Abstract
The discrete regulation of supercoiling, catenation and knotting by DNA topoisomerases is well documented both in vivo and in vitro, but the interplay between them is still poorly understood. Here we studied DNA catenanes of bacterial plasmids arising as a result of DNA replication in Escherichia coli cells whose topoisomerase IV activity was inhibited. We combined high-resolution two-dimensional agarose gel electrophoresis with numerical simulations in order to better understand the relationship between the negative supercoiling of DNA generated by DNA gyrase and the DNA interlinking resulting from replication of circular DNA molecules. We showed that in those replication intermediates formed in vivo, catenation and negative supercoiling compete with each other. In interlinked molecules with high catenation numbers negative supercoiling is greatly limited. However, when interlinking decreases, as required for the segregation of newly replicated sister duplexes, their negative supercoiling increases. This observation indicates that negative supercoiling plays an active role during progressive decatenation of newly replicated DNA molecules in vivo.
INTRODUCTION
Supercoiling, catenation and knotting are the three best-known aspects of DNA topology (1). Understanding how they change during DNA replication is hindered, though, by the fact that the geometry of DNA replication intermediates (RIs) is significantly altered upon their isolation for analysis in vitro. Once DNA is purified and all the proteins bound are eliminated, the structure of RIs do not represent anymore their situation in vivo. Within living cells a protein complex known as the replisome binds to RIs at the replication forks (2). Some authors propose that replisomes get anchored to the cell membrane (3) and this would preclude rotation of the sister duplexes at the forks preventing the diffusion of unremoved linking of the parental strands from the unreplicated to the replicated portion. However, other authors provide experimental evidence suggesting that forks can rotate in vivo and sister duplexes do form precatenanes (4–7).
The early studies by Sundin and Varshavsky established that freshly replicated circular DNA molecules are intertwined forming torus-type catenanes where the two sister duplexes wind around each other in a right-handed fashion (8,9). In cells with fully active type II DNA topoisomerases, these intertwined molecules are progressively decatenated. However, when type II DNA topoisomerases are inhibited, catenanes with high catenation numbers accumulate (3,7,10–12), although the occurrence of catenanes can be evidenced even without the inhibition of topoisomerases, using chase-labelling techniques (11,13,14). Studies of catenanes formed by site specific recombination (15) revealed that in bacterial cells the rate of decatenation decreases when DNA supercoiling is reduced (16). In vitro studies measuring the equilibrium fraction of DNA catenanes revealed that this fraction decreases when one of the rings is supercoiled thus indicating that DNA supercoiling should favour DNA decatenation (17). However, there were no dedicated studies investigating the correlation between the level of supercoiling and the progression of decatenation in freshly replicated DNA molecules. Here, we used high-resolution two-dimensional (2D) agarose gel electrophoresis to analyze the correlation between interlinking and supercoiling produced in vivo in the RIs of bacterial plasmids. We also performed numerical simulations of DNA catenanes with varying levels of interlinking and supercoiling to better understand how the structure of these catenanes is affected by the interplay between the negative (−) supercoiling generated by DNA gyrase and the interlinking that results from replication of circular DNA molecules.
MATERIALS AND METHODS
Bacterial strains, plasmids and culture medium
The E. coli strain used in this study was DH5αF′ {F′/gyrA96(Nalr) recA1 relA1 endA1 thi-1 hsdR17 (rk−mk+) glnV44 deoR Δ(lacZYA-argF)U169[F80dΔ(lacZ)M15]}. Competent cells were transformed with monomeric forms of pBR18 (4383 bp), a derivative of pBR322 where the tetracycline resistance promoter has been replaced with the polylinker of pU18. Cells were grown and DNA isolated as described earlier (18,19).
Cell and DNA treatments
To preferentially inhibit topoisomerase IV (Topo IV), cells were treated with 15 µM norfloxacine for 30–60 min before harvest. To induce single-stranded DNA breaks, DNA was exposed to Nb-BsmI (BioLabs) at 12.6 u/µg of DNA for 5 min at 37°C. The reaction was blocked with 100 µg/ml proteinase K for 30 min at 37°C. For a partial digestion the concentration of the enzyme was lowered to 9 u/µg of DNA.
Two-dimensional (2D) agarose gel electrophoresis and southern transfer
The first dimension was in a 0.4% agarose gel in TBE buffer at 0.9 V/cm at room temperature for 25 h. The second dimension was in a 1% agarose gel in TBE buffer containing none or different concentrations of chloroquine run perpendicular with respect to the first dimension. The dissolved agarose was poured around the excised agarose lane from the first dimension and electrophoresis was at 4.5 V/cm in a 4°C cold chamber for 10–13 h. Southern transfer was performed as described before (18,19).
Non-radioactive hybridization
Probes were labelled with the Random Primer Fluorescein kit (NEN Life Sciences Products). Membranes were prehybridized in a 20 ml prehybrization solution (2× SSPE, 0.5% Blotto, 1% SDS, 10% dextran sulphate and 0.5 mg/ml sonicated and denaturated salmon sperm DNA) at 65°C for 4–6 h. Labelled DNA was added and hybridization lasted for 12–16 h. Hybridized membranes were sequentially washed with 2× SSC and 0.1% SDS, 0.5× SSC and 0.1% SDS, 0.1× SSC and 0.1% SDS for 15 min each at room temperature except for the last wash, which took place at 65°C. Detection was performed with an antifluorescein-AP conjugate and CDP-Star (NEN) according to the instructions provided by the manufacturer.
Simulations methods
Two component catenanes were modelled as worm-like chains of discrete segments (20). Each chain has a total length of 1.5 µm (which corresponds to ∼4410 bp) and is composed of 75 segments. The segments are hard-wall cylinders with a diameter of 3 nm, which corresponds to the effective diameter of DNA under conditions where the electrostatic charge is largely screened by the counterions, such as under conditions found in living cells (21,22). The configuration space of the molecules is explored with standard crankshaft moves where the only crankshafts considered are those that do not result in interpenetrations of hard-wall cylinders that represent any of non-consecutive segments of the chain. The probability of acceptance (P) of a new configuration (Cnew) depends on the energy difference between Cold and Cnew according to the Metropolis–Monte Carlo rule, i.e. P(ColdΠCnew) = 1, if Eold > Enew and P(ColdΠCnew) = exp[(Eold – Enew)/kBT], if Eold < Enew (23). For the energy calculation, we take into account the elastic energy connected to bending and torsional deformations of modelled molecules. The bending energy is given by
where kB is the Boltzman constant, T the temperature, g a bending rigidity constant (24) fixing the persistence length at 50 nm, and θi the angles between successive segments. Knowing that ΔLk = ΔTw + Wr, we can relate the change in linking number ΔLk to the change in twist ΔTw and to the writhe Wr. Therefore, the torsional energy is given by
where C is the torsional rigidity constant fixed at 3 × 10−19 J nm, and L the total length of one chain. The choice of ΔLk fixes the level of supercoiling and permits to simulate supercoiled molecules (25). In addition to the energetic acceptance rules, we also rejected all moves that resulted in a change of the topology of the modelled catenanes as detected by change of the Alexander polynomial for knots Aknot(s) and for catenanes Acat(s, t). In addition to the Metropolis–Monte Carlo approach permitting us to model and analyze thermally equilibrated catenanes with different catenation numbers and different levels of DNA supercoiling, we also were interested in the free energy difference between catenanes with different levels of DNA supercoiling. The free energy difference allowed us to estimate the work done by DNA gyrase while increasing the level of (−) supercoiling in catenanes with various catenation numbers. To this aim we calculated the differences in free energy between states separated by ΔLk = 2, since gyrase modifies Lk in steps of two. These calculations required that we measured the average writhe 〈Wr(ΔLk)〉 of relevant states that were obtained in the Metropolis–Monte Carlo simulation procedure described above, and the free energy of supercoiling G(ΔLk) could then be evaluated numerically (26) with
Since catenanes in torsionally relaxed configurations have already an intrinsic writhe Wr0 (20,27), the integral was in fact not evaluated between 0 and ΔLk, but between Wr0 and Wr0 + ΔLk. To achieve a good sampling, 100 independent simulation runs were performed. In each one, ΔLk was decreased by steps of one from 0 to −12. First, 106 moves were performed at ΔLk = 0 to achieve thermal equilibration. After that, 2 × 105 moves were performed and used to calculate the average value of the writhe for each ΔLk. At each change of ΔLk, 2 × 105 moves were done to equilibrate the system. Finally, the writhe values were averaged over the 100 independent runs.
