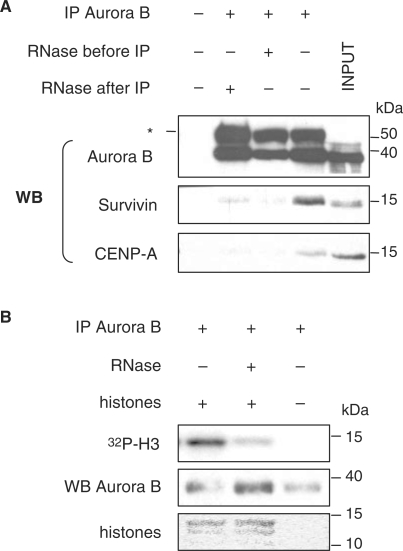
Figure 4.
Interactions of Aurora B with Survivin and CENP-A and its kinase activity are sensitive to RNA depletion. (A) Co-IP experiment using an anti-Aurora B antibody. RNaseA treatment was performed before or after IP of Aurora B. Input lysates (INPUT) and precipitated proteins in the absence of immunoprecipitating antibody (first line) are shown as positive and negative controls, respectively. Co-precipitated Survivin and CENP-A were visualized by western blotting (WB). Asterisk indicates IgG heavy chain which is detected by the secondary antibody. (B) In vitro kinase activity of immunoprecipitated Aurora B from MEL nuclear extracts, treated (+) or not (−) with RNaseA, using core histones as exogenous substrate. Specific histone H3 phosphate incorporation (32P-H3) was revealed by autoradiography, Aurora B protein levels were determined by western blotting and histones visualized by Ponceau red staining.
We then asked whether the enzymatic activity of Aurora B kinase was also sensitive to RNA depletion. Aurora B was immunoprecipitated from MEL cells extracts and examined for its ability to phosphorylate histone H3 in vitro, a physiological substrate of this kinase, in presence or absence of RNaseA treatment. The results showed that endogenous Aurora B complex displayed high specific kinase activity when assayed against purified histones and that its activity significantly decreased after RNaseA treatment (Figure 4B).
Thus, our data revealed that, in the context of the native immunoprecipitated Aurora B complex, RNA/protein interactions are crucial for interactions of Aurora B with its protein partners CENP-A and Survivin. In addition, Aurora B kinase activity appeared to be sensitive to RNaseA treatment.
Kinase activity of Aurora B is enhanced in presence of minor satellite RNA
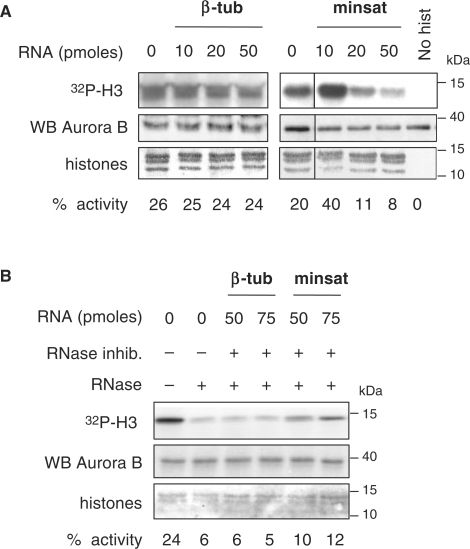
Based on the findings that Aurora B associates with minor satellite transcripts and that its kinase activity is sensitive to RNaseA treatment, we performed experiments in which the kinase assay was realized in the presence of increasing amounts of in vitro transcribed minor satellite RNA. Interestingly, Aurora B kinase activity showed a two-fold increase in presence of minor satellite RNA, in conditions where the estimated amounts of RNA and kinase were equivalent (10 pmol), whereas addition of higher amounts (20–50 pmol) of minor satellite RNA impaired the activity of the endogenous kinase (Figure 5A, right panel). These effects were specific to minor satellite RNA since an unrelated control RNA showed no such effects under the same conditions (Figure 5A, left panel). Importantly, the decreased Aurora B kinase activity after RNaseA treatment was substantially rescued by addition of large amounts of RNase inhibitors combined to addition of increasing amounts of in vitro transcribed minor satellite RNA (Figure 5B). By contrast, in the same conditions, addition of increasing amount of control RNA did not have any effect on Aurora B kinase activity (Figure 5B).