RESULTS
Two-Dimensional (2D) agarose gel electrophoresis
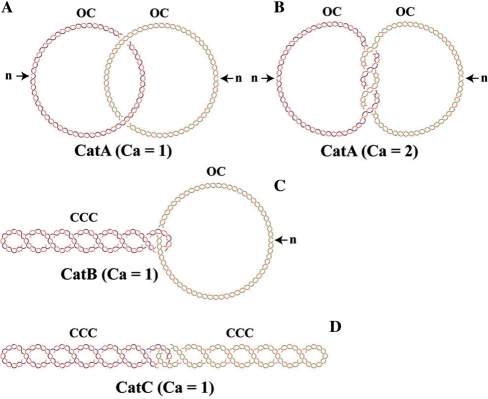
It is well established (8,9) that catenanes occur in three different forms (Figure 1): those formed by two nicked rings (CatAs); those formed by one nicked ring and the other covalently closed (CatBs); and those formed by two covalently closed rings (CatCs). In addition, the rings can be catenated just once (Ca = 1) or multiple times (Ca ≥ 2). Ca is the catenation number and equals half of the signed sum of intermolecular nodes (1). It is well known that in prokaryotes catenanes accumulate in vivo after the inhibition of Topo IV (3,10). Actually, this is the main feature that allowed the identification of Topo IV as the specialized decatenase in E. coli (10,16,28,29).
Figure 1.
Cartoon representing different forms of catenanes after projection on a plane. (A) One pair of nicked DNA rings catenated once. (B) One pair of nicked DNA rings catenated twice. (C) One pair of DNA rings catenated once where one ring is nicked and the other covalently closed and supercoiled. (D) One pair of DNA rings catenated once where both rings are covalently closed and supercoiled. n = nick; OC = open circle; CCC = covalently closed circle; Ca = catenation number. Parental DNA strands are depicted in blue and green while newly synthesized strands are depicted in red.
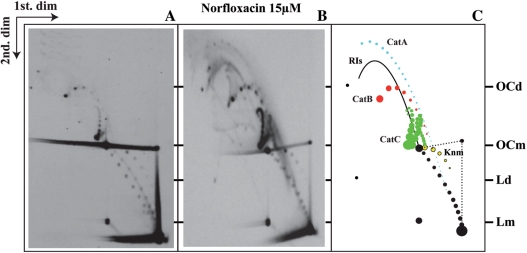
We chose to study supercoiling and catenation of a bacterial plasmid, pBR18 (30–32), in a strain of E. coli, DH5αF′, carrying a mutation that makes DNA gyrase but not Topo IV resistant to quinolones (33,34). To identify catenated molecules we used high-resolution 2D agarose gel electrophoresis (7,35,36). This approach allowed us to confirm that all three forms of pBR18 catenanes accumulated when the transfected DH5αF′ E. coli cells were exposed to 15 µM norfloxacin for 30–60 min before harvest (Figure 2). It is worth noting that the most abundant form corresponded to CatCs followed by CatBs. CatAs were barely visible.
Figure 2.
Preferential inhibition of Topo IV leads to the accumulation of DNA catenanes. Autoradiograms of 2D gels corresponding to pBR18 isolated from DH5αF′ E. coli cells untreated (A) and after exposure to norfloxacin (B). A diagrammatic interpretation of the different signals in the autoradiograms is shown to the right (C) where CatAs are depicted in blue, CatBs in red and CatCs in green. RIs = nicked replication intermediates; OCd = open circles corresponding to dimers; OCm = open circles corresponding to monomers; Knm = nicked knotted monomers; Ld = linearized dimers; Lm = linearized monomers. Monomeric topoisomers are depicted in black.
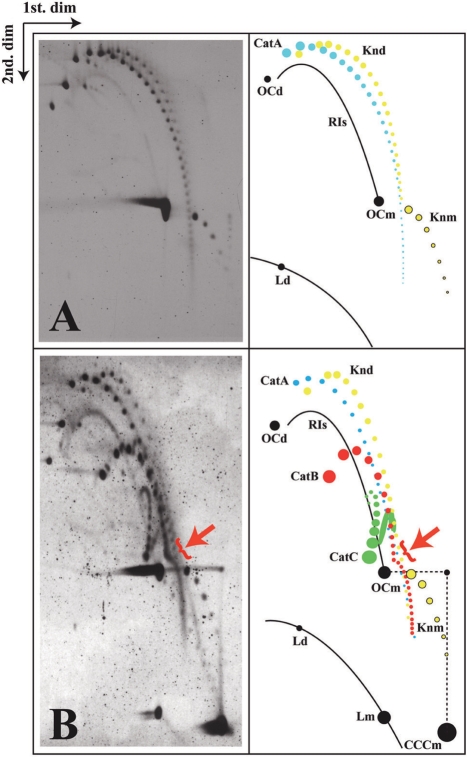
There are two ways to confirm the nature of the signals generated by catenated rings in 2D gels (36). First, the induction of single-stranded breaks or nicks converts all three families of catenanes into a single one: CatAs. Secondly, an intercalating agent can be added during the second dimension of the 2D gel (7,18,36,37). Such intercalating agents change the DNA twist (Tw), which in covalently closed circles (CCCs) induces positive (+) writhe (Wr). This dramatically alters the electrophoretic mobility of covalently closed DNA molecules. Nicked forms, on the other hand, are not affected in a significant manner, as a change of twist in nicked DNA molecules does not induce writhing. To introduce single-stranded breaks we used the nicking endonuclease Nb-BsmI (38). As clearly shown in Figure 3A, 2D gels revealed that after Nb-BsmI nicking those signals that were identified as CatBs and CatCs disappeared while the intensity and complexity of the signals corresponding to CatA forms were enhanced notably. Catenated rings with Ca ≈ 40 were readily identified in highly exposed autoradiograms (data not shown). Upon nicking, two other families of stereoisomers became prominent as well (Figure 3A). As demonstrated earlier, these families are composed of knotted DNA with increasing complexity that has molecular masses corresponding to monomers and dimers, respectively (36,39,40). These knotted forms became clearly visible only after nicking eliminated the structural heterogeneity of knotted DNA molecules of a given type caused by the natural variance of their supercoiling level.
Figure 3.
Partial and complete nicking allows identification of all forms of catenanes. Autoradiograms of 2D gels corresponding to pBR18 isolated from DH5αF′ E. coli cells after exposure to norfloxacin and complete (A) or partial (B) digestion with the single-stranded DNA nicking enzyme Nb-BsmI. A diagrammatic interpretation of the autoradiograms is shown to their right where CatAs are depicted in blue, CatBs in red and CatCs in green. A red arrow in (B) points to a distinct inflection that was evidenced for CatBs when Ca reached 10–12. RIs = nicked replication intermediates; Knd = nicked knotted dimers; OCd = open circles corresponding to dimers; OCm = open circles corresponding to monomers; Knm = nicked knotted monomers; Ld = linearized dimers; Lm = linearized monomers. Monomeric topoisomers are depicted in black.
The experiments with partial nicking allowed the additional verification that CatC species contained two supercoiled rings while CatB species just one. After partial nicking with the Nb-BsmI nuclease, we observed a significant reduction of CatCs, the amount of CatBs remained almost unchanged while CatA forms increased (Figure 3B). Since we knew from the complete nicking experiment that CatB forms are converted into CatA forms, we concluded that the quasi-constant amount of CatB forms was due to the partial nicking of CatC forms that replenished CatB forms and this compensated for the nicking-induced transitions of CatB into CatA forms. Therefore, Nb-BsmI nicking and partial nicking allowed us to identify and characterize all forms of catenanes.
To facilitate the understanding of the gels, let us consider first the CatA family and address the question why the species showing the slowest mobility in the first dimension was in fact not the slowest one in the second dimension. The conditions of the first dimension, i.e. low voltage and low concentration of agarose, made that the interaction of DNA with the gel matrix was weak and the molecules behaved similarly to sedimenting DNA molecules in solution where their speed of motion increases with their overall compaction (41). For this reason, in the first dimension, the analyzed DNA molecules migrated with the order reflecting their overall dimensions in relatively unperturbed configurations, i.e. those forms that on average have open configurations migrate as the slowest. In the second dimension the voltage and the concentration of agarose was elevated. Under such conditions passage of DNA molecules through the gel becomes not only dependent on the overall dimensions of unperturbed DNA molecules but also on the deformability of the molecules that allow them to pass through rather small pores in the gel with the elevated concentration of the agarose. Apparently, the singly linked catenanes were deformed easier than doubly linked catenanes despite the fact that the unperturbed overall dimensions of the former ones are bigger than for the latter. This interplay between overall dimensions and deformability of catenated DNA molecules explains the initial, raising portions of arcs that are visible for CatAs and are even more pronounced for CatB and CatC families (see Figures 2C and 3B). For molecules with higher catenation numbers corresponding to CatAs, we observed that the progressive compaction apparently compensated for the possible decrease of deformability since the deformations needed to pass through the gel pores are smaller for those molecules whose overall dimensions were already strongly reduced.
Earlier theoretical works that have considered the energy of DNA catenanes proposed that introduction of negative supercoiling into strongly catenated DNA rings should be energetically very costly (42). The veracity of that proposal was not yet tested on catenanes formed in vivo. Our 2D gels, however, offer an interesting insight into the competition between catenation and supercoiling occurring in vivo. As already mentioned, for DNA species with the same total mass, as was the case for all the catenated species analyzed here, their electrophoretic mobility in low concentration agarose gels at low voltage grew with their overall compactness and this in turn steadily increased with the average number of crossings that the molecules revealed upon projection (43). For this reason, during the first direction run the electrophoretic mobility of CatAs increased monotonically with the number of crossings resulting from their catenation (20,27) since those crossings augmented their average number of nodes. In the case of CatCs, however, in addition to the crossings due to the catenation of both rings there were also crossings that resulted from supercoiling of the component rings (see Figure 1). If increasing catenation makes it more difficult for DNA gyrase to introduce DNA supercoiling, then one would expect that, as catenation increases, each of the component rings should decrease its supercoiling. As a result, the average number of crossings for CatCs could decrease with increasing catenation, and this would manifest itself in a ‘reverse’ order of electrophoretic mobility during the first dimension for CatC species with increasing catenation numbers. In fact, a closer inspection of the gel shown in Figure 3 (and also gels where the second dimension occurred in the presence of 10 and 20 µg/ml chloroquine, see Figure 4), revealed that, starting from Ca ≈ 5, the electrophoretic mobility of CatCs during the first dimension progressively decreased with increasing extents of DNA catenation. This effect stopped though upon reaching Ca ≈ 12 (see Figure 3). Since the first few interlinks do not impose strong mechanical constraint into catenated DNA molecules, we could expect that those interlinks would not diminish significantly the level of DNA supercoiling that could be introduced by DNA gyrase into these molecules. This could explain why the ‘reverse’ order of electrophoretic mobility only started when Ca ≈ 5. However, why does the ‘reverse’ migration effect stop when Ca ≈ 12. For DNA molecules with so many interlinks the increase of mechanical constraint due to each new interlink is significant and this should clearly diminish the number of negative supercoils that DNA gyrase could introduce into those molecules. We initially thought that DNA gyrase was not able to introduce DNA supercoiling for catenanes where Ca > 12, and therefore, from this point on, further increases of DNA catenation would not be compensated anymore by a decrease in the level of DNA supercoiling. However, our modelling studies (see below) indicated that, component rings in catenanes with Ca > 12 should be still able to accept and store DNA gyrase-generated supercoils, although to a lower level than less catenated DNA. Therefore, the competition between supercoiling and catenation should also concern species with Ca > 12. In the part describing simulation of the catenanes’ configurations, we provide an explanation why during the first dimension the reverse order of electrophoretic migration of CatC species does not extend beyond Ca > 12.
Figure 4.
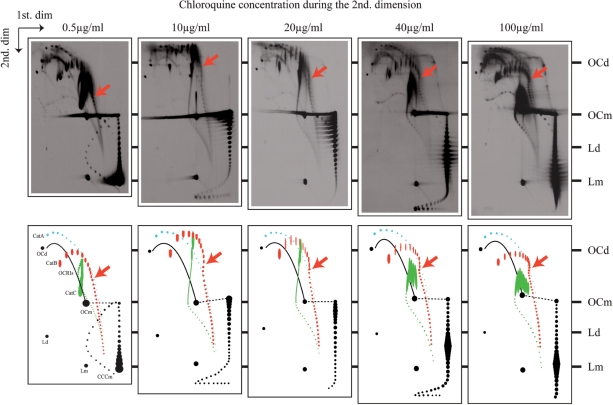
In CatBs and CatCs interlinking and supercoiling compete with each other. Autoradiograms of 2D gels corresponding to pBR18 isolated from DH5αF′ E. coli cells after exposure to norfloxacin to accumulate catenated forms. The second dimension occurred in the presence of different concentrations of chloroquine and the autoradiograms were aligned so that OCs and Ls, whose electrophoretic mobility is not significantly affected by chloroquine, coincided. A diagrammatic interpretation of each autoradiogram is shown below where CatAs are depicted in blue, CatBs in red and CatCs in green. In the diagrams corresponding to 10 and 40 µg/ml chloroquine, CatCs where Ca = 3–12 were drawn as a continuous line as individual spots were difficult to identify at the resolution of these photographs. Monomeric topoisomers are depicted in black. OCRIs = nicked replication intermediates; OCd = open circles corresponding to dimers; OCm = open circles corresponding to monomers; Ld = linearized dimers; Lm = linearized monomers. Red arrows point to CatBs with the lowest Ca value (10–12) that chloroquine was unable to decompose in topoisomers during the second dimension.