Figure 5.
Minor satellite RNA potentiate endogenous Aurora B kinase activity. (A) Kinase activity of immunoprecipitated Aurora B in presence of increasing amounts (10, 20, 50 pmol) of in vitro transcribed minor satellite RNA (minsat). Control reactions were performed in presence of increasing amounts (10, 20, 50 pmol) of in vitro transcribed β-tubulin RNA (β-tub) or in absence of histones (No hist). (B) Kinase activity of endogenous Aurora B after RNaseA treatment and rescue assay with addition of in vitro transcribed minor satellite RNA (50, 75 pmol), in presence of RNase inhibitors. As a negative control, the same amount of in vitro transcribed β-tubulin RNA (50, 75 pmol) was used in the rescue assay. Specific histone H3 phosphate incorporation (32P-H3) was quantified by phosphorimager, Aurora B protein levels were determined by western blotting and histones visualized by Ponceau red staining.
Together, these data show that decreased Aurora B kinase activity after RNA depletion can be specifically rescued by restitution of minor satellite transcripts in the reaction, and suggest that minor satellite transcripts may regulate Aurora B enzymatic activity.
DISCUSSION
We report a functional role for RNA transcribed from minor satellite repeats as part of ribonucleoprotein complexes of murine centromeric chromatin, key factors in mediating interactions between protein components of the CPC and in potentiating the activity of the mitotic kinase Aurora B.
We have previously characterized RNA transcribed from minor satellite repeats found at murine centromeres, and showed by RNA-FISH (15) or using biochemically purified chromocenters (this study) that they associate with chromocenters. Native ChIP experiments aimed to selectively precipitate centromeric chromatin fractions, based on their unique content in the histone H3 variant CENP-A, revealed that murine minor satellite RNA specifically precipitated with CENP-A-associated chromatin fractions. Interestingly, another case of centromere-encoded RNA co-precipitated with CENP-A exists in maize (10). Other examples of non-coding RNA specifically associated with defined chromatin domains from which they are transcribed have been reported. For example, Xist RNA is transcribed from and associates with the inactive X-chromosome (24). Worth mentioning, Xist also associates with a histone variant, macroH2A, deposited at facultative heterochromatin formed on the inactive X-chromosome (25). A more recent example described telomeric-repeat-containing RNA enriched at telomeric heterochromatin and involved in the maintenance of telomere integrity (26,27). Thus, transcripts emanating from specific chromosomal domains in conjunction with definite epigenetic marks may contribute to the specification of their higher-order chromatin organization. However, whether transcription across these regions, or transcripts themselves, define distinct chromatin domains with differential histone modifications or variants remains to be clarified.
Although several experimental evidence support that transcription across centromeric repeats, and/or remodelling of nucleosomes, contributes to centromere formation and deposition of the histone variant CENP-A (7,10,28), other data uncovered the implication of non-coding RNA in stabilizing the binding of structural non-histone proteins to chromatin (29). For example, binding of HP1 to heterochromatin is sensitive to digestion with RNaseA (13) and requires the contribution of the hinge domain, known to bind RNA in vitro (14). Therefore, minor satellite transcripts themselves, and not transcription per se, may be involved in formation of ribonucleoprotein complexes located at centromeres and in centromere specification. CENP-A may serve as a docking platform for centromeric RNA-dependent loading of centromeric complexes. Consistent with this, we found that minor satellite RNA associate with proteins of the CPC and that interactions between endogenous passenger proteins Aurora B and Survivin within CENP-A chromatin require an RNA component.