As previously mentioned, the electrophoretic mobility during the second dimension is the result of a complex interplay between compaction and the deformability of DNA molecules. Therefore, we interpreted a transient decrease of electrophoretic mobility in the second dimension for CatCs where Ca > 12 (see Figure 3) as an indication that those species were progressively more rigid and resisted the deformations needed to thread through small pores in the gel during the second dimension. Such an effect, however, became less important when the compaction due to high interlinking was sufficient to allow the molecules to pass through small pores in the gel.
If the ‘reverse order’ observed during the first dimension for CatC catenanes with Ca ranging between 5 and 12 indeed resulted from a decrease of supercoiling in both covalently closed rings, then a less dramatic effect should be observed for CatB catenanes where the increasing catenation would only be able to decrease DNA supercoiling in one of the rings. Therefore, for CatBs one should expect a compression zone where the increasing interlinking would be associated with the decreasing supercoiling of just one of the rings. Indeed, it is readily visible in Figures 3 and 4 that, during the first dimension, a compression zone formed for CatB species with catenation numbers ranging between 6 and 12, while thereafter CatBs increased their separation during the first dimension.
Uncatenated supercoiled molecules isolated from living cells usually show Gaussian distribution of topoisomers. For this reason one would expect that supercoiled rings in CatBs and CatCs should also show Gaussian distribution of topoisomers, and this would introduce heterogeneity within CatBs and CatCs with a given catenation number. In spite of this, though, in our gels where both first and second dimensions were performed without the addition of chloroquine, CatBs and CatCs with a given catenation number migrated as single bands. This was best visualized for CatBs. This behaviour was most likely the consequence of the fact that electrophoresis in agarose gels without the addition of an intercalant does not resolve individual topoisomers in (−) supercoiled DNA molecules that have a torsional tension approaching that of native supercoiled DNA.
To better understand the contribution of DNA supercoiling of the component rings of CatBs and CatCs to their electrophoretic mobility, we investigated the effects of an intercalating agent during the second dimension. To this aim, we run the second dimension of 2D gels in the presence of different concentrations of chloroquine (Figure 4), while separation in the first dimension occurred without chloroquine. Of course, all the supercoiled monomeric plasmids in the sample were also affected by the presence of the intercalating agent and their electrophoretic behaviour was used as an internal control indicating the extent of DNA underwinding at a given concentration of chloroquine (18,44). Also, covalently closed partially replicated DNA molecules (RIs) generated a distinct pattern that appeared at the top right corner of those gels where the second dimension occurred in the presence of high concentrations of chloroquine (7,36). In the corresponding schematic diagrams in Figure 4, though, we did not indicate the position of these partially replicated DNA molecules simply to help the readers to focus on catenated DNA species.
As expected for molecules composed of nicked DNA rings, the electrophoretic mobility of CatAs remained almost constant regardless of the concentration of chloroquine used during the second dimension. For CatCs, their migration was significantly affected but the formed patterns were somewhat difficult to discern due to overlapping of the bands. Notwithstanding, in some of these gels it was clearly visualized that for CatCs their increasing catenation, up to Ca ≈ 12, was associated with a decrease of electrophoretic mobility during the first dimension (as discussed earlier in case of native gels). The most informative case corresponded to CatBs as for this family individual members were identified all through. The electrophoretic mobility of CatBs with low catenation numbers decreased during the second dimension as the concentration of chloroquine increased from 0.5 to 10 µg/ml. In the gel corresponding to the latter, the upper part of the oblong CatB signals where Ca = 2–6 showed almost the same mobility during the second dimension as their CatA counterparts indicating that for these species, 10 µg/ml chloroquine have eliminated almost all their native (−) supercoiling. The same CatB species, however, recovered some electrophoretic mobility during the second dimension when exposed to 20 µg/ml chloroquine. It is important to note that, using this concentration of the intercalating agent, some CatB species that migrated as a single band during the first dimension were separated into a number of distinct bands during the second dimension. In particular, CatBs where Ca = 2–5 were decomposed into 10–12 separate bands forming a characteristic pattern where the most intense band was in the centre flanked above and below by bands with progressively decreasing intensities (see Figure 4, 20 µg/ml for a high-resolution example). Such a pattern indicated that, at this concentration of chloroquine, individual topoisomers of the covalently closed ring present in each of these CatB species were resolved, and formed a characteristic pattern revealing a Gaussian family of topoisomers. Although, it was expected that component rings of catenated species should show a Gaussian distribution of topoisomers similar to that one observed for supercoiled uncatenated plasmid molecules, this was not demonstrated before. Interestingly, while CatBs with low catenation numbers clearly showed a broad spread of topoisomers (with 10–12 members for each Gaussian family), this spread progressively decreased with increasing catenation numbers. The contraction of those Gaussian families with increasing catenation confirmed that the energetic cost of introducing DNA supercoils into covalently closed rings grew as their catenation increased. When the energetic cost of supercoiling is high, the energy differences between different topoisomers increases and this contracts the Gaussian families (1). Starting from CatB species where Ca ≈ 12, the presence of chloroquine was unable to reveal topoisomers with different linking numbers. Based on our modelling studies (see below), we interpret this observation as an indication that, for highly interlinked catenanes that already have a reduced level of DNA supercoiling, the differences in the linking number of component rings do not result in significant differences of the electrophoretic migration of these catenated species.
The three observations: (i) the ‘reverse order’ of migration of CatCs with Ca ranging between 5 and 12; (ii) the compression zone observed for corresponding CatB species and (iii) the progressive shrinking of the Gaussian families of topoisomers in component rings with increasing catenation numbers, all indicated that DNA supercoiling and DNA catenation compete with each other during the process leading to the separation of newly replicated DNA molecules. Of course, in vivo the natural sequence of events is from highly catenated molecules with low supercoiling to poorly catenated species with high supercoiling.
Numerical simulations of DNA catenanes
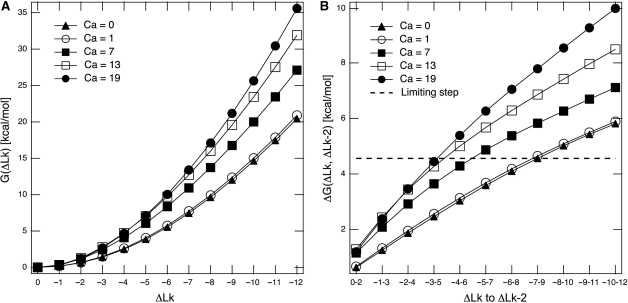
To get further insights into structural transitions induced by DNA catenation and DNA supercoiling into DNA catenanes, and to quantify the energetic cost of introducing (−) supercoiling into highly intertwined DNA molecules, we performed numerical simulations of CatC DNA catenanes. CatC species are the natural forms of freshly replicated DNA molecules in vivo and in the 2D gels experiments described above they showed the strongest manifestation of the interplay between DNA catenation and supercoiling. We modelled DNA molecules of 4400 bp with different levels of supercoiling and catenation. Figure 5A shows the values of free energy of DNA supercoiling for CatCs with increasing catenation numbers (profiles displaced vertically) and increasing level of negative supercoiling. It is well visible that the free energy of DNA supercoiling increases with increasing catenation number, indicating that DNA gyrase would find it harder and harder to introduce negative supercoiling to catenanes with increasing catenation number. In addition, as shown in Figure 5B, the free energy difference between sequential topoisomers within CatC species with a given catenation number increases quicker for CatCs with high catenation numbers as compared to those with lower ones. Let us consider therefore at which point DNA gyrase could get stalled while acting on various CatC species. It is well known that DNA molecules isolated from normal bacterial cells are supercoiled to a density of σ ≈ −0.05 (−1 ΔLk/20 Lk). However, it is less known that 50% of this supercoiling is introduced due to the binding of HU proteins. Therefore, in mutant cells without HU proteins, the density of supercoiling introduced by DNA gyrase amounts only to σ ≈ −0.022 (45). Independent topological measures of DNA supercoiling in living bacterial cells have demonstrated that the steady state level of DNA supercoiling in vivo is about twice lower than observed after DNA deproteinization (46). Therefore, we can assume that under physiological conditions in vivo, DNA gyrase can furnish just enough energy to supercoil DNA to σ ≈ −0.022 and that the final level will be doubled by the fact that binding of histone-like proteins, which constrain DNA supercoiling, decreases the torsional stress and allows further action of DNA gyrase. Possibly, a less favourable ATP/ADP ratio or other factors make that DNA gyrase in vivo stops DNA supercoiling at the lover level that it could be achieved in vitro (47). For DNA molecules with 4400 bp, assuming that the critical σ value is close to −0.022, we would expect that DNA gyrase should be able to supercoil DNA up to a topoisomer with a ΔLk = −9, but would not be able to further supercoil this topoisomer to a ΔLk = −11 (gyrase changes the linking number in steps of two). Therefore, for DNA of this size we can consider the free energy difference between the topoisomers with ΔLk = −7 and −9 as the maximal energy barrier that can be surmounted by DNA gyrase under physiological conditions. Looking at Figure 5 (profile for supercoiled circles that are not catenated, i.e. where Ca = 0) we can see that this barrier corresponds to ∼4.6 kcal/mol. Now, we can check when this barrier is reached in the case of CatCs with increasing catenation numbers. Supercoiled rings in singly linked catenanes (Ca = 1) behaved almost indistinguishable from not catenated rings. This observation agrees with several earlier studies showing that the supercoiling of singly linked rings is not affected by catenation as interlinking involves mainly apical loops of supercoiled DNA molecules and thus can be easily accommodated without distortion of the superhelix (48). Also, snapshots of our simulations showed that catenation involves preferentially apical loops of supercoiled DNA molecules (see Figure 6D). For catenanes with increasing catenation numbers, however, we observed that the energy barrier blocking any further action of DNA gyrase moved towards lower levels of (−) supercoiling. For example, for catenanes with Ca = 7, DNA gyrase would be expected to stop at a topoisomer with a ΔLk = −6, while for those with Ca = 13, supercoiling would proceed only to a ΔLk = −5. A further increase of catenation number still increases the energetic costs of introducing (−) supercoils.
Figure 5.
The free energy of DNA supercoiling increases with the extent of DNA catenation. (A) Profiles of the free energy of DNA supercoiling i.e. the energy that would be liberated upon nicking of the catenated DNA with various levels of catenation (Ca) and various extent of supercoiling (ΔLk) for modelled DNA molecules of ∼4400 bp. Notice that the free energy of catenation is not included in the graph since we are not considering here the energy that would be liberated upon progressive decatenation of modelled catenanes by Topo IV. (B) Free energy costs that DNA gyrase would need to overcome to perform sequential supercoiling cycles in catenated DNA molecules with ∼4400 bp that have different catenation numbers. Notice that during each catalytic cycle DNA gyrase reduces the linking number by two, notice also that each sequential cycle requires more energy and that in more complex catenanes this increase is more rapid than in less complex ones. The dashed line indicates the estimated level to which DNA gyrase would be able to supercoil protein-free DNA rings with different extents of catenation.
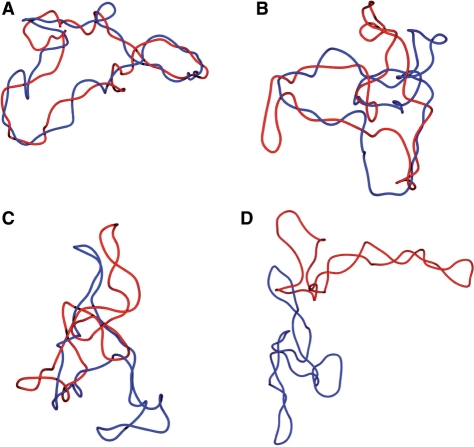
Figure 6.
Structure of DNA catenanes with decreasing catenation numbers and expected levels of DNA supercoiling that DNA gyrase could introduce. Smoothed snapshots from Metropolis–Monte Carlo simulations of DNA catenanes representing progressive stages of decatenation process under conditions where the action of DNA gyrase maintains the DNA at supercoiling level set by a quasi-constant energy input that DNA gyrase can use for every catalytic cycle under in vivo conditions. (A) Stage with Ca = 19. Winding of the two molecules around each other is distributed along the entire length while the intramolecular plectonemes due to supercoiling do not form yet. (B) Stage with Ca = 13. Regions with intramolecular plectonemic supercoiling start to be visible and they correspond to regions where the two rings do not wind around each other. (C) Stage with Ca = 7. The region where the two rings wind around each other becomes confined while the regions with plectonemic supercoiling appear like typical branches in supercoiled DNA (25). (D) Stage with Ca = 1. Component rings have the typical appearance of plectonemically supercoiled DNA molecules while catenation preferentially involves apical loops of supercoiled DNA (48).
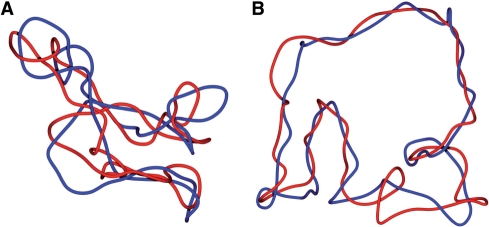
The snapshots from numerical simulations shown in Figure 6 allow us to get more insights into the structure of DNA catenanes than the energy profiles shown in Figure 5 or the pattern of DNA bands observed in 2D gels (see Figures 3 and 4). Those snapshots show catenanes with various catenation numbers and with the highest attainable supercoiling level for any given catenation number, assuming that DNA gyrase stalls when energetic costs of introducing additional supercoil exceed 4.6 kcal/mol (see Figure 5B). We can see that for highly catenated rings, their right-handed winding around each other extends over the entire length of the two rings while supercoiling of individual rings is not yet visible (Figure 6A). However, as the catenation number decreases, the regions of toroidal winding of the two rings decrease and the regions visible as plectonemic supercoiling increase (Figure 6B–D). Interestingly, according to our simulations, the DNA gyrase-induced supercoiling would be hardly sufficient to induce formation of plectonemic supercoils in DNA catenanes with Ca > 12. As discussed earlier, our gels indicated that CatCs, which most likely reached similar energetic limits of DNA supercoiling, were decreasing their compactness with increasing catenation numbers but this effect stopped after reaching Ca ≈ 12. Analyzed configurations permitted us to propose that up to Ca ≈ 12 the molecules ‘trade’ their plectonemic supercoils for the interlinking of component rings and this decreases their overall compactness. As Ca increases further, though, the DNA gyrase-induced supercoiling should get further limited but our simulations indicated that this would rather compact than decompact the concerned catenanes. Figure 7A and B show the effect of introducing (−) supercoiling into highly interlinked catenanes where Ca = 19. In Figure 7A, the modelled configuration is free of torsional stress while, in Figure 7B, each ring has a ΔLk = −5. It is well visible that configurations free of torsional stress are significantly more compacted than those with torsional stress due to (−) supercoiling. Our simulations revealed thus that the first few (−) supercoils introduced into strongly catenated CatCs do not form regions with plectonemic winding but just act to untwist the higher order winding of entire catenanes (see Figure 7). Therefore, when increasing catenation limits the possibility of these molecules to acquire DNA supercoiling, this manifests itself in a faster electrophoretic migration, which is in contrast to the effect of elimination of plectonemic supercoils in less catenated DNA molecules. In addition, the electrophoretic migration seems to be affected more strongly by differences in the number of plectonemic supercoils than by differences in the number of supercoils that are not transformed into plectonemes. That would then explain why Gaussian families are not visible when the (−) supercoiling is not strong enough to induce the formation of plectonemes. Our simulations suggest that this would concern catenanes with Ca > 12 since natural supercoiling is not sufficient to induce the formation of plectonemes in these catenanes.