A particular feature of centromeric chromatin domains is that many of their associated proteins display dynamic changes in distribution patterns during cell cycle (30). In particular, the chromosomal passenger proteins such as Aurora B and Survivin, and to a lesser extent INCENP, accumulate near centromeres in early G2 (31–33). In correlation with this dynamic organization of centromeric domains, our analysis of minor satellite RNA levels revealed that the pool of centromeric RNA greatly varies during cell-cycle progression. Indeed, minor satellite transcripts accumulated gradually during the course of S phase, reached a maximum at G2/M phase and became undetectable early after mitosis, when cells re-enter the next G1 phase. Likewise, a cell-cycle regulation has also been reported for pericentromeric heterochromatin transcription both in mouse (34) and in fission yeast (35), resulting in accumulation of major transcripts in late G1/S phase. In contrast, we detected low levels of minor satellite RNA in G1 and S phase. Together with the observations that they associate with CENP-A-associated chromatin domains, and reach high levels in G2/M phase, this favours the hypothesis that minor satellite transcripts are key players in the assembly of protein complexes at the centromere, before mitosis, rather than in CENP-A deposition that was suggested to occur in G1 phase (36).
Indeed, we found that minor satellite transcripts levels greatly increased during G2/M phase, concomitant with assembly of Aurora B and Survivin to centromeres (32). At this phase, our NChIP experiments demonstrated that centromeric transcripts associate with proteins of the CPC, Aurora B and Survivin, as well as with their mitotic substrate histone H3 phosphorylated on its serine 10 (37). In agreement with the recent observations that human centromeric α-satellite transcripts are components of protein complexes located at centromeres of metaphase chromosomes (38), our data suggest that non-coding minor satellite RNA are implicated in the formation of specific centromeric complexes during G2/M transition. In addition, we provided the first experimental evidence that an RNA component favours interactions between Aurora B and its partner Survivin, as well as with CENP-A, supporting a structural role for centromeric RNA in stabilizing centromeric-associated complexes. We propose that interaction with minor satellite transcripts may represent an additional mechanism for both Aurora B proper association within CENP-A chromatin domains and enzymatic function. Minor satellite RNA could play an indirect role by providing a permissive chromatin environment for the productive interaction of Aurora B and Survivin with centromeric domains. Alternatively, minor satellite RNA could favour the interaction between Aurora B and Survivin required for the function of this complex (39,40).
We previously reported that sustained expression of minor satellite transcripts resulted in dramatic changes in localization of Aurora B, which failed to target centromeric regions of mitotic chromosome (15). In addition, deregulated accumulation of minor satellite transcripts led to impaired centromeric function and abnormal chromosome segregation (15). Consistent with this, our present data suggest a critical role for centromeric transcripts in centromere identity and function. Their implication in formation of centromere-associated complexes at specific phases of the cell cycle together with centromere failure when centromere satellite transcripts accumulate in stressed cells (15–17) argues that levels of these transcripts must be tightly regulated.
More importantly, our data put forward a functional relationship between Aurora B kinase activity and minor satellite RNA. We demonstrated that Aurora B kinase activity is sensitive to RNA depletion, probably due to disruption of the Aurora B/Survivin complex, required for Aurora B activity (39,40). Of importance, Aurora B kinase activity can be potentiated by addition of minor satellite transcripts, in conditions where the estimated amounts of RNA and kinase are equivalent. In addition, its decreased activity after RNA depletion can be specifically rescued by restitution of minor satellite RNA in the kinase assay. Recently, RNA molecules have emerged as active participants in regulating, catalysing and controlling biological processes, a role first ascribed to proteins. Several examples described RNA molecules bound to proteins and regulating their activity, subcellular location and interactions with protein partners (41). Thus, it is conceivable that specific functions and enzymatic activity of Aurora B are controlled not only by its proteins partners but also by interactions with non-coding minor satellite transcripts.
Until now, posttranslational modifications and protein components have been postulated to be the targeting actors of the CPC (42,43). We now added the evidence that both interaction of Aurora B with CENP-A-associated chromatin and kinase activity might be controlled by non-coding RNA transcribed from the domain where this enzyme is recruited and active at a specific stage of the cell cycle.