Figure 7.
Limited levels of (−) supercoiling decompacts highly interlinked catenanes. Smoothed snapshots of equilibrated configurations of catenanes with Ca = 19, which component rings are either free of torsional tension (ΔLk = 0) (A) or are mildly (−) supercoiled (ΔLk = −5) (B). (A) Highly interlinked catenane with component rings that are free of torsional tension adopt a twisted configuration since this lowers their elastic energy. The same conclusion was reached in simulation studies of highly linked nicked DNA circles (20). (B) Introduction of torsional stress, which would have resulted from two catalytic cycles of DNA gyrase on each of torsionally relaxed but covalently closed component DNA rings, brings the catenanes into a more open configuration with a more regular toroidal structure.
DISCUSSION
Although in this study Topo IV activity was inhibited to slow down the resolution of postreplicative catenanes, there is experimental evidence that highly catenated forms in untreated cells also arise but are difficult to detect due to quick decatenation (11,13,14). As soon as the replication forks approach the terminus region and unreplicated DNA shrinks to 200 bp or less, there are sterical difficulties for type II topoisomerases to reduce the linking number in this region (8,9). Both, DNA gyrase and Topo IV perform their sign inversion reactions by forming a DNA loop and such a loop requires a DNA fragment of more than 100 bp long (27,49). In addition, replisomes also extend ahead of the point of strand separation and this additionally increases the length of the terminus region for which it is not possible to eliminate the remaining linking number before the completion of replication. If this duplex region is in the order of 300 bp then freshly replicated circles will have a catenation number where Ca ≈ 30. For relatively small plasmids, such as the one studied here, such an extent of catenation introduces a strong mechanical constrain that interferes with DNA gyrase ability to introduce supercoiling into freshly replicated DNA molecules. For much longer circular molecules, like large plasmids or entire bacterial chromosomes, the 30 or so interlinks would spread along the molecules and would not result in a mechanical constrain that could significantly limit the action of DNA gyrase.
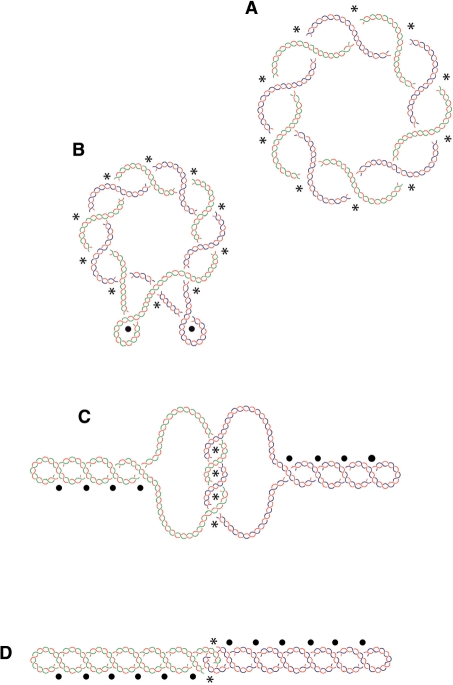
Based on our results presented here and earlier literature data, we propose that postreplicative decatenation of small plasmids in healthy bacterial cells proceeds in steps as idealized in the cartoon shown in Figure 8. Just after the completion of replication, catenanes would have high interlinking numbers that could even preclude them to acquire (−) supercoiling (Figure 8A). The mechanical tension in such highly catenated molecules could drive their decatenation. However, as decatenation progresses the mechanical constrain due to interlinking decreases, this diminishes the driving force for decatenation but permits DNA gyrase to introduce increasing amounts of (−) supercoils that re-establish the free energy gradient leading to decatenation (42) (Figure 8B–D).
Figure 8.
Cartoon illustrating the transition from a highly intertwined non-supercoiled catenane (A) to a singly catenated and highly supercoiled one (D). Just after completion of DNA replication both sister duplexes are highly intertwined and share a common axis. These type C torus catenanes accept a low level of DNA supercoiling that does not yet manifest itself in form of extruded plectonemes (A). As decatenation progresses the mechanical stress in DNA molecules decreases and this allows DNA gyrase to introduce more (−) supercoiling that start to manifest itself as extruded plectonemes (B). As the degree of catenation decreases, supercoiling builds-up and type C torus catenanes tend to have winding and supercoiling clearly segregated (C). Finally, intertwining becomes frequently confined to apical portions (48) and the sister duplexes appear highly supercoiled just before their segregation (D). Asterisks denote intermolecular crossings and dots intramolecular crossings. Parental DNA strands are depicted in blue and green while newly synthesized strands are depicted in red.
In the case of large plasmids or entire bacterial chromosomes the mechanical constrain due to catenation would be very low and this would allow the immediate build up of (−) supercoiling, which then would provide the free energy gradient leading to decatenation. The decatenation process would proceed then as shown in Figure 8C and D.
As already mentioned, our model is in agreement with experimental results obtained in vivo and in vitro showing that decatenation of catenanes with low catenation numbers is actually stimulated by DNA supercoiling (50,51). Also, theoretical studies considering the energy of supercoiled catenanes proposed an active role of DNA supercoiling in the process of DNA decatenation (42).
Based on theoretical considerations, it was shown that DNA supercoiling can provide the free energy gradient that drives Topo IV-mediated DNA decatenation (42) and unknotting (52). If that is the case, what would then be the function of Topo IV ability to reduce DNA catenation and knotting levels of nicked and thus non-supercoiled DNA molecules much below the thermodynamic equilibrium? (53). Possibly, very efficient unknotting and unlinking of nicked DNA is a consequence of a complex mechanism that ensures that, in case of DNA molecules forming CatC catenanes, Topo IV should preferentially recognize DNA–DNA juxtapositions between segments of two catenated DNA molecules and avoid acting on DNA–DNA juxtapositions between opposing segments of intramolecular plectonemes. The model presented in Figure 8 would be efficient only if Topo IV preferentially recognizes intermolecular juxtapositions and neglects intramolecular juxtapositions. Otherwise, (−) supercoiling would be progressively eliminated and the favourable energy gradient for decatenation would be lost if not replenished again by ATP-requiring DNA gyrase. Several papers established that Topo IV is sensitive to the geometry of DNA–DNA juxtaposition and that explains why it preferentially relaxes (+) supercoiled DNA while hardly acts on (−) supercoiled DNA (50,54,55). Of course, the (−) supercoiled DNA should be immune to Topo IV as otherwise cells would enter a vicious cycle of Topo IV relaxing the DNA and DNA gyrase supercoiling it, where both enzymes would use a lot of ATP. The fact that (−) supercoiled DNA is hardly a substrate for Topo IV is essential for the model presented in Figure 8. However, the geometry of right-handed winding of catenated rings around each other is to a great extent very similar to the geometry of intramolecular winding in (−) supercoiled DNA and therefore it is difficult to understand how Topo IV could efficiently catalyze decatenation without inducing relaxation of DNA supercoiling. This puzzle is known as the Topo IV decatenation paradox (2,50).
More studies intended to understand the topologically complex structure of supercoiled catenanes are needed to reveal what are the structural elements that could guide Topo IV to perform an efficient decatenation while preserving (−) supercoiled DNA from relaxation (50,54,56,57).
FUNDING
Spanish Ministerio de Ciencia e Innovación (BIO2005-02224 and BFU2008-00408/BMC to J.B.S. and BFU2007-62670 to P.H.); and the Swiss National Science Foundation (3100A0-116275 to A.S.). Funding for open access charge: Spanish Ministerio de Ciencia e Innovación (BIO2005-02224) and BFU2008-00408/BMC to J.B.S. and BFU2007-62670 to P.H.); and the Swiss National Science Foundation (3100A0-116275 to A.S.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We acknowledge María José Fernández-Nestosa, Marta Fierro-Fernández, Doris Gómez, Virginia López, Estefanía Monturus de Carandini, Leonor Rodríguez, María Rodríguez, María Tenorio and Zaira García for their suggestions and support during the course of this study. We also thank Prof. Lynn Zechiedrich and Prof. Kenneth Marians for bacterial strains.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Bates AD, Maxwell A. DNA Topology. Oxford: Oxford University Press; 2005. [Google Scholar]
- 2.Schvartzman JB, Stasiak A. A topological view of the replicon. EMBO Rep. 2004;5:256–261. doi: 10.1038/sj.embor.7400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine C, Hiasa H, Marians KJ. DNA gyrase and topoisomerase IV: Biochemical activities, physiological roles during chromosome replication, and drug sensitivities. BBA Gene Struct. Express. 1998;1400:29–43. doi: 10.1016/s0167-4781(98)00126-2. [DOI] [PubMed] [Google Scholar]
- 4.Champoux JJ, Been MD. In: Mechanistic Studies of DNA Replication and Genetic Recombination. Alberts B, editor. New York: Academic Press; 1980. pp. 809–815. [Google Scholar]
- 5.Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ. Independent positioning and action of Escherichia coli replisomes in live cells. Cell. 2008;133:90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Reyes-Lamothe R, Sherratt DJ. Modulation of Escherichia coli sister chromosome cohesion by topoisomerase IV. Genes Dev. 2008;22:2426–2433. doi: 10.1101/gad.487508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lucas I, Germe T, Chevrier-Miller M, Hyrien O. Topoisomerase II can unlink replicating DNA by precatenane removal. EMBO J. 2001;20:6509–6519. doi: 10.1093/emboj/20.22.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundin O, Varshavsky A. Terminal stages of SV40 DNA replication proceed via multiply intertwined catenated dimers. Cell. 1980;21:103–114. doi: 10.1016/0092-8674(80)90118-x. [DOI] [PubMed] [Google Scholar]
- 9.Sundin O, Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers dissection of the final stages of SV40 DNA replication. Cell. 1981;25:659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- 10.Adams DE, Shekhtman EM, Zechiedrich EL, Schmid MB, Cozzarelli NR. The role of Topoisomerase-IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- 11.Hiasa H, Marians KJ. Two distinct modes of strand unlinking during theta-type DNA replication. J. Biol. Chem. 1996;271:21529–21535. doi: 10.1074/jbc.271.35.21529. [DOI] [PubMed] [Google Scholar]
- 12.Peng H, Marians KJ. Decatenation activity of Topoisomerase-IV during oriC and pBR322 DNA replication invitro. Proc. Natl Acad. Sci. USA. 1993;90:8571–8575. doi: 10.1073/pnas.90.18.8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leonard AC, Hucul JA, Helmstetter CE. Kinetics of minichromosome replication in Escherichia coli B/r. J. Bacteriol. 1982;149:499–507. doi: 10.1128/jb.149.2.499-507.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Germe T, Hyrien O. Topoisomerase II-DNA complexes trapped by ICRF-193 perturb chromatin structure. EMBO Rep. 2005;6:729–735. doi: 10.1038/sj.embor.7400465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spengler SJ, Stasiak A, Cozzarelli NR. The stereostructure of knots and catenanes produced by phage lambda integrative recombination: implications for mechanism and DNA structure. Cell. 1985;42:325–334. doi: 10.1016/s0092-8674(85)80128-8. [DOI] [PubMed] [Google Scholar]
- 16.Zechiedrich EL, Khodursky AB, Cozzarelli NR. Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes Dev. 1997;11:2580–2592. doi: 10.1101/gad.11.19.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rybenkov VV, Vologodskii AV, Cozzarelli NR. The effect of ionic conditions on the conformations of supercoiled DNA. II. Equilibrium catenation. J. Mol. Biol. 1997;267:312–323. doi: 10.1006/jmbi.1996.0877. [DOI] [PubMed] [Google Scholar]
- 18.Olavarrieta L, Martínez-Robles ML, Sogo JM, Stasiak A, Hernández P, Krimer DB, Schvartzman JB. Supercoiling, knotting and replication fork reversal in partially replicated plasmids. Nucleic Acids Res. 2002;30:656–666. doi: 10.1093/nar/30.3.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santamaría D, Hernández P, Martínez-Robles ML, Krimer DB, Schvartzman JB. Premature termination of DNA replication in plasmids carrying two inversely oriented ColE1 origins. J. Mol. Biol. 2000;300:75–82. doi: 10.1006/jmbi.2000.3843. [DOI] [PubMed] [Google Scholar]
- 20.Vologodskii AV, Cozzarelli NR. Monte carlo analysis of the conformation of DNA catenanes. J. Mol. Biol. 1993;232:1130–1140. doi: 10.1006/jmbi.1993.1465. [DOI] [PubMed] [Google Scholar]
- 21.Bednar J, Furrer P, Stasiak A, Dubochet J, Egelman EH, Bates AD. The twist, writhe and overall shape of supercoiled DNA change during counterion-induced transition from a loosely to a tightly interwound superhelix. Possible implications for DNA structure in vivo. J. Mol. Biol. 1994;235:825–847. doi: 10.1006/jmbi.1994.1042. [DOI] [PubMed] [Google Scholar]
- 22.Rybenkov VV, Cozzarelli NR, Vologodskii AV. Probability of DNA knotting and the effective diameter of the DNA double helix. Proc. Natl Acad. Sci. USA. 1993;90:5307–5311. doi: 10.1073/pnas.90.11.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E. Equation of state calculations by fast computing machines. J. Chem. Phys. 1953;21:1087–1092. [Google Scholar]
- 24.Frank-Kamenetskii MD, Lukashin AV, Anshelevich VV, Vologodskii AV. Torsional and bending rigidity of the double helix from data on small DNA rings. J. Biomol. Struct. Dyn. 1985;2:1005–1012. doi: 10.1080/07391102.1985.10507616. [DOI] [PubMed] [Google Scholar]
- 25.Vologodskii AV, Levene SD, Klenin KV, Frank-Kamenetskii M, Cozzarelli NR. Conformational and thermodynamic properties of supercoiled DNA. J. Mol. Biol. 1992;227:1224–1243. doi: 10.1016/0022-2836(92)90533-p. [DOI] [PubMed] [Google Scholar]