Altogether, our data added centromeric transcripts to the ever-growing list of functional non-coding RNA (44) and provide new insights into the implication of minor satellite RNA in the establishment of a functional centromere, by regulating Aurora B association with CENP-A-associated domains and enzymatic function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
INSERM and Association pour la Recherche sur le Cancer ‘ARECA’ network and Association Française contre les Myopathies for work in C.F. lab. Marie Curie Research fellowship from the EU fp6 program ‘Eurythron’ MRTN-CT-2004-005499 and Association pour la Recherche sur le Cancer (to F.F.); Agence Nationale pour la Recherche (to G.V.); Fondation pour la Recherche Médicale and Association Française contre les Myopathies (to F.H.). Funding for open access charge: INSERM.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Nathalie Beaujean for stimulating discussions and Slimane Ait-Si-Ali and Jonathan Weitzman for critical reading of the manuscript. We thank Brigitte Chanaud and Laurence Stouvenel, from the Cytometry facility of the Cochin Institute, for technical help.
REFERENCES
- 1.Vafa O, Sullivan KF. Chromatin containing CENP-A and alpha-satellite DNA is a major component of the inner kinetochore plate. Curr. Biol. 1997;7:897–900. doi: 10.1016/s0960-9822(06)00381-2. [DOI] [PubMed] [Google Scholar]
- 2.Guenatri M, Bailly D, Maison C, Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 2004;166:493–505. doi: 10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foltz DR, Jansen LE, Black BE, Bailey AO, Yates J.R., III, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 4.Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 5.Carone DM, Longo MS, Ferreri GC, Hall L, Harris M, Shook N, Bulazel KV, Carone BR, Obergfell C, O'Neill MJ, et al. A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma. 2009;118:113–125. doi: 10.1007/s00412-008-0181-5. [DOI] [PubMed] [Google Scholar]
- 6.Li F, Sonbuchner L, Kyes SA, Epp C, Deitsch KW. Nuclear non-coding RNAs are transcribed from the centromeres of Plasmodium falciparum and are associated with centromeric chromatin. J. Biol. Chem. 2008;283:5692–5698. doi: 10.1074/jbc.M707344200. [DOI] [PubMed] [Google Scholar]
- 7.Nakano M, Okamoto Y, Ohzeki J, Masumoto H. Epigenetic assembly of centromeric chromatin at ectopic alpha-satellite sites on human chromosomes. J. Cell Sci. 2003;116:4021–4034. doi: 10.1242/jcs.00697. [DOI] [PubMed] [Google Scholar]
- 8.Pezer Z, Ugarkovic D. RNA Pol II promotes transcription of centromeric satellite DNA in beetles. PLoS ONE. 2008;3:e1594. doi: 10.1371/journal.pone.0001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudert F, Bronner S, Garnier JM, Dolle P. Transcripts from opposite strands of gamma satellite DNA are differentially expressed during mouse development. Mamm. Genome. 1995;6:76–83. doi: 10.1007/BF00303248. [DOI] [PubMed] [Google Scholar]
- 10.Topp CN, Zhong CX, Dawe RK. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl Acad. Sci. USA. 2004;101:15986–15991. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volpe T, Schramke V, Hamilton GL, White SA, Teng G, Martienssen RA, Allshire RC. RNA interference is required for normal centromere function in fission yeast. Chromosome. Res. 2003;11:137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- 12.Zhang W, Yi C, Bao W, Liu B, Cui J, Yu H, Cao X, Gu M, Liu M, Cheng Z. The transcribed 165-bp CentO satellite is the major functional centromeric element in the wild rice species Oryza punctata. Plant Physiol. 2005;139:306–315. doi: 10.1104/pp.105.064147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 14.Muchardt C, Guilleme M, Seeler JS, Trouche D, Dejean A, Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 2002;3:975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouzinba-Segard H, Guais A, Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc. Natl Acad. Sci. USA. 2006;103:8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, Vourc'h C. Stress-induced transcription of satellite III repeats. J. Cell Biol. 2004;164:25–33. doi: 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valgardsdottir R, Chiodi I, Giordano M, Cobianchi F, Riva S, Biamonti G. Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol. Biol. Cell. 2005;16:2597–2604. doi: 10.1091/mbc.E04-12-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong AK, Rattner JB. Sequence organization and cytological localization of the minor satellite of mouse. Nucleic Acids Res. 1988;16:11645–11661. doi: 10.1093/nar/16.24.11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francastel C, Magis W, Groudine M. Nuclear relocation of a transactivator subunit precedes target gene activation. Proc. Natl Acad. Sci. USA. 2001;98:12120–12125. doi: 10.1073/pnas.211444898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 22.Jeppesen P, Mitchell A, Turner B, Perry P. Antibodies to defined histone epitopes reveal variations in chromatin conformation and underacetylation of centric heterochromatin in human metaphase chromosomes. Chromosoma. 1992;101:322–332. doi: 10.1007/BF00346011. [DOI] [PubMed] [Google Scholar]
- 23.Vader G, Medema RH, Lens SM. The chromosomal passenger complex: guiding Aurora-B through mitosis. J. Cell Biol. 2006;173:833–837. doi: 10.1083/jcb.200604032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brockdorff N. The role of Xist in X-inactivation. Curr. Opin. Genet. Dev. 1998;8:328–333. doi: 10.1016/s0959-437x(98)80090-7. [DOI] [PubMed] [Google Scholar]
- 25.Gilbert SL, Pehrson JR, Sharp PA. XIST RNA associates with specific regions of the inactive X chromatin. J. Biol Chem. 2000;275:36491–36494. doi: 10.1074/jbc.C000409200. [DOI] [PubMed] [Google Scholar]
- 26.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 27.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 28.Chen ES, Saitoh S, Yanagida M, Takahashi K. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol. Cell. 2003;11:175–187. doi: 10.1016/s1097-2765(03)00011-x. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Campos A, Azorin F. RNA is an integral component of chromatin that contributes to its structural organization. PLoS ONE. 2007;2:e1182. doi: 10.1371/journal.pone.0001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musacchio A, Hardwick KG. The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- 31.Beardmore VA, Ahonen LJ, Gorbsky GJ, Kallio MJ. Survivin dynamics increases at centromeres during G2/M phase transition and is regulated by microtubule-attachment and Aurora B kinase activity. J. Cell Sci. 2004;117:4033–4042. doi: 10.1242/jcs.01242. [DOI] [PubMed] [Google Scholar]
- 32.Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, Zhou H, Sen S, Allis CD, Sassone-Corsi P. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeitlin SG, Shelby RD, Sullivan KF. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 2001;155:1147–1157. doi: 10.1083/jcb.200108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu J, Gilbert DM. Proliferation-dependent and cell cycle regulated transcription of mouse pericentric heterochromatin. J. Cell Biol. 2007;179:411–421. doi: 10.1083/jcb.200706176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 36.Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeitlin SG, Barber CM, Allis CD, Sullivan KF. Differential regulation of CENP-A and histone H3 phosphorylation in G2/M. J. Cell Sci. 2001;114:653–661. doi: 10.1242/jcs.114.4.653. [DOI] [PubMed] [Google Scholar]
- 38.Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolton MA, Lan W, Powers SE, McCleland ML, Kuang J, Stukenberg PT. Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and phosphorylation. Mol. Biol. Cell. 2002;13:3064–3077. doi: 10.1091/mbc.E02-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Jin S, Tahir SK, Zhang H, Liu X, Sarthy AV, McGonigal TP, Liu Z, Rosenberg SH, Ng SC. Survivin enhances Aurora-B kinase activity and localizes Aurora-B in human cells. J. Biol. Chem. 2003;278:486–490. doi: 10.1074/jbc.M211119200. [DOI] [PubMed] [Google Scholar]
- 41.Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006;15(Spec No 1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 42.Ainsztein AM, Kandels-Lewis SE, Mackay AM, Earnshaw WC. INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J. Cell Biol. 1998;143:1763–1774. doi: 10.1083/jcb.143.7.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vader G, Kauw JJ, Medema RH, Lens SM. Survivin mediates targeting of the chromosomal passenger complex to the centromere and midbody. EMBO Rep. 2006;7:85–92. doi: 10.1038/sj.embor.7400562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barrandon C, Spiluttini B, Bensaude O. Non-coding RNAs regulating the transcriptional machinery. Biol. Cell. 2008;100:83–95. doi: 10.1042/BC20070090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.