- 26.Gebe JA, Allison SA, Clendenning JB, Schurr JM. Monte Carlo simulations of supercoiling free energies for unknotted and trefoil knotted DNAs. Biophys. J. 1995;68:619–633. doi: 10.1016/S0006-3495(95)80223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laurie B, Katritch V, Sogo J, Koller T, Dubochet J, Stasiak A. Geometry and physics of catenanes applied to the study of DNA replication. Biophys. J. 1998;74:2815–2822. doi: 10.1016/S0006-3495(98)77988-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khodursky AB, Zechiedrich EL, Cozzarelli NR. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc. Natl Acad. Sci. USA. 1995;92:11801–11805. doi: 10.1073/pnas.92.25.11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zechiedrich EL, Osheroff N. Eukaryotic topoisomerases recognize nucleic acid topology by preferentially interacting with DNA crossovers. EMBO J. 1990;9:4555–4562. doi: 10.1002/j.1460-2075.1990.tb07908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santamaría D, delaCueva G, Martínez-Robles ML, Krimer DB, Hernández P, Schvartzman JB. DnaB helicase is unable to dissociate RNA–DNA hybrids—its implication in the polar pausing of replication forks at ColE1 origins. J. Biol. Chem. 1998;273:33386–33396. doi: 10.1074/jbc.273.50.33386. [DOI] [PubMed] [Google Scholar]
- 31.Fierro-Fernandez M, Hernandez P, Krimer DB, Schvartzman JB. Replication fork reversal occurs spontaneously after digestion but is constrained in supercoiled domains. J. Biol. Chem. 2007;282:18190–18196. doi: 10.1074/jbc.M701559200. [DOI] [PubMed] [Google Scholar]
- 32.Fierro-Fernandez M, Hernandez P, Krimer DB, Stasiak A, Schvartzman JB. Topological locking restrains replication fork reversal. Proc. Natl Acad. Sci. USA. 2007;104:1500–1505. doi: 10.1073/pnas.0609204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith SS, Michael MZ, Graham MW. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida H, Kojima T, Yamagishi J, Nakamura S. Quinolone-resistant mutations of the gyrA gene of Escherichia coli. Mol. Gen. Genet. 1988;211:1–7. doi: 10.1007/BF00338386. [DOI] [PubMed] [Google Scholar]
- 35.Brewer BJ, Sena EP, Fangman WL. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Cancer Cells. 1988;6:229–234. [Google Scholar]
- 36.Martín-Parras L, Lucas I, Martínez-Robles ML, Hernández P, Krimer DB, Hyrien O, Schvartzman JB. Topological complexity of different populations of pBR322 as visualized by two-dimensional agarose gel electrophoresis. Nucleic Acids Res. 1998;26:3424–3432. doi: 10.1093/nar/26.14.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baxter J, Diffley JF. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol. Cell. 2008;30:790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 38.Xu SY, Zhu Z, Zhang P, Chan SH, Samuelson JC, Xiao J, Ingalls D, Wilson GG. Discovery of natural nicking endonucleases Nb.BsrDI and Nb.BtsI and engineering of top-strand nicking variants from BsrDI and BtsI. Nucleic Acids Res. 2007;35:4608–4618. doi: 10.1093/nar/gkm481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olavarrieta L, Hernández P, Krimer DB, Schvartzman JB. DNA knotting caused by head-on collision of transcription and replication. J. Mol. Biol. 2002;322:1–6. doi: 10.1016/s0022-2836(02)00740-4. [DOI] [PubMed] [Google Scholar]
- 40.Olavarrieta L, Martínez-Robles ML, Hernández P, Krimer DB, Schvartzman JB. Knotting dynamics during DNA replication. Mol. Microbiol. 2002;46:699–707. doi: 10.1046/j.1365-2958.2002.03217.x. [DOI] [PubMed] [Google Scholar]
- 41.Vologodskii AV, Crisona NJ, Laurie B, Pieranski P, Katritch V, Dubochet J, Stasiak A. Sedimentation and electrophoretic migration of DNA knots and catenanes. J. Mol. Biol. 1998;278:1–3. doi: 10.1006/jmbi.1998.1696. [DOI] [PubMed] [Google Scholar]
- 42.Marko JF. Coupling of intramolecular and intermolecular linkage complexity of two DNAs. Phys. Rev. E. 1999;59:900–912. [Google Scholar]
- 43.Stasiak A, Katrich V, Bednar J, Michoud D, Dubochet J. Electrophoretic mobility of DNA knots. Nature. 1996;384:122. doi: 10.1038/384122a0. [DOI] [PubMed] [Google Scholar]
- 44.Mayan-Santos MD, Martinez-Robles ML, Hernandez P, Krimer D, Schvartzman JB. DNA is more negatively supercoiled in bacterial plasmids than in minichromosomes isolated from budding yeast. Electrophoresis. 2007;28:3845–3853. doi: 10.1002/elps.200700294. [DOI] [PubMed] [Google Scholar]
- 45.Bowater RP, Chen D, Lilley DM. Elevated unconstrained supercoiling of plasmid DNA generated by transcription and translation of the tetracycline resistance gene in eubacteria. Biochemistry. 1994;33:9266–9275. doi: 10.1021/bi00197a030. [DOI] [PubMed] [Google Scholar]
- 46.Bliska JB, Cozzarelli NR. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J. Mol. Biol. 1987;194:205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- 47.Bates AD, Maxwell A. DNA gyrase can supercoil DNA circles as small as 174 base pairs. EMBO J. 1989;8:1861–1866. doi: 10.1002/j.1460-2075.1989.tb03582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levene SD, Donahue C, Boles TC, Cozzarelli NR. Analysis of the structure of dimeric DNA catenanes by electron microscopy. Biophys. J. 1995;69:1036–1045. doi: 10.1016/S0006-3495(95)79978-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sogo JM, Stahl H, Koller T, Knippers R. Structure of replicating Simian Virus 40 minichromosomes. J. Mol. Biol. 1986;189:189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- 50.Charvin G, Bensimon D, Croquette V. Single-molecule study of DNA unlinking by eukaryotic and prokaryotic type-II topoisomerases. Proc. Natl Acad. Sci. USA. 2003;100:9820–9825. doi: 10.1073/pnas.1631550100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ullsperger C, Cozzarelli NR. Contrasting enzymatic activities of topoisomerase IV and DNA gyrase from Escherichia coli. J. Biol. Chem. 1996;271:31549–31555. doi: 10.1074/jbc.271.49.31549. [DOI] [PubMed] [Google Scholar]
- 52.Burnier Y, Dorier J, Stasiak A. DNA supercoiling inhibits DNA knotting. Nucleic Acids Res. 2008;36:4956–4963. doi: 10.1093/nar/gkn467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rybenkov VV, Ullsperger C, Vologodskii AV, Cozzarelli NR. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science. 1997;277:690–693. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- 54.Stone MD, Bryant Z, Crisona NJ, Smith SB, Vologodskii A, Bustamante C, Cozzarelli NR. Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases. Proc. Natl Acad. Sci. USA. 2003;100:8654–8659. doi: 10.1073/pnas.1133178100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neuman KC, Charvin G, Bensimon D, Croquette V. Mechanisms of chiral discrimination by topoisomerase IV. Proc. Natl Acad. Sci. USA. 2009;106:6986–6991. doi: 10.1073/pnas.0900574106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klenin K, Langowski J, Vologodskii A. Computational analysis of the chiral action of type II DNA topoisomerases. J. Mol. Biol. 2002;320:359–367. doi: 10.1016/S0022-2836(02)00447-3. [DOI] [PubMed] [Google Scholar]
- 57.Trigueros S, Salceda J, Bermúdez I, Fernández X, Roca J. Asymmetric removal of supercoils suggests how topoisomerase II simplifies DNA topology. J. Mol. Biol. 2004;335:723–731. doi: 10.1016/j.jmb.2003.11.011. [DOI] [PubMed] [Google Scholar]